Abstract
Although Kingella kingae is the most common etiology of osteoarticular infections in young children, is a frequent cause of bacteremia in those younger than 4 years, and has been involved in clusters of invasive infections among daycare center attendees, the population structure of the species has not been systematically studied. Using multilocus sequence typing, we investigated the genetic diversity of the largest intercontinental collection of K. kingae strains to date. To facilitate typing of bacterial isolates, we developed a novel genotyping tool that targets the DNA uptake sequence (DUS). Among 324 strains isolated from asymptomatic carriers and patients from Israel, Europe, North America, and Australia with various invasive forms of the disease from 1960 to 2013, we identified 64 sequence types (STs) and 12 ST complexes (STcs). Five predominant STcs, comprising 72.2% of all strains, were distributed intercontinentally. ST-6 was the most frequent, showing a worldwide distribution, and appeared genotypically isolated by exhibiting few neighboring STs, suggesting an optimal fitness. ST-14 and ST-23 appeared to be the oldest groups of bacteria, while ST-25 probably emerged more recently from the highly evolutive ST-23. Using the DUS typing method, randomly chosen isolates were correctly classified to one of the major STcs. The comprehensive description of K. kingae evolution would help to detect new emerging clones and decipher virulence and fitness mechanisms. The rapid and reproducible DUS typing method may serve in the initial investigation of K. kingae outbreaks.
INTRODUCTION
Kingella kingae is a fastidious Gram-negative coccobacillus and a normal component of the oropharyngeal microbiota in young children, with a prevalence reaching to 10% to 12% among 12- to 24-month-old children (1, 2). The optimization of conventional culture techniques and the development of current molecular techniques, such as real-time PCR, led to the recognition of K. kingae as the major pathogen causing osteoarticular infections (OAIs) in children younger than 4 years in many countries (3–6) and as a common etiology of occult bacteremia (7) and, more rarely, of endocarditis in children and adults (3, 7, 8). Moreover, six outbreaks of invasive K. kingae infections in daycare centers in the United States, Israel, and France were recently described (8–12), with an average infection rate of around 20% (9–11) and up to an 85% carriage rate among healthy attendees (11), suggesting the increased colonization fitness, transmissibility, and invasiveness of some K. kingae clones.
Genetic typing of K. kingae isolates was recently performed with different molecular methods, including multilocus sequence typing (MLST), pulsed-field gel electrophoresis (PFGE), single-locus polymorphisms of the rtxA gene encoding the RTX toxin (13, 14), and the por gene that encodes the bacterial porin (15). The typing results revealed noticeable genomic heterogeneity in the species. To date, 40 MLST sequence types (STs) and 73 PFGE clones have been identified, as well as 18 rtxA and 12 different por alleles (13–18). Remarkable congruence has been observed between the results obtained using different typing methods (16, 18). Some MLST/PFGE groups have been shown to be positively or negatively associated with specific clinical syndromes, such as osteoarticular infections, occult bacteremia, or endocarditis (16). In a previous study of the genetic relationship between the K. kingae strains as determined by MLST, only 103 isolates derived from a few countries, mostly Israel and France, were included (13). This pioneer investigation revealed that several clones isolated from geographically distant locations were genotypically undistinguishable (13).
However, the PFGE and MLST methods, which have been used for K. kingae typing, are time-consuming, labor intensive, and costly; therefore, they are inadequate for conducting large-scale epidemiological studies. Moreover, single-gene typing methods that target virulence genes, such as rtxA, cannot be used because they are potentially subjected to selective pressure by the host's immune response, as well as horizontal transfers, with the same rtxA and por alleles observed in distantly related STs (13, 18). Therefore, the development of a rapid and cost-effective method is still pending. The DNA uptake sequence (DUS) is a short sequence required by certain bacteria, notably Neisseriaceae, for the acquisition of extracellular DNA (19, 20). The DUS, recently identified in K. kingae, consists of 12 nucleotides that are present on either the positive or negative DNA strand and are repeated in 2,787 copies in the ATCC 23330 K. kingae type strain genome (19). Given that the genome size of K. kingae is approximately 2 Mb (21, 22), the DUS should be randomly repeated, on average, every 500 to 1,000 bp; therefore, we hypothesized that these 12 nucleotides may serve as a potential PCR target for studying the genomic polymorphism of the strains using a one-primer amplification method.
In the current study, we provided a comprehensive description of the genetic diversity of K. kingae isolates by MLST on the largest intercontinental collection of 324 strains. Additionally, we developed a novel, rapid, and cost-effective molecular tool, based on DUS typing (DUST), to discriminate among the major ST complexes (STcs). This method may be used as a practical tool for rapidly investigating epidemiologically linked cases of K. kingae disease.
MATERIALS AND METHODS
Bacterial strains.
We collected 366 K. kingae isolates that have been isolated since the 1960s from different clinical and geographical origins. Among them, 42 isolates were epidemiologically related, derived from the same patient or from outbreaks of invasive disease. These 42 isolates were not utilized for the phylogenetic study, but a fraction of them was used to assess the performance of the DUST method.
Overall, 324 unrelated isolates were included in the phylogenetic study. The distributions of those isolates by country of origin, clinical syndrome, and year of isolation are indicated in Table S1 in the supplemental material. Some of these strains (n = 134) from Israel (n = 92), France (n = 29), the United States (n = 5), Iceland (n = 4), Norway (n = 3), and Russia (n = 1) were used previously in typing studies (13, 18).
Multilocus sequencing typing.
A detailed description of the MLST method has been published elsewhere (13). Briefly, fragments of 6 housekeeping genes (abcZ, adk, aroE, cpn60, gdh, and recA) were analyzed for the MLST. The STs and gene alleles identified are available at the Pasteur Institute of Paris website (http://www.pasteur.fr/recherche/genopole/PF8/mlst/Kingella_kingae.html).
The genomic relatedness within the population of strains (13, 18) was investigated by comparing allelic profiles using the minimum spanning tree method employing the BioNumerics software (version 7.1; Applied-Maths, Belgium). Isolates were grouped into ST complexes (STcs) if they differed at no more than one locus from at least one other member of the group. Founder genotypes of STcs were defined as the ST of the STc with the highest number of neighboring STs (single-locus variants).
DNA uptake sequence typing.
DNA was extracted from specimens with the BioRobot EZ1 workstation using the EZ1 DNA tissue kit (Qiagen, Courtaboeuf, France) according to the manufacturer's recommendations and stored at −80°C. The DUS, previously described for K. kingae (19), was used to design the primer king3DUS (5′-AAGCAGCCTGCA-3′). DNA PCR amplification was performed in a 50-μl reaction mixture that contained 25 μl of multiplex PCR master mix and 10 μl of Q-Solution (Qiagen), 1 μl of primer stock solution (50 μM), and 2 μl of DNA (5 ng/μl). Preliminary experiments were performed to determine the optimal amplification conditions and annealing temperatures (ranging from 40°C to 65°C) and different durations of the elongation step (30 s to 3 min) (data not shown). Optimal amplification was obtained with an iCycler (Bio-Rad, Marnes la Coquette, France), with an initial step of 15 min at 95°C followed by 30 cycles of 30 s at 95°C, 30 s at 40°C, 1 min 30 s at 72°C, and a final extension step of 7 min at 72°C. The amplification products were stained with ethidium bromide and visualized under UV light after a 70-min migration at 135 V in 2% high-resolution agarose gel. Bands below 300 bp and above 1,500 bp were excluded from the analysis because of the variability in their intensities, and the genetic relationship between the strains was determined by comparing fingerprint profiles using the curve-based Pearson correlation (optimization, 0.5%; curve smoothing, 0%). A dendrogram was constructed by using the unweighted-pair group method using average linkages (UPGMA) method (branch quality, cophenetic correlation) with BioNumerics software (version 7.1; Applied-Maths, Belgium).
Statistical analysis.
The potential of individual K. kingae STs to cause a specific syndrome was assessed using a previously described method (16). Briefly, an odds ratio (OR) was calculated as follows: the OR for ST-π is equal to (ad)/(bc), where a is the number of isolates belonging to ST-π causing a syndrome, b is the number of ST-π isolates causing other syndromes, c is the number of non-ST-π isolates causing the given syndrome, and d is the number of non-ST-π isolates causing other syndromes. For instance, an OR of >1 indicates an increased probability for a given ST to cause a specific syndrome, and an OR of <1 indicates a reduced probability for the ST. We computed 95% confidence intervals (CIs) by means. When the 95% CI did not include the unity, the observed OR was considered statistically significant.
Categorical variables were compared with the chi-square test or Fisher's exact test, as appropriate.
All tests were performed using the R statistical package version 3.1.0 (R Foundation for Statistical Computing, Vienna, Austria). A P value of <0.05 was considered significant for all comparisons.
RESULTS
Spatiotemporal distribution of major K. kingae STcs.
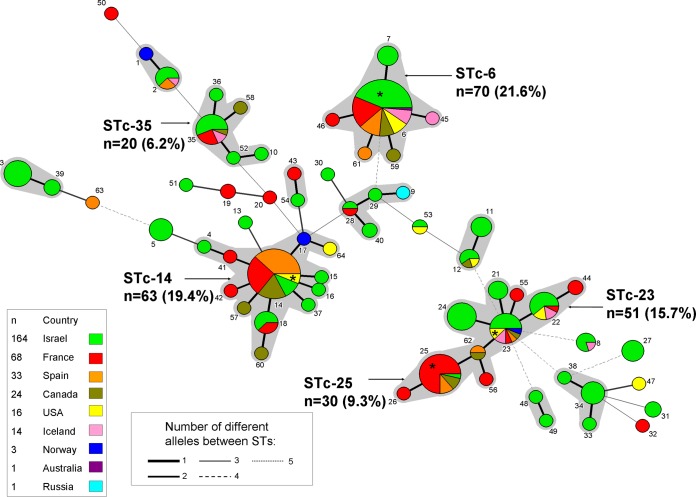
We performed MLST analysis on 324 epidemiologically unrelated strains isolated from patients with a variety of invasive K. kingae infections or from asymptomatic carriers, comprising 64 strains from France, 164 from Israel, 16 from the United States, 14 from Iceland, 33 from Spain, 24 from Canada, 1 from Russia, and 1 from Australia, as well as on 7 K. kingae strains available in the Pasteur Institute of Paris collection (4 from France and 3 from Norway) (Table 1). The MLST minimum spanning tree of all these strains, considering their country of isolation, is depicted in Fig. 1. A total of 64 STs and 12 STcs were identified. Five STcs (namely, STc-6, -14, -23, -25, and -35) were clearly predominant (each representing >5% of the entire strain population) and collectively represented 234 (72.2%) strains, including 70 (21.6%), 63 (19.4%), 51 (15.7%), 30 (9.3%), and 20 (6.2%) strains, respectively. STc-23 and STc-25 shared the ST-62, while the other 3 predominant STcs were distantly related (Fig. 1). The 5 main STcs showed remarkable intercontinental distributions (Fig. 1), although some STcs were overrepresented in some countries. In comparison to the distributions from all other countries, ST-25 and ST-14 were overrepresented in France (21/68, 30.9%) and in Spain (17/33, 51.5%) (P < 0.001, chi-square test), while they were both underrepresented in the Israeli set (0.6% were ST-25, and 3% were ST-14; P < 0.001) (Table 2).
TABLE 1.
Distribution of 324 Kingella kingae isolates by country of origin and clinical syndrome
| Clinical syndromea | No. of isolates causing the indicated clinical syndrome and originating from: |
Total no. of isolates | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Israel | France | Spain | Canada | United States | Iceland | Norway | Russia | Australia | ||
| Healthy carriage | 97 | 4 | 1 | 2 | 1 | 1 | 106 | |||
| OAI | 31 | 52 | 25 | 17 | 14 | 14 | 153 | |||
| SSTI | 3 | 3 | ||||||||
| Occult bacteremia | 26 | 3 | 7 | 2 | 2 | 40 | ||||
| Endocarditis | 10 | 2 | 1 | 13 | ||||||
| Unknown | 4 | 5 | 9 | |||||||
| Total | 164 | 68 | 33 | 24 | 16 | 14 | 3 | 1 | 1 | 324 |
OAI, osteoarticular infection; SSTI, skin and soft tissue infection.
FIG 1.
Minimum spanning tree analysis, using BioNumerics version 7.1, of the 324 Kingella kingae isolates based on allelic profiles of 6 housekeeping genes. Each circle corresponds to a sequence type (ST). The ST number is given beside the circle, and the size of the circle is related to the number of isolates found with that profile (from 1 for small circles [e.g., ST-1] to 60 [e.g., ST-6]). Each color inside the circles represents the geographical origin of the strains (green, Israel; red, France; orange, Spain; khaki, Canada; pink, Iceland; blue, Norway; yellow, United States; purple, Australia; and turquoise, Russia). Gray zones between some groups of circles indicate that these profiles belong to the same ST complex. Width of the line joining two STs indicates the number of alleles differing. *, One strain of the ST was responsible for an outbreak of K. kingae invasive infections at a daycare center in the country related to the colored background.
TABLE 2.
Distribution of the 5 most frequent sequence types by country of isolation
| STa | No. (%) of isolates with the indicated ST and originating from: |
Total no. (%) of isolates | ||||
|---|---|---|---|---|---|---|
| Israel | France | Spain | Canada | United States | ||
| ST-6 | 26 (15.9) | 11 (16.2) | 7 (21.2) | 5 (20.8) | 5 (31.3) | 60 (18.5) |
| ST-14 | 5 (3.0)b | 12 (17.6) | 17 (51.5)b | 8 (33.3) | 3 (18.8) | 45 (13.9) |
| ST-23 | 8 (4.9) | 1 (1.5) | 1 (3.0) | 3 (12.5) | 2 (12.5) | 16 (4.9) |
| ST-25 | 1 (0.6)b | 21 (30.9)b | 3 (9.0) | 1 (4.2) | 0 (0.0) | 28 (8.6) |
| ST-35 | 9 (5.5) | 4 (5.9) | 0 (0.0) | 1 (4.2) | 0 (0.0) | 16 (4.9) |
| Total | 164 (100) | 68 (100) | 33 (100) | 24 (100) | 16 (100) | 324 (100) |
Sequence type.
P < 0.001 compared to the all-other-countries distribution of the given ST by the chi-square test.
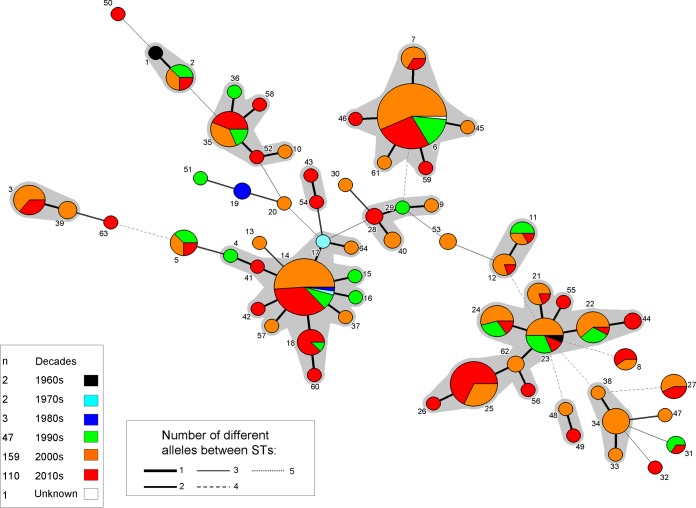
Some of the predominant STcs showed remarkable persistence over time (Fig. 2). For, instance, some strains belonging to STc-23 and STc-14 were isolated in the 1960s and the 1970s, respectively. In contrast, no isolates belonging to STc-25 were found before 2002, which may suggest that it is a newly emerged clone (Fig. 2). Of note, strain ATCC 23330, which has a noninvasive origin and is the oldest K. kingae strain available, is the sole isolate of ST-1 (Fig. 1). Our work indicates, on a large scale, that this strain is not representative of the major STcs or of K. kingae species as a whole.
FIG 2.
MLST minimum spanning tree analysis, using BioNumerics version 7.1, of the 324 Kingella kingae isolates depending on their year of isolation. Each color inside the circles represents the decade of isolation (white, unknown; black, 1960s; turquoise, 1970s; blue, 1980s; green, 1990s; orange, 2000s; and red, 2010s).
While studying the ratio between the number of strains belonging to a founder ST (defined as the ST with the highest number of neighboring STs within a given STc) and the total number of strains included in that STc, we observed significant differences. Indeed, ST-23 included only 16 of 50 (32%) isolates of STc-23, whereas ST-6 represented 85.7%; ST-14 included 71.4%, ST-25 included 96.6%, and ST-35 included 80% of their respective STcs (P < 0.001). Moreover, ST-23 was the founder of 3 important STs containing >5 strains each (ST-21, ST-22, and ST-24), while the other main STcs were composed of numerous minor STs (Fig. 1). These results may suggest that ST-23 is highly evolutive, giving origin to multiple and epidemiologically successful STs.
Relationship between STs and clinical syndromes.
Among 324 isolates, the clinical origins were known in 315 (97.2%) cases, including 106 (33.7%) derived from healthy carriers, 153 (48.6%) from patients with OAI, 40 (12.7%) with occult bacteremia, 13 with endocarditis (4.1%), and 3 (1.0%) with skin and soft tissue infections (Table 1). Three of the major STs were significantly associated with a specific syndrome (Table 3). ST-14 and ST-25 were positively associated with OAI and negatively associated with healthy carriage, while ST-24 (belonging to STc-23) was significantly associated with endocarditis (Table 3). Altogether, all other minor STs were significantly and positively associated with healthy carriage, especially ST-3, ST-34, and ST-5 (P < 0.001), as well as ST-11 and ST-27 (P = 0.003) (data not shown), whereas they were negatively associated with OAI (Table 3).
TABLE 3.
Distribution of the sequence types of Kingella kingae by clinical syndromes
| STa | Data by clinical syndromeb |
All syndromes (no.)c | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carriage |
OAI |
Occult bacteremia |
Endocarditis |
||||||||||
| n | OR (95% CI) | P | n | OR (95% CI) | P | n | OR (95% CI) | P | n | OR (95% CI) | P | ||
| 6 | 13 | 0.51 (0.24–1.02) | 0.047 | 30 | NS | 11 | NS | 4 | NS | 60 | |||
| 14 | 3 | 0.13 (0.02–0.41) | <0.001 | 32 | 3.66 (1.77–7.94)d | <0.001 | 6 | NS | 0 | NS | 45 | ||
| 22e | 4 | NS | 7 | NS | 2 | NS | 0 | NS | 13 | ||||
| 23e | 5 | NS | 6 | NS | 3 | NS | 1 | NS | 16 | ||||
| 24e | 4 | NS | 3 | 3 | NS | 3 | 9.89 (1.50–48.62) | 0.009 | 13 | ||||
| 25 | 4 | 0.33 (0.08–0.99) | 0.035 | 18 | 2.42 (1.02–6.09) | 0.03 | 0 | NS | 1 | NS | 28 | ||
| 35 | 4 | NS | 8 | NS | 3 | NS | 0 | NS | 16 | ||||
| Other | 72 | 5.54 (3.27–9.55) | <0.001f | 49 | 0.50 (0.31–0.81) | 0.003 | 12 | 4 | NS | 133 | |||
| Total | 106 | 153 | 40 | 13 | 324 | ||||||||
ST, sequence type.
OR, odds ratio; CI, confidence interval; OAI, osteoarticular infection; NS, not significant.
All represented strains from carriage, OAI, occult bacteremia, endocarditis, skin and soft tissue, and unknown origins.
Bold type represents significant positive association.
Belonged to ST-23.
Among these, other STs, including ST-3, ST-34, and ST-5 (P < 0.001) and ST-11 and ST-27 (P = 0.003), were positively associated with healthy carriage.
The strains responsible for four daycare center outbreaks reported worldwide (8–11) belonged to four different STs (Fig. 1). Indeed, isolates of the U.S. outbreaks belonged to ST-14 (8) and ST-23 (9), while ST-6 was involved in a cluster of disease at an Israeli facility (10), and ST-25 was involved in a French outbreak (11) (Fig. 1). The fact that the strains causing outbreaks, which had high infection rates and high colonization rates among healthy attendees, occurred in four different locations and belonged to four of the main invasive STs strengthens the hypothesis that these main predominant STs possess biological advantages that allow them to colonize the upper respiratory tract, disseminate from person to person, and cause invasive infections.
DUST is a rapid and efficient first-line typing tool.
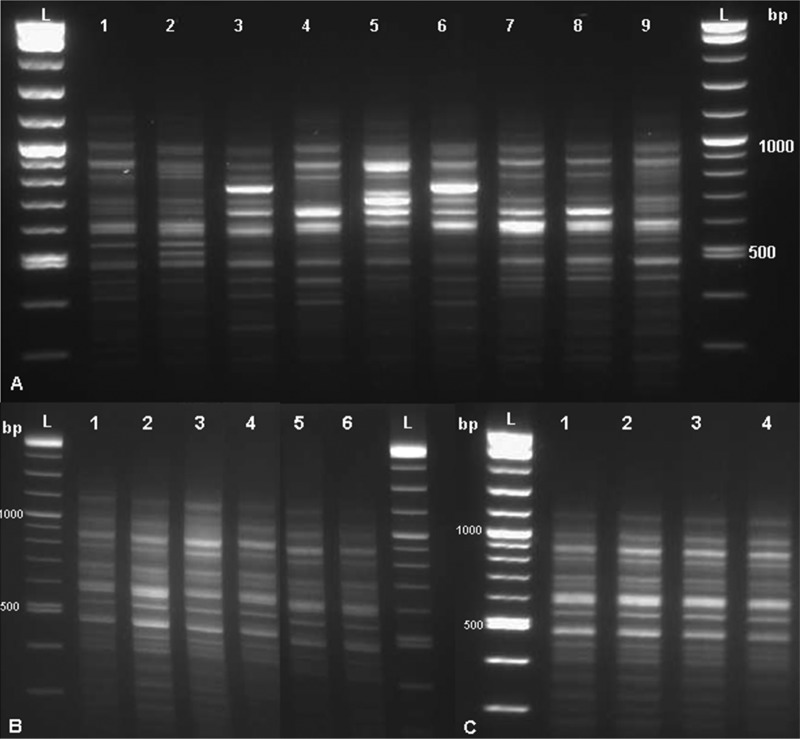
To develop a rapid and simple tool for epidemiological study, we hypothesized that the highly ubiquitous DUS, present in the K. kingae genome, may serve as a potential PCR target for studying genomic polymorphism (19). Of note, the DUS typing method amplifies sequences located between two DUS and not the DUS by itself. Therefore, each sequence between two DUS is unique. Initially, we performed DUST on strains belonging to each of the 9 major STcs (containing >5 isolates each) and observed 8 different clusters. Two highly related STcs (STc-23 and STc-25) exhibited similar patterns (Fig. 3A).
FIG 3.
Gel electrophoresis of the PCR targeting the Kingella kingae DNA uptake sequence. (A) One K. kingae strain was used for each of 9 studied sequence type complexes (STcs). Lane 1, STc-1; lane 2, STc-3; lane 3, STc-35; lane 4, STc-6; lane 5, STc-14; lane 6, STc-34; lane 7, STc-23; lane 8, STc-25; lane 9, STc-11; lanes L, DNA ladders. (B) Independent experiments using DNA extract of K. kingae type strain ATCC 23330 in quadruplicate using the same DNA extract (lanes 1 to 4) and with DNA derived from two different subcultures (lanes 5 and 6). Lanes L, DNA ladders. (C) Different dilutions of the DNA extract of K. kingae type strain ATCC 23330. Lane 1, pure; lane 2, 1/4; lane 3, 1/16; lane 4, 1/64; lane L, DNA ladders.
Then, to confirm the reproducibility of this PCR assay, we performed multiple experiments on the ATCC 23330 strain in quadruplicate using the same DNA extract, with DNA derived from 2 different subcultures, and using the same DNA extract at 1/4, 1/16, and 1/64 dilutions. No significant differences between experimental results were observed (Fig. 3B and C), indicating the robustness of this technique.
Furthermore, we performed DUS amplification on 8 randomly chosen strains belonging to each of the 9 major STcs (STc-1, STc-3, STc-6, STc-11, STc-14, STc-23, STc-25, STc-34, and STc-35) and in all the strains if the STc contained fewer than 8 strains, with the results presented in the dendrogram depicted in Fig. 4. We observed clusters of similar patterns between strains belonging to a given STc with >95% similarity, independent of the geographical origin of the strains (Fig. 4). All strains but one (ST-14-BOU) belonging to a given ST exhibited >97% similarity. Of particular interest, four epidemiologically related strains (STF5A, STF12A, STF15A, and STF16B), involved in the French daycare facility outbreak and belonging to ST-25 (11), were correctly linked by the DUST method (Fig. 4).
FIG 4.
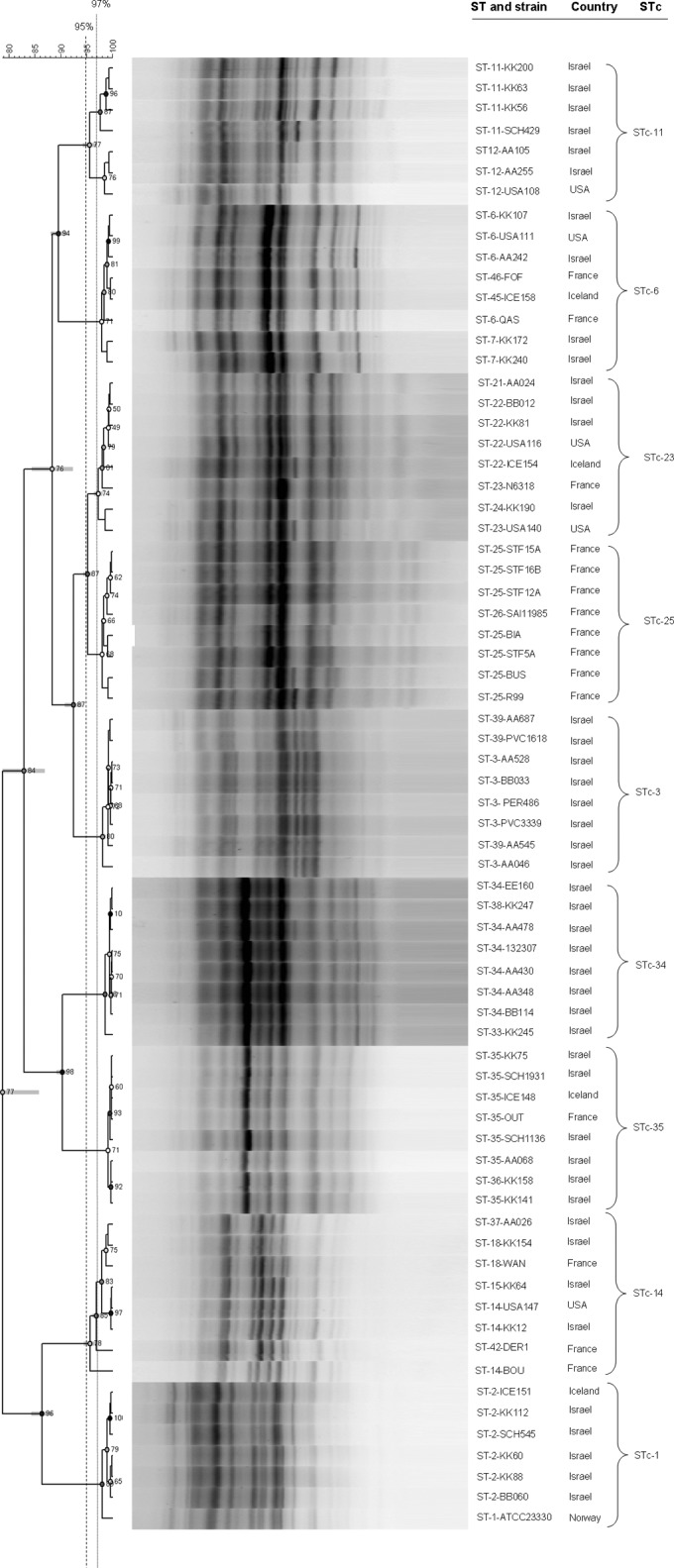
Dendrogram, using Pearson's correlation and constructed by using the UPGMA method, of PCR targeting the DNA uptake sequence on 7 to 8 randomly chosen Kingella kingae strains for each of the 9 main sequence type complexes (STcs). Fingerprint profiles are shown beside the strains name, their sequence type (ST), STc, and geographical origin. Strains exhibiting >97% similarity (dotted line) were considered undistinguishable, and a cluster of similar patterns was defined by >95% similarity (dashed line).
Finally, we performed DUST on one randomly chosen strain belonging to 9 minor STs (which included at least two isolates each), and we observed 8 different profiles (see Fig. S1 in the supplemental material).
DISCUSSION
In this study, we described the wide genetic diversity of the K. kingae species on the largest intercontinental strain collection to date. Interestingly, only 5 STcs represented 72.2% of the whole population, and each was intercontinentally distributed, suggesting a high dissemination potential.
Genotypic and spatiotemporal analyses allowed a step forward in acquiring knowledge about K. kingae genetic evolution. ST-23 appeared as a highly evolutive ST, giving origin to multiple and epidemiologically successful STs. ST-23 contained one of the oldest isolates, while ST-25 appeared to be a recently emerged ST. ST-23 and ST-25 were closely related, exhibiting 4 of 6 alleles in common, and shared the locus variant ST-62. Sequences of the two discordant alleles (abcZ and aroE) from the ST-23 and ST-25 strains differed by one nucleotide each, and these two nucleotides were identical in both ST-23 and Kingella oralis ATCC 51147, the most closely related species (data not shown). Altogether, these results suggest that ST-25 originally evolved from ST-23 and might have derived from ST-62 more recently.
Conversely, ST-6 appeared as an ST with optimal fitness. Indeed, it was quantitatively the predominant ST, represented in 18.5% (60 of 324) of all the typed strains; it was found in the four studied continents, it had very few neighboring STs, each containing a limited number of isolates, and it was distantly related to all other members of the species. Moreover, ST-6 was previously reported as common in Israel among healthy carriers and patients with invasive K. kingae infections (14, 16, 23). Thus, these results support the hypothesis that ST-6 has a successful evolutionary history and has reached an optimal equilibrium between enhanced colonization fitness, high transmissibility, and remarkable virulence.
Using the large intercontinental collection of K. kingae strains, we attempted to associate STs and clinical syndromes to confirm the results of our previous study on 181 Israeli invasive isolates (16). It was observed that clone K/ST-6 was associated with occult bacteremia, clone P/ST-24 with endocarditis, and clone N/ST-35 with OAI (16). In the current study, among 209 invasive strains with a known clinical source (67 from Israel and 142 from elsewhere), we found that ST-14 and ST-25 were also associated with OAI and confirmed that ST-24 is associated with endocarditis. However, as occult bacteremia strains are mainly from Israel, we were unable to confirm their association with ST-6 in our intercontinental collection. Similarly, interpretation of the association between STs and healthy carriage, which we highlighted, should be done with caution since 97/106 (91.5%) carriage strains were from Israel. Exploring the associations between STs and clinical syndromes may be useful to better understand the pathophysiology of K. kingae infections and to interpret the presence of K. kingae strains in the oropharynx of infected patients without osteoarticular sampling.
Genotyping studies using PFGE and MLST have allowed researchers to describe the genetic diversity of the K. kingae species (13, 14) and investigate the genetic relatedness between the strains, notably during outbreaks (8–11). However, these genotyping tools are time-consuming and labor-intensive. Based on the phylogenetic organization of the species that we described, we developed a novel and rapid molecular typing tool targeting the DUS. We observed that DUST was able to correctly classify randomly chosen isolates to 1 of the 9 major STcs that represented 86.4% (280/324) of all strains. DUST was easy to implement, suggesting that it may be a suitable method for a first-line investigation of outbreaks, because it enables reliable discrimination between genetically unrelated strains in <4 h and at a cost of <5 U.S. dollars per strain. However, further analysis using more discriminatory methods, such as MLST or PFGE, will be required to confirm the genetic linkage between strains. However, further experience and validation of the DUST method by using traditional methods with additional strains, such as MLST or PFGE, are needed.
In conclusion, K. kingae exhibits a wide genetic diversity, but only a few STcs are strongly predominant, internationally distributed, and responsible for the majority of diseases occurring worldwide. Our results suggest that these leading STcs possess some genetic determinants that give them the ability to disseminate from person to person and to cause invasive infections. Different evolutionary potentials were observed between the main STs; ST-23 is probably the most successfully evolving clone, whereas ST-6 appears to have optimal fitness. This comprehensive description of K. kingae evolution may help researchers to detect new emerging clones and determine strains that have to be studied to decipher virulence and fitness mechanisms. The novel DUST method may be very useful for providing rapid discrimination between the main STcs and may serve as an initial molecular tool for the epidemiological investigation of clusters of invasive K. kingae disease. This approach can probably be easily applied to other bacterial species typing, specifically to other members of the Neisseriaceae family.
Supplementary Material
ACKNOWLEDGMENTS
We thank Hoang Vu-Thien, Didier Moissenet, Jocelyne Cailllon, Philippe Ovetchkine, Raymond Ruimy, Philippe Lehours, Anne-Laure Robbe, Amadeu Gene, and Darren Hewson for providing K. kingae strains.
This work was supported in part by the Société Française de Pédiatrie. The funders had no role in the study design, data collection or analysis, publication decision, or preparation of the manuscript.
Footnotes
Published ahead of print 20 August 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.01609-14.
REFERENCES
- 1.Yagupsky P, Porsch E, St Geme JW., 3rd 2011. Kingella kingae: an emerging pathogen in young children. Pediatrics 127:557–565. 10.1542/peds.2010-1867. [DOI] [PubMed] [Google Scholar]
- 2.Ceroni D, Dubois-Ferriere V, Anderson R, Combescure C, Lamah L, Cherkaoui A, Schrenzel J. 2012. Small risk of osteoarticular infections in children with asymptomatic oropharyngeal carriage of Kingella kingae. Pediatr. Infect. Dis. J. 31:983–985. 10.1097/INF.0b013e31825d3419. [DOI] [PubMed] [Google Scholar]
- 3.Yagupsky P. 2004. Kingella kingae: from medical rarity to an emerging paediatric pathogen. Lancet Infect. Dis. 4:358–367. 10.1016/S1473-3099(04)01046-1. [DOI] [PubMed] [Google Scholar]
- 4.Ilharreborde B, Bidet P, Lorrot M, Even J, Mariani-Kurkdjian P, Liguori S, Vitoux C, Lefevre Y, Doit C, Fitoussi F, Pennecot G, Bingen E, Mazda K, Bonacorsi S. 2009. New real-time PCR-based method for Kingella kingae DNA detection: application to samples collected from 89 children with acute arthritis. J. Clin. Microbiol. 47:1837–1841. 10.1128/JCM.00144-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ceroni D, Cherkaoui A, Ferey S, Kaelin A, Schrenzel J. 2010. Kingella kingae osteoarticular infections in young children: clinical features and contribution of a new specific real-time PCR assay to the diagnosis. J. Pediatr. Orthop. 30:301–304. 10.1097/BPO.0b013e3181d4732f. [DOI] [PubMed] [Google Scholar]
- 6.Chometon S, Benito Y, Chaker M, Boisset S, Ploton C, Berard J, Vandenesch F, Freydiere AM. 2007. Specific real-time polymerase chain reaction places Kingella kingae as the most common cause of osteoarticular infections in young children. Pediatr. Infect. Dis. J. 26:377–381. 10.1097/01.inf.0000259954.88139.f4. [DOI] [PubMed] [Google Scholar]
- 7.Dubnov-Raz G, Ephros M, Garty BZ, Schlesinger Y, Maayan-Metzger A, Hasson J, Kassis I, Schwartz-Harari O, Yagupsky P. 2010. Invasive pediatric Kingella kingae infections: a nationwide collaborative study. Pediatr. Infect. Dis. J. 29:639–643. 10.1097/INF.0b013e3181d57a6c. [DOI] [PubMed] [Google Scholar]
- 8.Seña AC, Seed P, Nicholson B, Joyce M, Cunningham CK. 2010. Kingella kingae endocarditis and a cluster investigation among daycare attendees. Pediatr. Infect. Dis. J. 29:86–88. 10.1097/INF.0b013e3181b48cc3. [DOI] [PubMed] [Google Scholar]
- 9.Kiang KM, Ogunmodede F, Juni BA, Boxrud DJ, Glennen A, Bartkus JM, Cebelinski EA, Harriman K, Koop S, Faville R, Danila R, Lynfield R. 2005. Outbreak of osteomyelitis/septic arthritis caused by Kingella kingae among child care center attendees. Pediatrics 116:e206–e213. 10.1542/peds.2004-2051. [DOI] [PubMed] [Google Scholar]
- 10.Yagupsky P, Erlich Y, Ariela S, Trefler R, Porat N. 2006. Outbreak of Kingella kingae skeletal system infections in children in daycare. Pediatr. Infect. Dis. J. 25:526–532. 10.1097/01.inf.0000215243.42501.4f. [DOI] [PubMed] [Google Scholar]
- 11.Bidet P, Collin E, Basmaci R, Courroux C, Prisse V, Dufour V, Bingen E, Grimprel E, Bonacorsi S. 2013. Investigation of an outbreak of osteoarticular infections caused by Kingella kingae in a childcare center using molecular techniques. Pediatr. Infect. Dis. J. 32:558–560. 10.1097/INF.0b013e3182867f5e. [DOI] [PubMed] [Google Scholar]
- 12.Yagupsky P. 2014. Outbreaks of Kingella kingae infections in daycare facilities. Emerg. Infect. Dis. 20:746–753. 10.3201/eid2005.131633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Basmaci R, Yagupsky P, Ilharreborde B, Guyot K, Porat N, Chomton M, Thiberge JM, Mazda K, Bingen E, Bonacorsi S, Bidet P. 2012. Multilocus sequence typing and rtxA toxin gene sequencing analysis of Kingella kingae isolates demonstrates genetic diversity and international clones. PLoS One 7:e38078. 10.1371/journal.pone.0038078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yagupsky P, Weiss-Salz I, Fluss R, Freedman L, Peled N, Trefler R, Porat N, Dagan R. 2009. Dissemination of Kingella kingae in the community and long-term persistence of invasive clones. Pediatr. Infect. Dis. J. 28:707–710. 10.1097/INF.0b013e31819f1f36. [DOI] [PubMed] [Google Scholar]
- 15.Banerjee A, Kaplan JB, Soherwardy A, Nudell Y, Mackenzie GA, Johnson S, Balashova NV. 2013. Characterization of TEM-1 beta-lactamase producing Kingella kingae clinical isolates. Antimicrob. Agents Chemother. 57:4300–4306. 10.1128/AAC.00318-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amit U, Porat N, Basmaci R, Bidet P, Bonacorsi S, Dagan R, Yagupsky P. 2012. Genotyping of invasive Kingella kingae isolates reveals predominant clones and association with specific clinical syndromes. Clin. Infect. Dis. 55:1074–1079. 10.1093/cid/cis622. [DOI] [PubMed] [Google Scholar]
- 17.Yagupsky P, Slonim A, Amit U, Porat N, Dagan R. 2013. Beta-lactamase production by Kingella kingae in Israel is clonal and common in carriage organisms but rare among invasive strains. Eur. J. Clin. Microbiol. Infect. Dis. 32:1049–1053. 10.1007/s10096-013-1849-1. [DOI] [PubMed] [Google Scholar]
- 18.Basmaci R, Bonacorsi S, Bidet P, Balashova NV, Lau J, Munoz-Almagro C, Gene A, Yagupsky P. 13 June 2014. Genotyping, local prevalence, and international dissemination of beta-lactamase-producing Kingella kingae strains. Clin. Microbiol. Infect. 10.1111/1469-0691.12648. [DOI] [PubMed] [Google Scholar]
- 19.Frye SA, Nilsen M, Tonjum T, Ambur OH. 2013. Dialects of the DNA uptake sequence in Neisseriaceae. PLoS Genet. 9:e1003458. 10.1371/journal.pgen.1003458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duffin PM, Seifert HS. 2010. DNA uptake sequence-mediated enhancement of transformation in Neisseria gonorrhoeae is strain dependent. J. Bacteriol. 192:4436–4444. 10.1128/JB.00442-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaplan JB, Lo C, Xie G, Johnson SL, Chain PS, Donnelly R, Kachlany SC, Balashova NV. 2012. Genome sequence of Kingella kingae septic arthritis isolate PYKK081. J. Bacteriol. 194:3017. 10.1128/JB.00421-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fournier PE, Rouli L, El Karkouri K, Nguyen TT, Yagupsky P, Raoult D. 2012. Genomic comparison of Kingella kingae strains. J. Bacteriol. 194:5972. 10.1128/JB.01418-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Slonim A, Walker ES, Mishori E, Porat N, Dagan R, Yagupsky P. 1998. Person-to-person transmission of Kingella kingae among day care center attendees. J. Infect. Dis. 178:1843–1846. 10.1086/314488. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.