Abstract
A quantitative real-time PCR (qRT-PCR) assay with single-copy sensitivity targeting HIV-1 gag RNA (the gag single-copy assay [gSCA]) has been used widely to quantify plasma viremia below the limit of detection of clinical assays in patients on effective antiretroviral therapy (ART), but viral RNA in 15 to 30% of samples amplifies inefficiently because of primer/probe mismatches. We sought to develop improved single-copy assays with increased sensitivity by improving nucleic acid recovery, designing qRT-PCR primers and a probe for a highly conserved region of integrase in the HIV-1 pol gene (the integrase single-copy assay [iSCA]), and increasing the plasma volume tested (Mega-iSCA). We evaluated gSCA versus iSCA in paired plasma samples from 10 consecutive patients with viremia of >1,000 copies/ml and 25 consecutive patients on suppressive ART. Three of 10 viremic samples amplified inefficiently with gSCA compared to the Roche Cobas Ampliprep/TaqMan 2.0, whereas all 10 samples amplified efficiently with iSCA. Among 25 samples from patients on suppressive ART, 8 of 12 samples that were negative for HIV-1 RNA by gSCA had detectable HIV-1 RNA by iSCA, and iSCA detected 3-fold or higher HIV-1 RNA levels compared to gSCA in 10 of 25 samples. Large-volume plasma samples (>20 ml) from 7 patients were assayed using Mega-iSCA, and HIV-1 RNA was quantifiable in 6, including 4 of 5 that were negative by standard-volume iSCA. These improved assays with superior sensitivity will be useful for evaluating whether in vivo interventions can reduce plasma viremia and for assessing relationships between residual viremia and other virologic parameters, including the inducible proviral reservoir.
INTRODUCTION
Infection with human immunodeficiency virus type 1 (HIV-1) leads to high levels of cell-free virions in plasma within days following infection (1). Intensive study of HIV-1 plasma viremia has contributed to the current understanding of HIV-1 pathogenesis and has guided the development of effective antiretroviral therapy (ART). In chronically infected untreated patients, the amount of virus in plasma reflects the number of productively infected cells and is a strong independent predictor of HIV-1 disease progression to AIDS and death (2, 3). Following the discovery of potent antiretroviral drugs, determination of the decay dynamics of plasma viremia and productively infected cells was possible down to the limit of detection of available HIV-1 RNA assays (4–6). More sensitive qualitative assays revealed the persistence of viremia on combination ART (7), indicating that ART alone does not eliminate plasma viremia.
Further studies using a two-step quantitative real-time PCR (qRT-PCR) assay with single-copy sensitivity targeting HIV-1 gag (gag single-copy assay [gSCA]) (8), revealed that plasma viremia of between 1 and 3 copies of HIV-1 RNA per milliliter of plasma persists in most patients independent of the ART regimen used to suppress viremia (9). Additional studies revealed three phases of plasma HIV-1 RNA decay, reflective of HIV-infected populations of cells with different half-lives followed by a fourth phase of decay with an infinite half-life during which HIV-1 RNA levels average 1 copy per milliliter for at least 7 years following initiation of ART (10). Whether the persistent viremia on ART represents infectious virus remains unknown, but in some instances, viral sequences in plasma may match sequences of infectious virus recovered from resting CD4+ T cells (11).
gSCA has also been used in a number of settings to evaluate whether clinical interventions can affect residual viremia. Several investigators have used gSCA as an endpoint in ART intensification studies to evaluate whether addition of raltegravir or other antiretroviral agents to suppressive ART would reduce persistent low-level plasma virema and found that there was no effect on plasma viremia (12–14). In a recent study, incomplete adherence to ART was also found to be associated with higher levels of residual viremia (15). Another study evaluated cellular HIV-1 RNA and plasma viremia following in vivo administration of vorinostat (suberoylanilide hydroxamic acid [SAHA]), and found that cellular HIV-1 RNA was increased without a concomitant increase in plasma viremia as measured with gSCA (16). Levels of plasma viremia as measured by gSCA have also been evaluated as a potential biomarker of the size of the infectious reservoir in resting CD4+ T cells (17).
Despite its widespread use, the original gSCA has limitations. First, the extraction method to isolate HIV-1 RNA published in 2003 (8) may not maximize the recovery of nucleic acid from plasma. Second, the qRT-PCR primers and probe were designed to bind a conserved region of gag from HIV-1 subtype B variants from the United States based on sequences available in 2001 to 2002, but new sequences in the Los Alamos HIV Sequence Database indicate that there are more highly conserved regions elsewhere in the HIV-1 genome. Third, the volume of plasma extracted in the original description of the assay was limited to 7 ml. In theory, sensitivity could be improved by assaying larger plasma volumes, although this has not been shown. We sought to address these limitations of gSCA by improving nucleic acid recovery, designing new primers and a probe in a more conserved region of the HIV-1 genome, and increasing the volume of plasma assayed to potentially reduce censoring of data below the limit of detection, thereby improving the ability to detect an effect of interventions on residual viremia.
MATERIALS AND METHODS
Clinical specimens.
All blood donors were patients at the University of Pittsburgh AIDS Center for Treatment and provided written informed consent. The University of Pittsburgh Institutional Review Board approved the study. Donors were not specifically selected for subtype B infection and were recruited consecutively from the Center. Two groups of blood donors were recruited for this study: (i) donors with plasma viremia of >1,000 copies/ml as measured by Roche COBAS AmpliPrep/COBAS TaqMan, v2.0 (Roche TM2.0), and (ii) donors with viremia suppressed to <20 HIV RNA copies/ml on combination antiretroviral therapy (Roche TM2.0). Donors underwent phlebotomy (100 to 180 ml), and blood was processed within 4 h. Plasma was separated by centrifugation of whole blood at 400 × g for 10 min, followed by removal of the plasma layer and a second centrifugation of plasma at 400 × g. Aliquots of 1.5 ml of plasma were stored at −80°C.
Isolation of nucleic acid.
Total nucleic acid was isolated from plasma samples using the previously reported method (v1.0) (8) or using modifications (v2.0) reported by Venneti et al. (18). A total of 4 aliquots of plasma (∼6 ml) from each donor were thawed in a room temperature water bath. Samples from the same patient were pooled, split into two separate samples for gSCA and the improved SCA (iSCA), and a known quantity of internal standard, consisting of a replication-competent avian leukosis virus (ALV) long terminal repeat (LTR) with a splice adaptor (RCAS) (8, 19, 20), was spiked into each sample. The samples were then spun at 1,600 × g for 15 min at 4°C to pellet debris (e.g., fibrin and other insoluble complexes). (Control experiments showed that this centrifugation improved HIV-1 and RCAS recovery.) Plasma supernatants were then transferred to Seton Easy Seal tubes for ultracentrifugation, and the remaining volume in the tube was filled with Tris-buffered saline. Virions were then pelleted by ultracentrifugation at 170,000 × g for 30 min in a Sorvall T-1270 rotor. Supernatants were then removed, and 100 μl 3 M guanidinium hydrochloride (GuHCl) supplemented with 50 mM Tris-HCl, 1 mM CaCl2, and 1 mg/ml proteinase K was added, followed by incubation at 42°C for 1 h. Following this incubation, 400 μl of ∼6 M guanidinium thiocyanate (GuSCN) supplemented with 50 mM Tris-HCl, 1 mM EDTA, and 600 μg/ml of glycogen was added, followed by incubation for an additional 10 min at 42°C. Next, 500 μl of 100% isopropanol was added to precipitate nucleic acids, and the samples were centrifuged at 21,000 × g for 10 min. Supernatants were removed, and 750 μl of 70% ethanol wash was added, followed by centrifugation at 21,000 × g for 10 min. The supernatant was removed, and the pellets were air dried and resuspended in 55 μl of 5 mM Tris-HCl supplemented with 1 μM dithiothreitol and 1,000 U/ml of RNasin. Test plasma samples were always processed in parallel with HIV-negative plasma and low-copy-number plasma containing 5 HIV-1 RNA copies per milliliter (Rush University, Virology Quality Assurance Lab). HIV-1 RNA was not detected in any of the HIV-1-negative plasma samples (n > 50) throughout the study.
Reverse transcriptase qPCR.
A two-step qRT-PCR was used to quantify HIV-1 RNA in plasma. cDNA was reverse transcribed as previously described (8) from 10 μl of extract in triplicate in 30-μl reactions for HIV-1 RNA and duplicate reactions for RCAS; 5 μl of extract was used as input in the cDNA reaction mixture without reverse transcriptase to confirm that HIV-1 DNA was absent. For real-time PCR, the LightCycler 480 Probes Master ready-made master mix was used at a 0.7× concentration in a total volume of 50 μl with either 400 nM forward and reverse primers and 200 nM probe (iSCA) or 600 nM forward and reverse primers and 100 nM probe (gSCA and RCAS). The cycling parameters for real-time PCR were 95°C for 5 min, followed by 95°C for 10 s and 60°C for 1 min, for 45 cycles of amplification. A standard curve was generated for each assay by serial 3-fold dilution of HIV-1 RNA transcripts that were characterized by optical density at 260 nanometers and serial endpoint dilution to 1 copy per qRT-PCR. Consistent with a theoretical Poisson distribution, we observed 40 to 70% of reactions being positive at 1 input copy of HIV-1 RNA transcripts. HIV-1 RNA transcripts were transcribed in vitro from a T7 expression plasmid containing a 1.7-kb insert containing the 3′ end of pol (bases 4186 to 5913 of the HIV-1 reference sequence HXB2) generated by standard PCR from the plasmid xxLAI (21) (GenBank ascension no. K02013) using the forward primer 5′-AACAAGTAGATAAATTAGTCA-3′ and reverse primer 5′-TTACAATAGCAAGTGGTA-3′ at 600 nM each. Transcripts were purified, and unincorporated nucleotides were removed as previously described (22). The primer and probe sequences have been previously reported for gSCA and RCAS (8) and for iSCA (22).
Mega-iSCA.
To lower the limit of HIV-1 RNA detection, we assayed large volumes of plasma (>20 ml) by iSCA (Mega-iSCA). Plasma was divided into parallel replicate assays (4 to 5 ml of plasma being extracted for each replicate), and assayed as described above by iSCA. The total copy number in each sample was summed, multiplied by a correction factor to control for the proportion of the extract assayed for HIV-1 RNA, and divided by the total volume of plasma assayed to determine the number of copies of HIV-1 RNA per milliliter of plasma detected by Mega-iSCA.
Sequence and statistical analyses.
To identify nucleotide sequence conservation in particular loci of HIV-1, full-length nucleotide web alignments of all subtypes (A to K without recombinants) containing only 1 sequence per patient were downloaded in FASTA form from the HIV Sequence Database at Los Alamos National Laboratory (http://www.hiv.lanl.gov/). To manipulate alignments, identify conserved regions, and generate sequence logos, the statistical program R (23) and the packages seqinr (24), Biostrings (25), and seqLogo (26) were used. Average pairwise distance was calculated using the package APE (27) in R. Statistical analysis was performed using R, and all tests for significance were performed using two-sided α = 0.05 as significant. For comparisons of assays on a continuous scale, negative HIV-1 RNA results were assigned a value equal to 50% of the assay detection limit. Figures were generated in R using the packages ggplot2 (28) and scales (29).
RESULTS
Analysis of HIV-1 nucleotide sequences for highly conserved regions.
The previously reported gSCA primers and probe were originally designed to bind to a conserved region of gag in subtype B HIV-1 (8). Inefficient amplification of HIV-1 RNA in 15 to 30% of plasma samples, compared to FDA-cleared assays, led us to search for more conserved regions of HIV-1. Sequences (a total of 1,360) from all HIV-1 subtypes (A to K) available in the Los Alamos HIV Sequence Database as of 2012 were downloaded, and a consensus sequence for subtype B was built by selecting the most prevalent nucleotides at each position from the 675 subtype B sequences available.
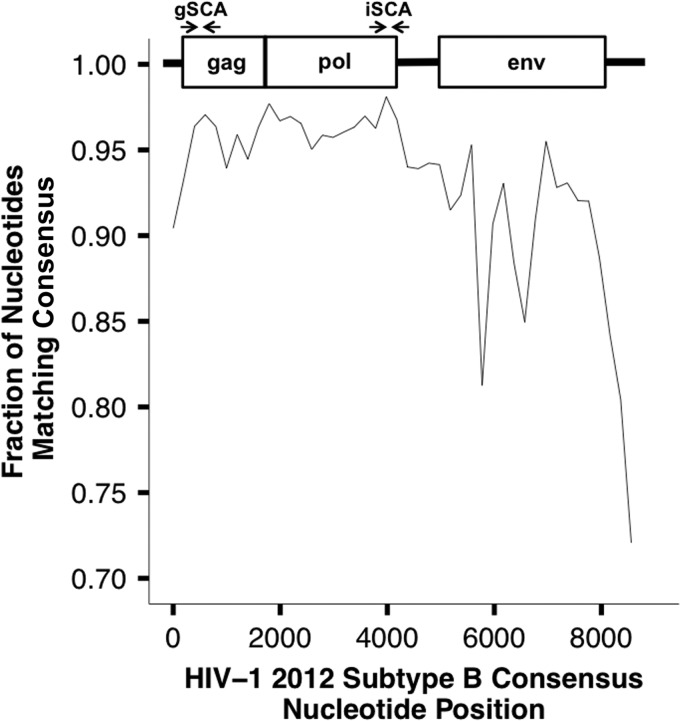
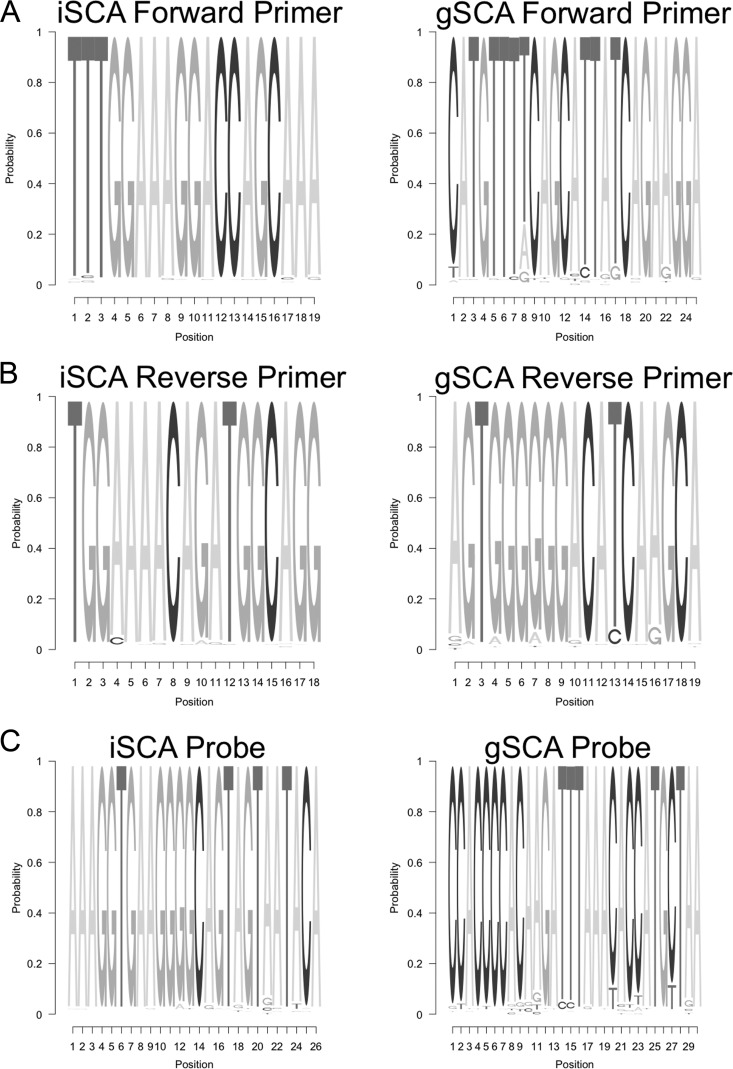
To identify the most highly conserved regions of the HIV-1 genome, the fraction of nucleotides at each position in 200-bp fragments of individual sequences that matched the consensus was determined (Fig. 1). The most highly conserved regions were in gag and at the 3′ end of pol (specifically, the region containing integrase), both of which had >95% sequence identity with the subtype B consensus. The integrase region (98.1%) was more highly conserved than the gag protein region (96.4%). Primer and probe binding sites in integrase were selected, and sequence logos were generated (Fig. 2) to compare the extent of mismatches for the new integrase primer/probe set versus the gag primer/probe set for their respective targets. The results of the sequence logos are in agreement with Fig. 1 in that the primer/probe binding sites in the selected integrase region are more highly conserved than in the previously selected gag site, especially for the probe. In addition, the diversity (average pairwise distance [APD]) calculated for the integrase primer/probe binding sites (2.6%) was significantly lower than for the gag primer/probe binding sites (3.1%; P value between APDs, <1 × 10−5). These analyses suggest that targeting the integrase region with a single-copy assay (iSCA) would result in a lower frequency of inefficient amplification than with the gSCA. When considering all subtypes, there was excellent agreement in sequence conservation in the integrase region, with the exception of a 1-nucleotide substitution present in subtype C present in the forward primer-binding site.
FIG 1.
Proportion of nucleotides in 200-bp fragments that match the HIV-1 2012 subtype B consensus sequence. A consensus sequence was generated from a total of 675 aligned full-length subtype B sequences from the Los Alamos HIV Sequence Database, and the number of bases in 200-bp fragments that matched the consensus sequence was calculated. The most highly conserved regions are present where the original gSCA primers were designed and in the 3′ region of pol, where the iSCA primers and probe target.
FIG 2.
Comparison of sequence logos for the forward (A), reverse (B), and probe (C) binding regions for iSCA and gSCA from 675 subtype B sequences from the Los Alamos HIV Sequence Database. The iSCA system has fewer mismatches than the gSCA system, suggesting that the primers and probe from the iSCA system would efficiently amplify HIV-1 RNA in a higher proportion of patient samples compared to the gSCA system.
Improvement of total nucleic acid recovery.
We next sought to improve the recovery of nucleic acid from plasma. The original gSCA nucleic acid extraction consisted of pelleting of virions by ultracentrifugation, digestion of proteins with proteinase K in water, and addition of the chaotropic agent guanidinium thiocyanate (GuSCN), followed by precipitation and washing of nucleic acid (8). We modified several steps in this extraction protocol as follows. (i) After ultracentrifugation, the sample was incubated with guanidinium hydrochloride (GuHCl) supplemented with proteinase K, Tris-HCl, and CaCl2 at 42°C as described in Materials and Methods. (ii) GuSCN supplemented with Tris-HCl, EDTA, and glycogen was then added. (iii) Centrifugation times following isopropanol precipitation and ethanol wash were shortened to 10 min.
To determine whether the modified extraction method (v2.0) improves recovery of HIV-1 RNA from plasma compared with the original extraction method (v1.0), we assessed recovery of HIV-1 RNA from each extraction method in parallel for paired samples. To generate paired samples, plasma from an HIV-1-positive viremic donor was diluted 100-fold and 1,000-fold with Tris-buffered saline containing 10% fetal bovine serum (Table 1). In replicate assays, HIV-1 RNA recovery was on average 3.0-fold higher at the 1:100 dilution and 2.0-fold higher at the 1:1,000 dilution with extraction method v2.0 compared with v1.0. We also evaluated the recovery of the RCAS internal standard spiked into HIV-1-negative plasma or plasma containing 5 copies of HIV-1 RNA per milliliter (Virology Quality Assurance Lab, Rush University). In paired samples, we found that v2.0 recovered 1.4- to 1.8-fold more RCAS RNA and 1.6-fold more HIV-1 RNA from the Virology Quality Assurance Lab standards.
TABLE 1.
Improved recovery of plasma HIV-1 RNA using a new extraction method
| Sample | Extraction method | No. of replicates | Mean no. of HIV-1 RNA copies/ml detected | Mean % of expected HIV-1 RNA detecteda |
|---|---|---|---|---|
| Viremic plasma diluted 1:100 | v1.0 | 4 | 98 | 36 |
| v2.0 | 4 | 292 | 108 | |
| Viremic plasma diluted 1:1,000 | v1.0 | 6 | 12 | 44 |
| v2.0 | 8 | 23 | 85 |
The numbers of expected HIV-1 RNA copies/ml detected were 271 for viremic plasma diluted 1:100 and 27.1 for viremic plasma diluted 1:1,000.
Performance of iSCA versus gSCA in viremic patients.
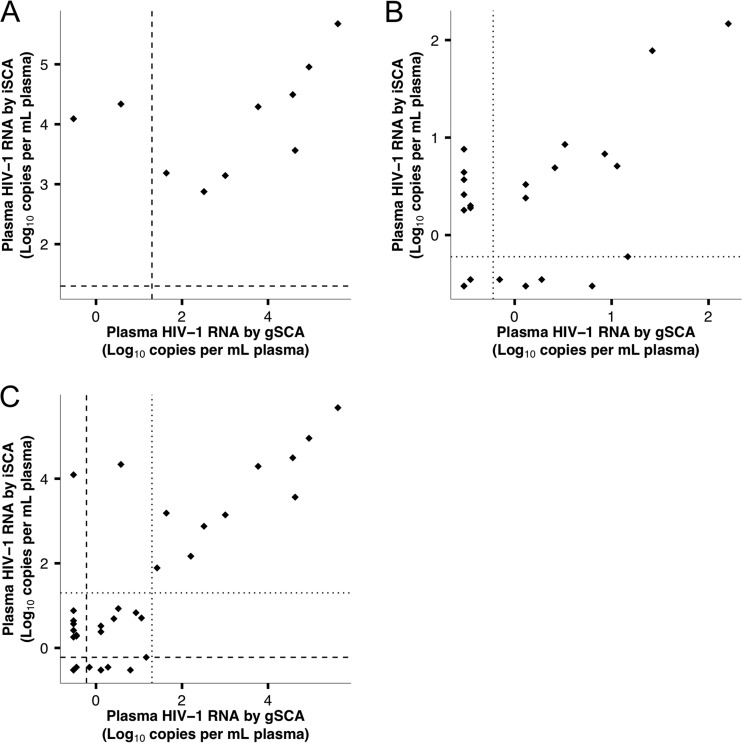
The performance of the gSCA and iSCA primer/probe systems was evaluated by quantifying HIV-1 RNA in duplicate plasma samples from 10 consecutive donors with plasma HIV-1 RNA values ranging from 1,202 to 382,189 copies/ml by Roche TM2.0. By gSCA, 1 of 10 patients had undetectable HIV-1 RNA, and a total of 3 of 10 patients had HIV-1 RNA levels less than 10% of the value determined by Roche TM2.0 (Table 2), consistent with prior reports that HIV-1 RNA from 15 to 30% of patient samples does not amplify efficiently. By iSCA, all 10 patients had HIV-1 RNA levels within 10-fold of the Roche TaqMan platform (P = 0.038, exact binomial test for difference in amplification efficiency between iSCA and gSCA). Levels of plasma viremia as measured with iSCA and gSCA did not correlate (Fig. 3A) (ρ = 0.54, P = 0.11), largely due to the 3 samples that were not efficiently amplified by gSCA.
TABLE 2.
Quantification of plasma viremia with gSCA and iSCA in 10 patients with untreated HIV-1 infection
| Patient ID no. | Plasma HIV-1 RNA (log10 HIV-1 RNA copies/ml plasma) |
||
|---|---|---|---|
| gSCA | iSCA | Roche TM2.0a | |
| 1 | −0.52b,c | 4.1 | 4.4 |
| 2 | 3.8 | 4.3 | 4.5 |
| 3 | 0.58c | 4.3 | 4.5 |
| 4 | 1.6c | 3.2 | 3.3 |
| 5 | 4.6 | 4.5 | 4.9 |
| 6 | 3.0 | 3.1 | 3.3 |
| 7 | 5.6 | 5.7 | 5.6 |
| 8 | 5.0 | 5.0 | 4.8 |
| 9 | 4.6 | 3.6 | 3.8 |
| 10 | 2.5 | 2.9 | 3.1 |
| Median (IQR) | 3.4 (1.9–4.6) | 4.2 (3.3–4.5) | 4.2 (3.4–4.7) |
Roche COBAS Ampliprep/COBAS TaqMan version 2.0.
Sample below the limit of detection of the assay.
Sample with <10-fold recovery compared to Roche TaqMan.
FIG 3.
Relationship between plasma HIV-1 RNA measured with gSCA versus iSCA. (A) There is no significant correlation between HIV-1 RNA values measured by gSCA and iSCA in the 10 samples from viremic patients because of the 3 samples with inefficient amplification by gSCA. The dashed lines indicate the limit of quantification of Roche TM2.0 (20 cps/ml). (B) There is a nonsignificant trend toward a correlation between HIV-1 RNA values measured by iSCA versus gSCA in the 25 plasma samples from patients on suppressive ART. The dotted lines indicate the limit of detection of iSCA and gSCA (0.6 cps/ml). (C) HIV-1 RNA values are significantly correlated between iSCA and gSCA for all 35 patients evaluated in this study. The dashed lines indicate the limit of quantification of Roche TM2.0 (20 cps/ml), and the dotted lines indicate the limit of detection of iSCA and gSCA (0.6 cps/ml).
Evaluation of iSCA versus gSCA in patients on suppressive ART.
The goal of this study was to improve the ability to detect and quantify HIV-1 RNA in patients on suppressive ART with plasma HIV-1 RNA below the limit of detection of clinical assays. To this end, we tested a panel of duplicate plasma samples from 25 consecutive donors on suppressive ART (Table 3; see Table S1 in the supplemental material). HIV-1 RNA was detected in 17 of 25 samples by iSCA versus 13 of 25 samples by gSCA. A total of 8 samples had detectable HIV-1 RNA by iSCA alone, whereas 4 had detectable HIV-1 RNA by gSCA alone. HIV-1 RNA values were ≥3-fold by iSCA compared with gSCA in 10 of 25 samples, versus 5 of 25 being ≥3-fold higher by gSCA than iSCA (P = 0.021, exact binomial test comparing fractions of samples that were ≥3-fold higher by either assay). The median HIV-1 RNA level of all 25 samples determined by iSCA was 2.0 copies/ml (interquartile range [IQR], 0.35 to 4.9 copies/ml) versus 1.3 copies/ml (IQR, 0.30 to 3.3 copies/ml) with gSCA (Table 3; see Table S1). HIV-1 RNA values measured by iSCA and gSCA showed a trend toward a positive correlation, but this was not significant (Fig. 3B; ρ = 0.36, P = 0.077). However, when considering all patient samples assayed (viremic and on suppressive ART), there was a significant correlation between iSCA and gSCA (Fig. 3C; ρ = 0.64, P < 0.001).
TABLE 3.
Summary of results from iSCA versus gSCA for 25 patients on ART
| Assay | No. of samples/total with: |
Median no. of HIV-1 RNA copies/ml (IQR) | No. of samples with HIV-1 RNA level 3-fold higher than alternative assay | |
|---|---|---|---|---|
| HIV RNA detected | Positive result by alternative assay | |||
| gSCA | 13/25 | 8/12 | 1.3 (0.30–3.3) | 5/25 |
| iSCA | 17/25 | 4/8 | 2.0 (0.35–4.9) | 10/25 |
Development and validation of Mega-iSCA.
The input volume of plasma assayed currently limits the sensitivity of single-copy assays, making it uncertain whether patients with HIV-1 RNA below the SCA detection limit are truly aviremic. We hypothesized that by increasing the volume of plasma assayed and thereby lowering the limit of detection, we would be able to detect viremia in more patients, including those with HIV-1 RNA below the limit of detection by standard-volume iSCA. To test this hypothesis, we assayed large volumes of plasma (20 to 35 ml) from 7 donors, including 5 with undetectable HIV-1 by standard-volume (i.e., 3.0 ml of plasma) iSCA (Table 4). Assuming that 30 ml of plasma has been assayed, the theoretical limit of detection with large-volume iSCA (Mega-iSCA) would be 0.06 copy of HIV-1 RNA per milliliter, about 10-fold less than the standard iSCA. Using Mega-iSCA, HIV-1 RNA was detected in 6 of 7 donors, including 4 of 5 with standard-volume iSCA results below the limit of detection (Table 4).
TABLE 4.
Mega-iSCA can be used to increase assay sensitivity
| Patient ID no. | iSCA |
Mega-iSCA |
|||
|---|---|---|---|---|---|
| Plasma vol tested (ml) | No. of copies/ml | Plasma vol tested (ml) | Total no. of copies detected | No. of copies/ml | |
| 6 | 2.9 | <0.7 | 23.4 | 0 | <0.08 |
| 11 | 2.9 | 5.1 | 25.6 | 19 | 1.4 |
| 14 | 2.8 | <0.7 | 25.5 | 71 | 5.1 |
| 18 | 2.7 | <0.7 | 28.9 | 7 | 0.4 |
| 23 | 2.8 | 2 | 33.6 | 11 | 0.7 |
| 26 | 2.9 | <0.6 | 29.0 | 4 | 0.3 |
| 27 | 2.7 | <0.7 | 29.4 | 15 | 0.9 |
DISCUSSION
We have developed improved HIV-1 single-copy assays (SCAs) with superior sensitivity to quantify the persistence of plasma viremia in patients on suppressive ART. The original single-copy assay targeting HIV-1 gag sequences (gSCA) has been successfully applied in many studies to gain insight into the persistence of viremia on ART, although inefficient amplification of viral sequences in 15 to 30% of patients and consequent false-negative results have been limitations. We have evaluated these limitations and modified the assay to improve amplification efficiency and lower the limit of detection.
First, we found that isolation of nucleic acid by sequential GuHCl/proteinase K incubation and GuSCN addition followed by precipitation of nucleic acid recovered more RCAS and HIV-1 RNA in test panels. The higher recovery observed is likely due to the use of GuHCl/proteinase K during the initial incubation, after the virions have been pelleted, rather than using the original extraction method with water and proteinase K. Use of the chaotropic agent GuHCl with proteinase K likely increases the digestion of proteins in protein/nucleic acid complexes compared with proteinase K in water alone. GuHCl and EDTA also likely inactivate RNases.
Second, to improve amplification efficiency, we targeted a more conserved region of the HIV-1 genome. The high rate of nucleotide substitution in HIV-1 has been extensively studied (30) and contributes to difficulties with sensitivity of nucleic acid-based diagnostics for HIV-1 (31). Using sequences in the Los Alamos HIV Sequence Database, we uncovered a region at the 3′ end of the pol gene that was more highly conserved than gag and that was also amenable to primer and probe design for qRT-PCR. Selection and use of this integrase region of the HIV-1 genome for qRT-PCR were effective, in that we eliminated primer/probe mismatches in the test panel of 10 viremic patients.
With the improved performance of iSCA for samples from patients with viremia detectable by Roche TM2.0, we proceeded with a head-to-head comparison of iSCA versus gSCA in samples from patients on suppressive ART, and generally with HIV-1 RNA <20 copies/ml by Roche TM2.0. In the suppressed patients, iSCA again had better performance than gSCA as evidenced by (i) detecting viremia in more patients (17 of 25 detectable by iSCA versus 13 of 25 detectable by gSCA with 8 of 12 not detected by gSCA detected by iSCA), (ii) measuring ≥3-fold higher HIV-1 RNA values in 10 of 25 samples with iSCA versus 5 of 25 samples with gSCA; and (iii) trending toward measuring a higher mean level of residual viremia (2.0 cps/ml for iSCA versus 1.3 cps/ml for gSCA). The median levels of HIV-1 RNA detected in samples from suppressed patients were low enough that stochastic variation in the number of virions between samples may have increased the variability in each assay. Other limitations of our study are the relatively small number of patients evaluated and the likelihood that only subtype B samples were tested. Nevertheless, the combination of a new extraction method and new primers and probe improved assay performance for iSCA compared with gSCA.
Finally, we assessed whether the sensitivity of iSCA could be improved by increasing the volume of plasma assayed. The typical limit of detection for gSCA/iSCA is 0.6 copy of HIV-1 RNA per milliliter of plasma, assuming that 3.0 ml of plasma has been assayed. It is possible that patients have levels of residual viremia that are below this limit of detection and thus score as undetectable by iSCA using the standard volume of plasma. Our data support this hypothesis, as we have shown that 4 of 5 patients with undetectable viremia by standard iSCA had detectable viremia by Mega-iSCA. Mega-iSCA is ≥10-fold more sensitive than iSCA and will be useful for future evaluations of residual viremia and changes in residual viremia following interventions. Plasma volumes of 20 to 30 ml can be easily obtained from large-volume blood draws (75 to 120 ml), which is a smaller blood volume than that required for an assay of inducible HIV-1 reservoirs from resting CD4+ T cells (32). Furthermore, the same whole-blood sample can be used for both assays.
Two studies have reported a positive correlation between the levels of persistent viremia measured by gSCA and infectious virus recovery from resting CD4+ T cells (17, 33), but another study found no relationship between gSCA and infectious virus recovery (32). The correlation between gSCA and levels of infectious virus suggests that HIV-1 RNA in the plasma may originate from stochastic reactivation of latently infected cells and that changes in levels of HIV-1 RNA in plasma following an intervention may be reflective of changes in the size of the latent reservoir. Future studies should seek to further elucidate the relationship between persistent plasma viremia and the infectious virus reservoir.
In summary, we have developed and validated an improved single-copy assay (iSCA) with an improved ability to detect and quantify low levels of plasma viremia. iSCA will undoubtedly be useful for future studies, such as an endpoint of clinical trials with interventions aimed at reducing the size of the HIV-1 reservoir. As the field of HIV-1 cure research continues to evolve, the ability to accurately and precisely quantify residual plasma viremia will be important to evaluate whether novel treatments have caused a change in viremia and whether HIV-1 RNA in plasma remains detectable by the most sensitive means possible after potentially curative interventions.
Supplementary Material
ACKNOWLEDGMENTS
J.W.M. is a consultant for Gilead Sciences and owns shares of RFS Pharma. A.R.C., D.V., M.A.B., E.M.A., M.F.K., E.F., D.K., J.M.C., and M.P. have no conflicts of interest.
This study was supported by the Pittsburgh AIDS Research Training Program 5 T32 AI065380-08 and Leidos contracts 25XS119 and 33XS110 through the National Cancer Institute. J.M.C. was a Research Professor of the American Cancer Society, with support from the F. M. Kirby Foundation.
We thank the volunteers for donating their blood for this study.
Footnotes
Published ahead of print 3 September 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.02060-14.
REFERENCES
- 1.Fiebig EW, Wright DJ, Rawal BD, Garret PE, Schumacher RT, Peddada L, Heldebrandt C, Smith R, Conrad A, Kleinman SH, Busch MP. 2003. Dynamics of HIV viremia and antibody seroconversion in plasma donors: implications for diagnosis and staging of primary HIV infection. AIDS 17:1871–1879. 10.1097/00002030-200309050-00005. [DOI] [PubMed] [Google Scholar]
- 2.Mellors JW, Rinaldo CR, Jr, Gupta P, White RM, Todd JA, Kingsley LA. 1996. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science 272:1167–1170. 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- 3.Mellors JW, Muñoz A, Giorgi JV, Margolick JB, Tassoni CJ, Gupta P, Kingsley LA, Todd JA, Saah AJ, Detels R, Phair JP, Rinaldo CR., Jr 1997. Plasma viral load and CD4+ lymphocytes as prognostic markers of HIV-1 infection. Ann. Intern. Med. 126:946–954. 10.7326/0003-4819-126-12-199706150-00003. [DOI] [PubMed] [Google Scholar]
- 4.Ho DD, Neumann AU, Perelson AS, Chen W, Leonard JM, Markowitz M. 1995. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature 373:123–126. 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 5.Perelson AS, Neumann AU, Markowitz M, Leonard JM, Ho DD. 1996. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science 271:1582–1586. 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- 6.Perelson AS, Essunger P, Cao Y, Vesanen M, Hurley A, Saksela K, Markowitz M, Ho DD. 1997. Decay characteristics of HIV-1-infected compartments during combination therapy. Nature 387:188–191. 10.1038/387188a0. [DOI] [PubMed] [Google Scholar]
- 7.Dornadula G, Zhang H, VanUitert B, Stern J, Livornese L, Jr, Ingerman MJ, Witek J, Kedanis RJ, Natkin J, DeSimone J, Pomerantz RJ. 1999. Residual HIV-1 RNA in blood plasma of patients taking suppressive highly active antiretroviral therapy. JAMA 282:1626–1632. [DOI] [PubMed] [Google Scholar]
- 8.Palmer S, Wiegand AP, Maldarelli F, Bazmi H, Mican JM, Polis M, Dewar RL, Planta A, Liu S, Metcalf JA, Mellors JW, Coffin JM. 2003. New real-time reverse transcriptase-initiated PCR assay with single-copy sensitivity for human immunodeficiency virus type 1 RNA in plasma. J. Clin. Microbiol. 41:4531–4536. 10.1128/JCM.41.10.4531-4536.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maldarelli F, Palmer S, King MS, Wiegand A, Polis MA, Mican J, Kovacs JA, Davey RT, Rock-Kress D, Dewar R, Liu S, Metcalf JA, Rehm C, Brun SC, Hanna GJ, Kempf DJ, Coffin JM, Mellors JW. 2007. ART suppresses plasma HIV-1 RNA to a stable set point predicted by pretherapy viremia. PLoS Pathog. 3:e46. 10.1371/journal.ppat.0030046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palmer S, Maldarelli F, Wiegand A, Bernstein B, Hanna GJ, Brun SC, Kempf DJ, Mellors JW, Coffin JM, King MS. 2008. Low-level viremia persists for at least 7 years in patients on suppressive antiretroviral therapy. Proc. Natl. Acad. Sci. U. S. A. 105:3879–3884. 10.1073/pnas.0800050105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anderson JA, Archin NM, Ince W, Parker D, Wiegand A, Coffin JM, Kuruc J, Eron J, Swanstrom R, Margolis DM. 2011. Clonal sequences recovered from plasma from patients with residual HIV-1 viremia and on intensified antiretroviral therapy are identical to replicating viral RNAs recovered from circulating resting CD4+ T cells. J. Virol. 85:5220–5223. 10.1128/JVI.00284-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dinoso JB, Kim SY, Wiegand AM, Palmer SE, Gange JS, Cranmer L, O'Shea A, Callender M, Spivak A, Brennan T, Kearney MF, Proschan MA, Mican JM, Rehm CA, Coffin JM, Mellors JW, Siliciano RF, Maldarelli F. 2009. Treatment intensification does not reduce residual HIV-1 viremia in patients on highly active antiretroviral therapy. Proc. Natl. Acad. Sci. U. S. A. 106:9403–9408. 10.1073/pnas.0903107106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McMahon D, Jones J, Wiegand A, Gange SJ, Kearney M, Palmer S, McNulty S, Metcalf JA, Acosta E, Rehm C, Coffin JM, Mellors JW, Maldarelli F. 2010. Short-course raltegravir intensification does not reduce persistent low-level viremia in patients with HIV-1 suppression during receipt of combination antiretroviral therapy. Clin. Infect. Dis. 50:912–919. 10.1086/650749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gandhi RT, Zheng L, Bosch RJ, Chan ES, Margolis DM, Read S, Kallungal B, Palmer S, Medvik K, Lederman MM, Alatrakchi N, Jacobson JM, Wiegand A, Kearney M, Coffin JM, Mellors JW, Eron JJ, AIDS Clinical Trials Group A5244 Team 2010. The effect of raltegravir intensification on low-level residual viremia in HIV-infected patients on antiretroviral therapy: a randomized controlled trial. PLoS Med. 7:e1000321. 10.1371/journal.pmed.1000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li JZ, Gallien S, Ribaudo H, Heisey A, Bangsberg DR, Kuritzkes DR. 2014. Incomplete adherence to antiretroviral therapy is associated with higher levels of residual HIV-1 viremia. AIDS 28:181–186. 10.1097/QAD.0000000000000123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Archin NM, Liberty AL, Kashuba AD, Choudhary SK, Kuruc JD, Crooks AM, Parker DC, Anderson EM, Kearney MF, Strain MC, Richman DD, Hudgens MG, Bosch RJ, Coffin JM, Eron JJ, Hazuda DJ, Margolis DM. 2012. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature 487:482–485. 10.1038/nature11286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gandhi RT, Bosch RJ, Aga E, Bedison MA, Bastow B, Schmitz JL, Siliciano JD, Siliciano RF, Eron JJ, Mellors JW, ACTG A5173 Team. 2013. Residual plasma viremia and infectious HIV-1 recovery from resting memory CD4 cells in patients on antiretroviral therapy: results from ACTG A5173. Antivir. Ther. 18:607–613. 10.3851/IMP2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Venneti S, Bonneh-Barkay D, Lopresti BJ, Bissel SJ, Wang G, Mathis CA, Piatak M, Jr, Lifson JD, Nyaundi JO, Murphey-Corb M, Wiley CA. 2008. Longitudinal in vivo positron emission tomography imaging of infected and activated brain macrophages in a macaque model of human immunodeficiency virus encephalitis correlates with central and peripheral markers of encephalitis and areas of synaptic degeneration. Am. J. Pathol. 172:1603–1616. 10.2353/ajpath.2008.070967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schaefer-Klien J, Givol I, Barsov EV, Whitcomb JM, Van Brocklin M, Foster DN, Federspiel MJ, Hughes SH. 1998. The EV-O-derived cell line DF-1 supports the efficient replication of avian leukosis-sarcoma viruses and vectors. Virology 248:305–311. 10.1006/viro.1998.9291. [DOI] [PubMed] [Google Scholar]
- 20.Hughes SH. 2004. The RCAS vector system. Folia Biol. (Praha) 50:107–119. [DOI] [PubMed] [Google Scholar]
- 21.Wain-Hobson S, Vartanian JP, Henry M, Chenciner N, Cheynier R, Delassus S, Martins LP, Sala M, Nugeyre MT, Guétard D, Klatzmann D, Gluckman JC, Willy Rozenbaum, Françoise Barré-Sinoussi, Montagnier L. 1991. LAV revisited: origins of the early HIV-1 isolated from Institut Pasteur. Science 252:961–965. 10.1126/science.2035026. [DOI] [PubMed] [Google Scholar]
- 22.Cillo AR, Krishnan A, Mitsuyasu RT, McMahon DK, Li S, Rossi JJ, Zaia JA, Mellors JW. 2013. Plasma viremia and cellular HIV-1 DNA persist despite autologous hematopoietic stem cell transplantation for HIV-related lymphoma. J. Acquir. Immune Defic. Syndr. 63:438–441. 10.1097/QAI.0b013e31828e6163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Core Team R. 2014. A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org/. [Google Scholar]
- 24.Charif D, Lobry JR. 2007. Seqin{R} 1.0–2: a contributed packaged to the {R} project for statistical computing devoted to biological sequences retrieval and analysis. p 207–232 In Bastolla U, Porto M, Roman HE, Vendruscolo M. (ed), Structural approaches to sequence evolution: molecules, networks and populations, Springer Verlag, New York, NY. [Google Scholar]
- 25.Pages H, Aboyoun P, Gentleman R, DebRoy S. 2014. Biostrings: string objects representing biological sequences, and matching algorithms. R package version 2.32.1. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 26.Bembom O. 2014. seqLogo: sequence logos for DNA sequence alignments. R package version 1.30.0. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 27.Paradis E, Claude J, Strimmer K. 2004. APE: analysis of phylogenetics and evolution in R language. Bioinformatics 20:289–290. 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 28.Wickham H. 2009. ggplot2: elegant graphics for data analysis. Springer, New York, NY. [Google Scholar]
- 29.Wickham H. 2014. scales: scale functions for graphics. R package, version 0.2.4. R Foundation for Statistical Computing, Vienna, Austria. http://CRAN.R-project.org/package=scales. [Google Scholar]
- 30.Mansky LM, Temin HM. 1995. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J. Virol. 69:5087–5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rouet F, Chiax ML, Nerrient E, Hgo-Giang-Huong N, Plantier JC, Burgard M, Peeters M, Damond F, Ekouevi DK, Msellati P, Ferradini L, Rukobo S, Maréchal V, Schvachsa N, Wakrim L, Rafalimanana C, Rakotoambinina B, Viard JP, Seigneurin JM, Rouzioux C. 2007. Impact of HIV-1 genetic diversity on plasma HIV-1 RNA quantification: usefulness of the Agence Nationale de Recherches sur le SIDA second-generation long terminal repeat-based real-time reverse transcriptase polymerase chain reaction test. J. Acquir. Immune Defic. Syndr. 45:380–388. 10.1097/QAI.0b013e3180640cf5. [DOI] [PubMed] [Google Scholar]
- 32.Eriksson S, Graf EH, Dahl V, Strain MC, Yukl SA, Lysenko ES, Bosch RJ, Lai J, Chioma S, Emad F, Abdel-Mohsen M, Hoh R, Hecht F, Hunt P, Somsouk M, Wong J, Johnston R, Siliciano RF, Richman DD, O'Doherty U, Palmer S, Deeks SG, Siliciano JD. 2013. Comparative analysis of measures of viral reservoirs in HIV-1 eradication studies. PLoS Pathog. 9:e1003174. 10.1371/journal.ppat.1003174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Archin NM, Vaidya NK, Kuruc JD, Liberty AL, Weigand A, Kearney MF, Cohen MS, Coffin JM, Bosch RJ, Gay CL, Eron JJ, Margolis DM, Perelson AS. 2012. Immediate antiviral therapy appears to restrict resting CD4+ cell HIV-1 infection without accelerating the decay of latent infection. Proc. Natl. Acad. Sci. U. S. A. 109:9523–9526. 10.1073/pnas.1120248109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.