Abstract
Clostridium difficile is a well-known enteric pathogen of humans and the causative agent of high-morbidity enteritis in piglets aged 1 to 7 days. C. difficile prevalence in Australian piglets is as high as 70%. The current diagnostic assays have been validated only for human infections, and there are no published studies assessing their performance in Australian piglets. We evaluated the suitability of five assays for detecting C. difficile in 157 specimens of piglet feces. The assays included a loop-mediated isothermal amplification (LMIA)-PCR for tcdA (illumigene C. difficile; Meridian), a real-time PCR for tcdB (GeneOhm Cdiff; Becton Dickinson), two-component enzyme immunoassays (EIA) for C. difficile glutamate dehydrogenase (GDH) (EIA-GDH) and TcdA/TcdB (EIA-TcdA/TcdB) (C. diff Quik Chek; Alere), and direct culture (DC) (C. difficile chromID agar; bioMérieux). The assays for detection of the organism were compared against enrichment culture (EC), and assays for detection of toxins/toxin genes were compared against EC followed by PCR for toxin genes (toxigenic EC [TEC]). The recovery of C. difficile by EC was 39.5% (n = 62/157), and TEC revealed that 58.1% (n = 36/62) of isolates were positive for at least one toxin gene (tcdA/tcdB). Compared with those for EC/TEC, the sensitivities, specificities, positive predictive values, and negative predictive values were, respectively, as follows: DC, 91.9, 100.0, 100.0, and 95.0%; EIA-GDH, 41.9, 92.6, 78.8, and 71.0%; EIA-TcdA/TcdB, 5.6, 99.2, 66.7, and 77.9%; real-time PCR, 42.9, 96.7, 78.9, and 85.4% and LMIA-PCR, 25.0, 95.9, 64.3, and 81.1%. The performance of the molecular methods was poor, suggesting that the current commercially available assays for diagnosis of C. difficile in humans are not suitable for use in piglets. C. difficile recovery by the DC provides a cost-effective alternative.
INTRODUCTION
Clostridium difficile is reported outside Australia as a major cause of preweaning scour in neonatal pigs aged 1 to 7 days (1, 2). In the United States, C. difficile infection (CDI) is the most commonly diagnosed cause of enteritis in neonatal pigs, with outbreak-associated mortality reaching 50% (1). Good stockmanship can reduce mortality; however, the morbidity remains high, and piglets that recover from CDI remain on average 10 to 15% underweight and take longer to wean (2).
Like human neonates, piglets are gnotobiotic at birth, and the normal microflora does not establish in the gastrointestinal tract until around 5 days of age (3). This lack of colonization resistance means that piglets are particularly susceptible to C. difficile colonization after parturition (up to 72% prevalence at 2 days) (4). All piglets in an affected farrowing facility may be colonized soon after birth, most likely as a result of ingestion of spores from the already contaminated piggery environment (4, 5). Porcine CDI is characterized by profuse nonhemorrhagic, yellow, pasty-to-watery scour; however, diarrhea alone is not a good predictor of CDI in individual piglets (6, 7). In some cases, the piglets are nondiarrheic, constipated, or obstipated yet reveal colitis, typhlitis, or edema upon necropsy (8–10).
Toxigenic enrichment culture (TEC) is the preferred “gold standard” for diagnosing human CDI in the laboratory as it has a high level of sensitivity. It also has the advantage of recovering isolates for further characterization and epidemiological analysis (11). Enzyme immunoassay (EIA) kits targeting the C. difficile toxins TcdA and TcdB or the glutamate dehydrogenase (GDH) antigen are used in both human and veterinary settings (12, 13). However, these assays have relatively low sensitivity and the limitation of cross-reactivity with the GDH of Clostridium sordellii (12). Consequently, a two-step algorithm, such as an initial GDH screening EIA with a confirmatory toxin EIA or nucleic acid amplification test (NAAT) performed on all positive assays, is currently recommended in human health (14, 15). NAAT-based assays for detection of the genes encoding TcdA and TcdB (tcdA and tcdB, respectively), with their rapid turnaround times and high sensitivities, have significantly improved the detection of CDI (12). While many commercially available assays for the detection of C. difficile have been systematically evaluated for use in humans, their performance with stool samples of animal origin has not been validated. Currently no guidelines are available for diagnosing CDI or detecting C. difficile in animals. The literature on this topic is scarce and limited to a few studies in Europe and North America which have reported varied performance of assays for detecting C. difficile in feces of equine (16), canine (17), and porcine origin (10, 13, 14, 18).
Investigations in our laboratory have confirmed that toxigenic C. difficile is present in many pig herds in Australia (D. R. Knight, M. M. Squire, and T. V. Riley, submitted for publication). Unlike the Northern Hemisphere where a single PCR ribotype (RT), RT078, predominates in swine herds (19), in Australia there are many different RTs circulating among livestock (sheep, cattle, and pigs), including RT033, RT126, RT127, and RT237 (20, 21). To understand the role of C. difficile in piglet disease in Australia, it is essential that veterinary diagnostic laboratories are able to detect the organism in a timely and cost-effective manner.
The aim of this investigation was to evaluate the suitability of five commercially available assays for detecting C. difficile in specimens of piglet feces. (Preliminary results of this investigation were presented at the 8th International Conference on the Molecular Biology and Pathogenesis of the Clostridia [CLOSTPATH], Palm Cove, Australia, October 2013.)
MATERIALS AND METHODS
Sample collection.
A total of 157 fecal samples were obtained by rectal swabs from piglets aged <14 days during the period of June 2012 to March 2013. Sampling was performed by attending veterinarians. At the time of sampling, 49 piglets (31.2%) were actively scouring. The test population originated from 16 farms (piggeries) across five Australian states: New South Wales (NSW), n = 2; Queensland (QLD), n = 6; Victoria (VIC), n = 4; South Australia (SA), n = 1; and Western Australia (WA), n = 3. Farms varied in facility type (e.g., farrow to finish, growers, and breeders) and were geographically distinct. All samples were transported under ambient conditions in Amies transport medium with charcoal (Thermo Fisher Scientific, Waltham, MA, USA) to The University of Western Australia. The mean transport time from the farm to the laboratory was 8 days (range, 2 to 20 days). All samples were stored at 4°C and prepared for analysis within 24 h.
Sample preparation.
Upon receipt of the samples in the laboratory, slurries were prepared by suspension of the fecal swabs in 800 μl of phosphate-buffered saline (PBS). The samples were vortexed briefly to create a homogeneous suspension and split into 200-μl aliquots. Two aliquots were used for the NAATs and stored at −20°C until use, at which point a single freeze-thaw cycle was implemented according to the assay recommendations. One aliquot each was immediately used for enrichment culture (EC), EC followed by PCR for toxin genes (toxigenic EC [TEC]), and direct culture (DC) on chromID. Finally, one aliquot was stored at 2 to 8°C for use with the enzyme immunoassay and processed within 48 h.
EC, TEC, and isolate identification.
Isolation of C. difficile was based on our previously described EC methods (22) with some modifications. Fecal specimens were cultured in a selective enrichment broth containing gentamicin (5 mg/liter), cycloserine (200 mg/liter), and cefoxitin (10 mg/liter) (GCC) (PathWest Laboratory Medicine Excel Media, Mt. Claremont, WA) (23, 24). After 48 h of incubation, 1 ml of broth was alcohol shocked by the addition of an equal volume of absolute ethanol to enhance spore selection, and 10 μl was subcultured onto prereduced selective agar plates (cycloserine cefoxitin fructose agar [CCFA]) containing 0.1% sodium taurocholate (TCCFA) (PathWest Laboratory Medicine Excel Media). Plates were incubated in an anaerobic chamber (Don Whitley Scientific Ltd., Shipley, West Yorkshire, United Kingdom) at 37°C in an atmosphere containing 80% nitrogen, 10% hydrogen, and 10% carbon dioxide and examined after 24 and 48 h. The putative C. difficile colonies were subcultured onto prereduced blood agar. No plate spent more than 15 min outside the anaerobic chamber during the examination. Confirmatory identification of C. difficile was made on the basis of characteristic colony morphology on CCFA (yellow, ground-glass appearance), odor (horse dung smell), and chartreuse fluorescence under long-wave UV light (∼360 nm). The identity of uncertain isolates was confirmed by presence of l-proline aminopeptidase activity (Remel Inc., Lenexa, KS, USA) and Gram staining. The ability of all C. difficile isolates to produce toxin A and/or toxin B was determined by PCR (see below) to give a positive TEC result.
DC on chromogenic agar.
C. difficile chromID agar (bioMérieux, Marcy l'Etoile, France) is a chromogenic medium, containing sodium taurocholate and a proprietary chromogen mix that allows rapid and reliable isolation and presumptive identification of C. difficile strains in 24 h (25, 26). Most C. difficile isolates appear as black colonies on a clear background (26). Cultures were performed according to the manufacturer's recommendations. Plates were incubated in an anaerobic chamber and identified as described above.
EIA for GDH and TcdA/TcdB.
The C. diff Quik Chek Complete (Alere North America, Inc., Orlando, FL) is a rapid membrane EIA for the simultaneous detection of C. difficile toxins TcdA and TcdB and GDH in a single reaction well. All EIAs were carried out according to the manufacturer's recommendations, and results were recorded as either positive or negative for GDH and/or toxins A/B.
Loop-mediated isothermal amplification-PCR for tcdA.
The illumigene C. difficile amplification assay (Meridian Bioscience, Inc., Cincinnati, OH) is a NAAT based upon the principle of loop-mediated isothermal amplification (LMIA)-PCR. This assay detects toxigenic C. difficile strains by targeting a conserved 5′ 204-bp sequence of tcdA (27). LMIA-PCR assays were performed on the illumipro-10 instrument according to the manufacturer's instructions, and results were recorded as positive or negative using the illumipro-10 software.
Real-time PCR assay for tcdB.
The GeneOhm Cdiff Assay (Becton Dickinson, La Jolla, CA) is a NAAT using real-time PCR technology to amplify a conserved region of tcdB. The amplified products are detected by using fluorogenic target-specific hybridization probes (molecular beacons). Real-time PCR assays were performed on a SmartCycler (Cepheid, United Kingdom; supplied by Becton Dickinson at the time of the study) according to the manufacturer's instructions. The SmartCycler software recorded the results of the PCR assay as positive, negative, or unresolved.
Toxin profiling and PCR ribotyping of C. difficile isolates.
All isolates were screened by PCRs for the presence of tcdA and tcdB, binary toxin genes (cdtA and cdtB) and the repetitive region of tcdA using previously described methodology (28–30). PCR ribotyping was performed as previously described (31). The PCR ribotyping reaction products were concentrated using a Qiagen MinElute PCR purification kit (Qiagen, Venlo, Limburg, the Netherlands) and run on the QIAxcel capillary electrophoresis platform (Qiagen). The PCR products were visualized on QIAxcel ScreenGel software v1.0.2.0 (Qiagen). The PCR ribotyping products were analyzed using the Dice coefficient within BioNumerics software package v.6.5 (Applied Maths, Saint-Martens-Latem, Belgium). RTs were identified by comparison of banding patterns with those in our reference library, consisting of a collection of the most prevalent RTs currently circulating in humans and animals in Australia and a collection of 15 reference strains from the European Centre for Disease Prevention and Control (ECDC). Isolates that could not be identified with the reference library were designated with internal nomenclature beginning with the prefix QX.
Concordant, equivocal, and discrepant results.
For an assay detecting organisms (DC and EIA-GDH), a result was considered a true positive if positive by EC. For an assay detecting toxin or toxin genes (EIA-TcdA/TcdB, real-time PCR, and LMIA-PCR), a result was considered a true positive if positive by TEC. Discrepant results (false positives and false negatives) with respect to EC/TEC were repeated as were any equivocal or unresolved results. The percentage of concordance with EC/TEC was calculated for each assay.
Statistical analysis.
The sensitivity and specificity were calculated for each kit against the gold standard assay (EC/TEC). Sensitivity and specificity data were used to calculate the positive (PPVs) and negative predictive values (NPVs). Fisher's exact test was used to compare the recovery of C. difficile in the test systems with the recovery of C. difficile by the EC/TEC.
RESULTS
C. difficile detection.
Overall, C. difficile was isolated by EC from 39.5% (n = 62/157) of samples and by TEC from 22.9% (n = 36/157) of samples (Table 1). The recoveries from piggeries in different states ranged from 26.0 to 54.5% (data not shown). The recoveries of C. difficile isolates from piglets with (36.7%) and without (40.7%) diarrhea were not significantly different (P = 0.141). C. difficile was detected in 36.3% (n = 57/157) of samples by DC, 21.0% (n = 33/157) of samples by EIA-GDH, 1.9% (n = 3/157) of samples by EIA-TcdA/TcdB, 12.1% (n = 19/156) of samples by real-time PCR, and 8.9% (n = 14/157) of samples by LMIA-PCR (Table 1).
TABLE 1.
Detection of C. difficile in Australian piglet feces (n = 157)
| Target | Assaya |
C. difficile assay results: |
Assay concordance (no. [%]) |
|||
|---|---|---|---|---|---|---|
| No. (%) positive | No. (%) negative | P | With EC | With TEC | ||
| C. difficile | EC | 62 (39.5) | 95 (60.5) | |||
| DC | 57 (36.3) | 100 (63.7) | 0.56 | 152/157 (96.8) | ||
| EIA-GDH | 33 (21.0) | 124 (79.0) | <0.001 | 114/157 (72.6) | ||
| Toxin/toxin gene | TEC | 36 (22.9) | 121 (77.1) | |||
| EIA-TcdA/TcdB | 3 (1.9%) | 154 (98.1) | <0.001 | 122/157 (77.7) | ||
| Real-time PCR | 19 (12.1) | 138 (87.9) | 0.01 | 132/156 (84.1)b | ||
| LMIA-PCR | 14 (8.9) | 143 (91.1) | <0.001 | 125/157 (79.6) | ||
DC, direct culture (C. difficile chromID agar; bioMérieux); EIA-TcdA/TcdB, enzyme immunoassay (EIA) for TcdA and TcdB (C. diff Quik Chek; Alere); EIA-GDH, EIA for C. difficile glutamate dehydrogenase (GDH) (C. diff Quik Chek; Alere); real-time PCR, real-time PCR for tcdB (GeneOhm Cdiff; Becton Dickinson); LMIA-PCR, loop-mediated isothermal amplification-PCR for tcdA (illumigene C. difficile; Meridian); EC, enrichment culture; TEC, enrichment culture with PCR for toxin genes.
There was a single unresolved result for real-time PCR.
Characterization of C. difficile.
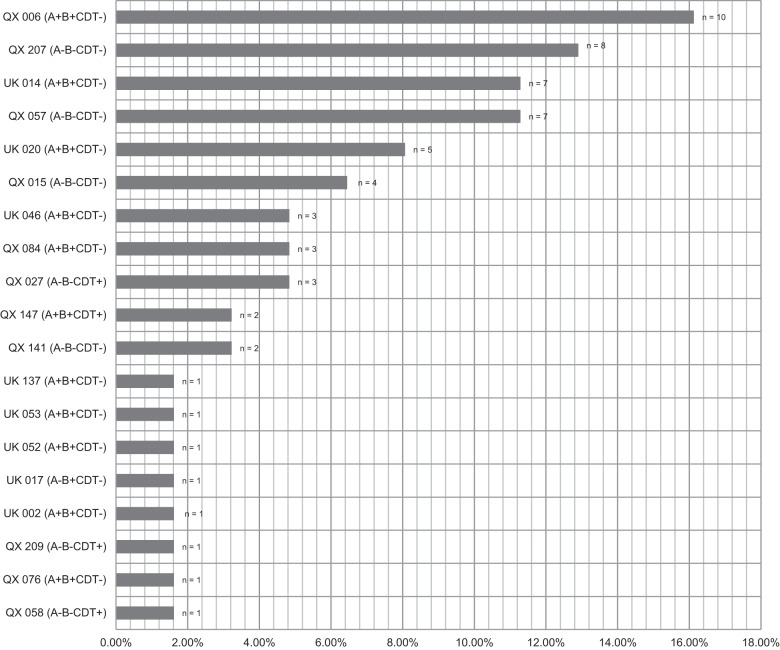
PCRs revealed that 58.1% (n = 36) of isolates were positive for at least one toxin gene (tcdA/tcdB). Overall, five toxin profiles were observed, the most common being A positive, B positive, CDT negative (A+ B+ CDT−) (n = 33, 53.2%). Two isolates (3.2%) were A+ B+ CDT+, one (1.6%) was A− B+ CDT−, and five (8.1%) had the uncommon genotype of A− B− CDT+. The remainder (n = 21, 33.9%) were negative for any toxin genes. Multiple RTs were identified (Fig. 1). Of the 62 isolates obtained from neonatal pigs, 32.3% (n = 20) were assigned one of eight internationally recognized RTs. The remaining isolates were assigned the prefix QX and given an internal number. No RT027 or RT078 was identified. QX006 (A+ B+ CDT−) was the most common RT found overall, representing 16.1% (10/62) of isolates. After QX006, the next four most prevalent RTs were QX207 (12.9%), QX057 (11.3%), UK014 (11.3%), and QX020 (8.1%).
FIG 1.
Summary of PCR ribotypes and toxin profiles from recovered C. difficile isolates (n = 62).
Concordant and discordant results.
DC and EIA-GDH concordances with EC were 96.8% (152/157) and 72.6% (114/157), respectively. The combined concordance for both assays with EC was 77% (121/157) (Table 1). Real-time PCR, EIA-TcdA/TcdB, and LMIA-PCR concordances with TEC were 84.1% (132/157), 77.7% (122/157), and 79.6% (125/157), respectively. The combined concordance for all three assays with TEC was 73.9% (116/157) (Table 1). There were a high number of discordant results, principally false negatives but also a few false positives (data not shown). There was a single equivocal result for real-time PCR that could not be resolved after repeat testing, resulting in a reduced total of samples for this assay (n = 156).
Sensitivities, specificities, PPVs, and NPVs of all assays compared to EC or TEC.
The prevalence of nontoxigenic (A− B−) strains of C. difficile in this study was high (42%). This observation raised the possibility of a population bias favoring strain types that do not have the targets (toxin or toxin genes) that the nonculture toxin-based methods (EIA-TcdA/TcdB, real-time PCR, and LMIA-PCR) are designed to detect. To fairly assess these three assays, they were evaluated against TEC, while assays to detect organisms (DC and EIA-GDH) were evaluated against EC (Table 2). Of all the comparator assays, DC had the highest sensitivity and specificity (91.9% and 100.0%, respectively). The sensitivity of EIA-GDH was 41.9% (Table 2). For the other three assays (EIA-TcdA/TcdB, real-time PCR, and LMIA-PCR), sensitivities were low, ranging from 5.6 to 42.9%, and predictive values for all assays varied widely (PPV range, 64.3 to 100.0%; NPV range, 71.0 to 95.0%) (Table 2).
TABLE 2.
Performance of DC and EIA-GDH and EIA-TcdA/TcdB, LMIA-PCR and real-time PCR, compared to EC and TEC, respectivelya
| Comparator test | Parameterb | Performance (95% confidence interval) |
||||
|---|---|---|---|---|---|---|
| DC | EIA-GDH | EIA-TcdA/TcdB | RT-PCR | LMIA-PCR | ||
| EC | % sensitivity | 91.9 (82.2–97.3) | 41.9 (29.5–55.2) | |||
| % specificity | 100.0 (96.2–100.0) | 92.6 (85.4–97.0) | ||||
| % PPV | 100.0 (93.7–100.0) | 78.8 (61.1–91.0) | ||||
| % NPV | 95.0 (88.7–98.3) | 71.0 (62.1–78.8) | ||||
| TEC | % sensitivity | 5.6 (0.8–18.7) | 42.9 (26.3–60.6) | 25.0 (12.2–42.2) | ||
| % specificity | 99.2 (95.5–99.9) | 96.7 (91.7–99.1) | 95.9 (90.6–98.6) | |||
| % PPV | 66.7 (11.6–94.5) | 78.9 (54.4–93.8) | 64.3 (35.2–87.1) | |||
| % NPV | 77.9 (70.5–84.2) | 85.4 (78.4–90.9) | 81.1 (73.7–87.2) | |||
DC, direct culture (C. difficile chromID agar; bioMérieux); EIA-TcdA/TcdB, enzyme immunoassay (EIA) for TcdA and TcdB (C. diff Quik Chek; Alere); EIA-GDH, EIA for C. difficile glutamate dehydrogenase (GDH) (C. diff Quik Chek; Alere); real-time PCR, real-time PCR for tcdB (GeneOhm Cdiff; Becton Dickinson); LMIA-PCR, loop-mediated isothermal amplification-PCR for tcdA (illumigene C. difficile; Meridian); EC, enrichment culture; TEC, enrichment culture with PCR for toxin genes.
PPV, positive predictive value; NPV, negative predictive value.
DISCUSSION
In the present study, 157 specimens of piglet feces were assessed for the presence of C. difficile or its toxins by EC/TEC, two NAAT methods (real-time PCR and LMIA-PCR), DC using chromogenic agar, and an EIA for GDH and toxins A and B. This is the first evaluation of commercially available diagnostic assays for detection of C. difficile or its toxins in a diverse range of C. difficile strains from Australian neonatal pigs. We showed that compared to EC or TEC, EIAs and NAATs have much lower sensitivities, specificities, and predictive values when used to detect C. difficile or its toxins in porcine rather than human feces (Tables 1 and 2).
Of the 157 samples collected in this study, 22.9% and 39.5% were positive for C. difficile by TEC and EC, respectively. The recovery by EC is similar to that in reports from the United States (34.3%) (32), from the Netherlands (42 to 48%) (14), and from lesser pork-producing countries like Slovenia (50.9%) (33) and the Czech Republic (56.7%) (34). Our laboratory has previously found by EC that C. difficile infection in Australian neonatal pigs is widespread with a national prevalence of 67% (Knight et al, submitted).
Studies in other countries have evaluated different GDH- and toxin-based detection assays in animals, including piglets (10, 13, 14, 16–18). However, the diverse strain population, geographic distribution of sampling sites, and sample transport logistics involved in our study provide a unique scenario for assessing the performance of these assays for detecting CDI in piglets. To date, few studies have evaluated DC (25, 26, 35), and of those, only one included samples of animal origin; however, these were C. difficile isolates not fecal samples (35). This study presents the first reported data worldwide on the performance of a chromogenic medium for recovery of C. difficile from animal fecal samples. DC performed the best of all the comparator assays and had a high sensitivity (91.9%) and specificity (100.0%). The overall recovery of C. difficile by DC was high (36.3%) and comparable to that by EC (39.5%) (96.8% concordance). These findings are consistent with studies performed on human feces (25, 35).
The detection of toxigenic and nontoxigenic C. difficile in porcine feces by the toxin- and molecular-based assays (EIA-TcdA/TcdB, real-time PCR, and LMIA-PCR) was poor. The concordances of these assays with TEC were lower than expected (EIA-TcdA/TcdB, 77.7%; real-time PCR, 84.1%; and LMIA-PCR, 79.6%), and the sensitivities ranged from 5.6 to 42.9%. Surprisingly, given the high prevalence of C. difficile in the population, the PPVs and NPVs for the NAAT-based assays (real-time PCR and LMIA-PCR) were unacceptably low (PPVs, 78.9% and 64.3%, respectively; NPVs, 85.4% and 81.1%, respectively). This finding is in accordance with those of other studies which found that the performance of NAAT-based assays to detect C. difficile in fecal samples of pigs, horses, and dogs was less than in human fecal samples (10, 14, 17).
The poor performance of all assays except DC was primarily due to the high number of discordant results, principally false negatives, and this could be attributable to a number of factors. Several environmental and host factors thought to influence the performance of human diagnostic assays have been suggested and may be relevant in animal studies. First, a study by Lyerly found that low specificity in enzyme immunoassays may be attributed to toxin degradation due to multiple freeze-thaw cycles (36). This is unlikely to account for the discordant results seen in our study as samples were thawed only once, according to the manufacturer's recommendations. Second, Jure and colleagues suggested that nonspecific binding of host fecal proteins to toxin in the gastrointestinal tract results in low levels of free unbound toxin in the sample (affecting enzyme-linked immunosorbent assays [ELISA] and cytotoxic assays) (37). Third, the low specificity seen with commercial assays testing animal feces is potentially caused by the presence of inhibitory substances or inactivating enzymes (10, 36). To date there have been limited data in the literature to support this hypothesis; however, it is conceivable that inherent differences in the composition of the feces between animals and humans influence the binding of primers or antigens in the case of EIAs. If inhibitory substances are present, they may be specific for certain proteins and may explain the discordant EIA results seen in this study where samples that yielded toxin-positive isolates were recorded by EIAs as GDH positive or TcdA or TcdB negative. Finally, Gumerlock et al. proposed that fecal proteases degrade levels of toxins in the stool (38). It is possible that the length of time the samples spent in transit (mean transport time of 8 days) had a detrimental effect on toxin levels in the feces, reducing them below the level of detection of EIA-based assays. This could explain the difference between the poor results presented here for the EIA and those reported by Keessen and colleagues (80 to 90%) (14). In that study, porcine feces were collected in April (European spring) and transported under refrigeration from farms within the relatively small geographic area of the Netherlands (∼1/200 the size of Australia). In our study, samples were transported over large distances (mean distance from farm to laboratory of ∼3,600 km) and in suboptimal (ambient) storage conditions. This is an important observation and likely reflects the circumstances under which samples are routinely transported from the site of collection to the veterinary laboratory. The fact that DC worked so well under these conditions underscores its suitability as a diagnostic test for C. difficile in Australia.
It is important that diagnostic tests perform well, independent of the strain types present in the test population. This study identified numerous PCR ribotypes, some of which were internationally known strains, predominantly RTs associated with carriage and disease in humans. The most prevalent RT was QX006 (16.1%), followed by QX207 (12.9%), UK014 (11.3%), QX057 (11.3%), and UK020 (8.1%). These top five RTs comprised 60% of the isolates recovered by TEC. RT014 and RT020 are often grouped together due to their very similar PCR ribotype fingerprint. RT014/020 is the most common RT in many countries, including the Netherlands (39) and Australia (B. Elliott and T. V. Riley, unpublished data). RT014 is not only well established in nosocomial cases of CDI but also a leading cause of disease in the community (39) and has been found in a small number of livestock (40). RT046 comprised approximately 5% of isolates and has recently been described in piglet and human populations in Sweden (41). The RT distribution appeared clonal with many RTs unique to individual farms and states (data not shown) and is consistent with our findings in a recent study (Knight et al., submitted).
Overall, 58% (n = 36) of isolates were positive for tcdA or tcdB or both. Of the remaining isolates, 34% (n = 21), including about half of the isolates comprising the top five RTs, were nontoxigenic (A− B− CDT−) strains and 8.1% (n = 5) of isolates were positive only for binary toxin (CDT+). These data indicate heterogeneity in the test population and are consistent with our findings in a recent study (Knight, submitted).
Of the limited number of diagnostic studies performed in animals to date, a single study by Keessen et al. provided typing data that indicated homogeneity of C. difficile strains in their test population (14). In that study, 99% (70/71) of isolates recovered from samples of porcine feces were RT078, the predominant RT circulating in animals in Europe. The same study reported significantly higher sensitivity for the real-time PCR assay and a range of EIA platforms. It is possible that their results were biased because of the single strain population. In particular, there may be differences in the antigenic features, toxin expression, or pathogenicity locus (PaLoc) primer binding sites. This idea has also been suggested by Tenover et al., who found significantly lower sensitivity with real-time PCR and EIA for RT078 than for many other ribotypes, including the epidemic strain RT027 (42). No RT078 was found in this or any previous studies in Australian livestock, and RT078 is also not endemic in human populations in Australia (43).
In our study, DC performed consistently well across all of the 19 RTs. This result is in accordance with the findings of Eckert et al. (25), who found no relationship between the PCR ribotype and isolate recovery using chromogenic agar. Conversely, all of the nonculture methods evaluated in this study (EIA-GDH, LMIA-PCR, real-time PCR, and EIA-TcdA/TcdB) performed consistently poorly across all 12 toxigenic RTs.
The performance of any assay is ultimately influenced by the choice of reference method. EC/TEC have not only high sensitivities and specificities for C. difficile but also the benefit of recovery of the isolate, which can be used for future epidemiological typing and antimicrobial susceptibility testing (11). EC/TEC are, however, slow and laborious, often taking up to 5 days for completion and are unlikely to be adopted by veterinary laboratories as a standard practice for C. difficile testing. We have shown that DC presents a suitable presumptive identification method for veterinary laboratories. The chromID Cdiff plates are highly selective; they limit the growth of endogenous flora so that C. difficile colonies are easy to identify and subculture prior to toxin profiling and epidemiological typing. DC outperformed the molecular methods assessed in this study as well as EC on TCCFA by negating the need for prereduction, alcohol shock, or the additional 24 h of incubation time (26). Other benefits of this medium are its relatively low cost (plates are ∼AU$3 each), and the environment required for C. difficile growth can be achieved relatively easily using anaerobic jars.
This study highlights the high prevalence and unique strain types of C. difficile present in Australian neonatal pig populations. Despite the poor performance of commercially available nonculture-based diagnostic assays, this study shows that DC represents a viable option for detecting C. difficile in piglets. In the hospital setting, discordant test results have major implications for patient care. False positives can lead to unnecessary treatment and isolation, and false negatives increase the risk of delay in treatment and cross-infection (44). The diagnosis of CDI in piglets may not be as time critical, but our results underscore the importance of developing cost-effective, sensitive, and specific assays for the rapid, reliable detection of C. difficile and its toxins in porcine feces.
ACKNOWLEDGMENTS
Funding for this study was provided by the Co-operative Research Centre for High Integrity Australian Pork (Pork CRC, Willaston, South Australia). We thank Hugo Dunlop of Chris Richards and Associates (East Bendigo, Victoria) for coordinating the collection of samples used in this study. We are grateful to Meridian Bioscience, Alere, and Becton Dickinson for contributions of the assay kits. Finally, we thank Roger Campbell and Graeme Crook and colleagues at the Pork CRC and Pat Mitchell, Darryl D'Souza, and colleagues at Australian Pork Ltd. (Barton, Australian Capital Territory), for their continued support.
D.R.K. and M.M.S. declare no conflicts of interest. T.V.R. has received speaker fees, educational grants, and travel assistance to attend scientific meetings from Alere, Becton Dickinson, bioMérieux, Cubist, Meridian, and Sanofi Pasteur.
Footnotes
Published ahead of print 13 August 2014
REFERENCES
- 1.Songer JG, Anderson MA. 2006. Clostridium difficile: an important pathogen of food animals. Anaerobe 12:1–4. 10.1016/j.anaerobe.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 2.Songer JG, Uzal FA. 2005. Clostridial enteric infections in pigs. J. Vet. Diagn. Invest. 17:528–536. 10.1177/104063870501700602. [DOI] [PubMed] [Google Scholar]
- 3.Salminen S, Isolauri E, Onnela T. 1995. Gut flora in normal and disordered states. Chemotherapy 41(Suppl 1):5–15. 10.1159/000239391. [DOI] [PubMed] [Google Scholar]
- 4.Weese JS, Wakeford T, Reid-Smith R, Rousseau J, Friendship R. 2010. Longitudinal investigation of Clostridium difficile shedding in piglets. Anaerobe 16:501–504. 10.1016/j.anaerobe.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 5.Hopman NEM, Keessen EC, Harmanus C, Sanders JG, van Leegoed LAMG, Kuijper EJ, Lipman LJA. 2011. Acquisition of Clostridium difficile by piglets. Vet. Microbiol. 149:186–192. 10.1016/j.vetmic.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 6.Yaeger MJ, Kinyon JM, Songer JG. 2007. A prospective, case control study evaluating the association between Clostridium difficile toxins in the colon of neonatal swine and gross and microscopic lesions. J. Vet. Diagn. Invest. 19:52–59. 10.1177/104063870701900108. [DOI] [PubMed] [Google Scholar]
- 7.Alvarez-Perez S, Blanco JL, Bouza E, Alba P, Gibert X, Maldonado J, Garcia ME. 2009. Prevalence of Clostridium difficile in diarrhoeic and non-diarrhoeic piglets. Vet. Microbiol. 137:302–305. 10.1016/j.vetmic.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 8.Waters EH, Orr JP, Clark EG, Schaufele CM. 1998. Typhlocolitis caused by Clostridium difficile in suckling piglets. J. Vet. Diagn. Invest. 10:104–108. 10.1177/104063879801000122. [DOI] [PubMed] [Google Scholar]
- 9.Songer JG, Post KW, Larson DJ, Jost BH, Glock RD. 2000. Infection of neonatal swine with Clostridium difficile. Swine Health Prod. 8:185–189. [Google Scholar]
- 10.Anderson MA, Songer JG. 2008. Evaluation of two enzyme immunoassays for detection of Clostridium difficile toxins A and B in swine. Vet. Microbiol. 128:204–206. 10.1016/j.vetmic.2007.09.022. [DOI] [PubMed] [Google Scholar]
- 11.Planche T, Wilcox MH. 2011. Reference assays for Clostridium difficile infection: one or two gold standards? J. Clin. Pathol. 64:1–5. 10.1136/jcp.2010.080135. [DOI] [PubMed] [Google Scholar]
- 12.Burnham CD, Carroll KC. 2013. Diagnosis of Clostridium difficile Infection: an ongoing conundrum for clinicians and for clinical laboratories. Clin. Microbiol. Rev. 26:604–630. 10.1128/CMR.00016-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Post KW, Jost H, Songer JG. 2002. Evaluation of a test for Clostridium difficile toxins A and B for the diagnosis of neonatal swine enteritis. J. Vet. Diagn. Invest. 14:258–259. 10.1177/104063870201400314. [DOI] [PubMed] [Google Scholar]
- 14.Keessen EC, Hopman NEM, van Leengoed LAMG, van Asten AJAM, Hermanus C, Kuijper EJ, Lipman LJA. 2011. Evaluation of four different diagnostic tests to detect Clostridium difficile in piglets. J. Clin. Microbiol. 49:1816–1821. 10.1128/JCM.00242-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swindells J, Brenwald N, Reading N, Oppenheim B. 2010. Evaluation of diagnostic tests for Clostridium difficile infection. J. Clin. Microbiol. 48:606–608. 10.1128/JCM.01579-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Medina-Torres CE, Weese JS, Staempfli HR. 2010. Validation of a commercial enzyme immunoassay for detection of Clostridium difficile toxins in feces of horses with acute diarrhea. J. Vet. Intern. Med. 24:628–632. 10.1111/j.1939-1676.2010.00506.x. [DOI] [PubMed] [Google Scholar]
- 17.Chouicha N, Marks SL. 2006. Evaluation of five enzyme immunoassays compared with the cytotoxicity assay for diagnosis of Clostridium difficile-associated diarrhea in dogs. J. Vet. Diagn. Invest. 18:182–188. 10.1177/104063870601800207. [DOI] [PubMed] [Google Scholar]
- 18.Alvarez-Perez S, Alba P, Blanco JL, Garcia ME. 2009. Detection of toxigenic Clostridium difficile in pig feces by PCR. Vet. Med. 54:360–366. [Google Scholar]
- 19.Keessen EC, van Leengoed LAMG, Bakker D, van den Brink KM, Kuijper EJ, Lipman LJA. 2010. Prevalence of Clostridium difficile in swine thought to have Clostridium difficile infections (CDI) in eleven swine operations in the netherlands. Tijdschr. Diergeneeskd. 135:134–137 (In Dutch.) [PubMed] [Google Scholar]
- 20.Knight DR, Thean SK, Putsathit P, Fenwick S, Riley TV. 2013. Cross-sectional study reveals high prevalence of Clostridium difficile non-PCR ribotype 078 strains in Australian veal calves at slaughter. Appl. Environ. Microbiol. 79:2630–2635. 10.1128/AEM.03951-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Squire MM, Carter GP, Mackin KE, Chakravorty A, Norén T, Elliott B, Lyras D, Riley TV. 2013. Novel molecular type of Clostridium difficile in neonatal pigs, Western Australia. Emerg. Infect. Dis. 19:790–792. 10.3201/eid1905.121062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bowman RA, Riley TV. 1988. The laboratory diagnosis of Clostridium difficile-associated diarrhoea. Eur. J. Clin. Microbiol. Infect. Dis. 7:476–484. 10.1007/BF01962596. [DOI] [PubMed] [Google Scholar]
- 23.Carroll SM, Bowman RA, Riley TV. 1983. A selective broth for Clostridium difficile. Pathology 15:165–167. 10.3109/00313028309084706. [DOI] [PubMed] [Google Scholar]
- 24.Brazier JS. 1998. The diagnosis of Clostridium difficile-associated disease. J. Antimicrob. Chemother. 41(Suppl C):29–40. 10.1093/jac/41.suppl_3.29. [DOI] [PubMed] [Google Scholar]
- 25.Eckert C, Burghoffer B, Lalande V, Barbut F. 2013. Evaluation of the chromogenic agar chromID C. difficile. J. Clin. Microbiol. 51:1002–1004. 10.1128/JCM.02601-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carson KC, Boseiwaqa LV, Thean SK, Foster NF, Riley TV. 2013. Isolation of Clostridium difficile from faecal specimens—a comparison of ChromID C. difficile agar and cycloserine cefoxitin fructose agar. J. Med. Microbiol. 62:1423–1427. 10.1099/jmm.0.056515-0. [DOI] [PubMed] [Google Scholar]
- 27.Carson KC, Aseeri A, MacKenzie B, Riley TV. 2012. Comparison of illumigene C. difficile and GeneOhm Cdiff assays on glutamate dehydrogenase positive faecal samples, abstr P2275, p 181 In Proceedings of the 22nd Annual European Congress on Clinical Microbiology and Infectious Diseases, London, United Kingdom. [Google Scholar]
- 28.Kato N, Ou CY, Kato H, Bartley SL, Brown VK, Dowell VR, Jr, Ueno K. 1991. Identification of toxigenic Clostridium difficile by the polymerase chain reaction. J. Clin. Microbiol. 29:33–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kato H, Kato N, Watanabe K, Iwai N, Nakamura H, Yamamoto T, Suzuki K, Kim SM, Chong SY, Wasito EB. 1998. Identification of toxin A-negative, toxin B-positive Clostridium difficile by PCR. J. Clin. Microbiol. 36:2178–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stubbs SL, Rupnik M, Gibert M, Brazier JS, Duerden BI, Popoff MR. 2000. Production of actin-specific ADP-ribosyltransferase (binary toxin) by strains of Clostridium difficile. FEMS Microbiol. Lett. 186:307–312. 10.1111/j.1574-6968.2000.tb09122.x. [DOI] [PubMed] [Google Scholar]
- 31.Stubbs SL, Brazier JS, O'Neill GL, Duerden BI. 1999. PCR targeted to the 16S-23S rRNA gene intergenic spacer region of Clostridium difficile and construction of a library consisting of 116 different PCR ribotypes. J. Clin. Microbiol. 37:461–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Susick EK, Putnam M, Bermudez DM, Thakur S. 2012. Longitudinal study comparing the dynamics of Clostridium difficile in conventional and antimicrobial free pigs at farm and slaughter. Vet. Microbiol. 157:172–178. 10.1016/j.vetmic.2011.12.017. [DOI] [PubMed] [Google Scholar]
- 33.Avbersek J, Janezic S, Pate M, Rupnik M, Zidaric V, Logar K, Vengust M, Zemljic M, Pirs T, Ocepek M. 2009. Diversity of Clostridium difficile in pigs and other animals in Slovenia. Anaerobe 15:252–255. 10.1016/j.anaerobe.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 34.Goldová J, Malinová A, Indra A, Vítek L, Branny P, Jirásková A. 2012. Clostridium difficile in piglets in the Czech Republic. Folia Microbiol. (Praha) 57:159–161. 10.1007/s12223-012-0102-0. [DOI] [PubMed] [Google Scholar]
- 35.Boseiwaqa LV, Foster NF, Thean SK, Squire MM, Riley TV, Carson KC. 2013. Comparison of ChromID C. difficile agar and cycloserine-cefoxitin-fructose agar for the recovery of Clostridium difficile. Pathology 45:495–500. 10.1097/PAT.0b013e3283632680. [DOI] [PubMed] [Google Scholar]
- 36.Lyerly DM. 1992. The toxins of Clostridium difficile and their detection in the clinical laboratory. In: Clinical microbiology updates, vol 4, p 1–6 Hoechst-Roussel, Somerville, NJ. [Google Scholar]
- 37.Jure MN, Morse SS, Stark DM. 1988. Identification of nonspecific reaction in laboratory rodent specimens tested by Rotazyme rotavirus ELISA. Lab. Anim. Sci. 38:273–278. [PubMed] [Google Scholar]
- 38.Gumerlock PH, Tang YJ, Meyers FJ, Silva JJ. 1991. Use of the polymerase chain reaction for the specific and direct detection of Clostridium difficile in human feces. Rev. Infect. Dis. 13:1053–1060. 10.1093/clinids/13.6.1053. [DOI] [PubMed] [Google Scholar]
- 39.Bauer MP, Notermans DW, van Benthem BH, Brazier JS, Wilcox MH, Rupnik M, Monnet DL, van Dissel JT, Kuijper EJ. 2011. Clostridium difficile infection in Europe: a hospital-based survey. Lancet 377:63–73. 10.1016/S0140-6736(10)61266-4. [DOI] [PubMed] [Google Scholar]
- 40.Koene MGJ, Mevius D, Wagenaar JA, Harmanus C, Hensgens MPM, Meetsma AM, Putirulan FF, van Bergen MAP, Kuijper EJ. 2012. Clostridium difficile in Dutch animals: their presence, characteristics and similarities with human isolates. Clin. Microbiol. Infect. 18:778–784. 10.1111/j.1469-0691.2011.03651.x. [DOI] [PubMed] [Google Scholar]
- 41.Norén T, Johansson K, Unemo M. 2013. Clostridium difficile PCR ribotype 046 is common among neonatal pigs and humans in Sweden. Clin. Microbiol. Infect. 20:O2–O6. 10.1111/1469-0691.12296. [DOI] [PubMed] [Google Scholar]
- 42.Tenover FC, Novak-Weekley S, Woods CW, Peterson LR, Davis T, Schreckenberger P, Fang FC, Dascal A, Gerding DN, Nomura JH, Goering RV, Akerlund T, Weissfeld AS, Baron EJ, Wong EJ, Marlowe EM, Whitmore J, Persing DH. 2010. Impact of strain type on detection of toxigenic Clostridium difficile: comparison of molecular diagnostic and enzyme immunoassay approaches. J. Clin. Microbiol. 48:3719–3724. 10.1128/JCM.00427-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Foster NF, Collins DA, Ditchburn SL, Duncan CN, van Schalkwyk JW, Golledge CL, Keed ABR, Riley TV. 2014. Epidemiology of Clostridium difficile infection in two tertiary-care hospitals in Perth, Western Australia: a cross-sectional study. New Microb. New Infect. 2:64–71. 10.1002/nmi2.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rupnik M, Wilcox MH, Gerding DN. 2009. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat. Rev. Microbiol. 7:526–536. 10.1038/nrmicro2164. [DOI] [PubMed] [Google Scholar]