Abstract
Streptococcus agalactiae frequently colonizes the urogenital tract, and it is a major cause of bacterial septicemia, meningitis, and pneumonia in newborns. For typing purposes, a microarray targeting group B streptococcus (GBS) virulence-associated markers and resistance genes was designed and validated with reference strains, as well as clinical and veterinary isolates. Selected isolates were also subjected to multilocus sequence typing. It was observed that putative typing markers, such as alleles of the alpha-like protein or capsule types, vary independently of each other, and they also vary independently from the affiliation to their multilocus sequence typing (MLST)-defined sequence types. Thus, it is not possible to assign isolates to sequence types based on the identification of a single distinct marker, such as a capsule type or alp allele. This suggests the occurrence of frequent genomic recombination. For array-based typing, a set of 11 markers (bac, alp, pil1 locus, pepS8, fbsB, capsule locus, hylB, abiG-I/-II plus Q8DZ34, pil2 locus, nss plus srr plus rogB2, and rgfC/A/D/B) was defined that provides a framework for splitting the tested 448 S. agalactiae isolates into 76 strains that clustered mainly according to MLST-defined clonal complexes. There was evidence for region- and host-specific differences in the population structure of S. agalactiae, as well as an overrepresentation of strains related to sequence type 17 among the invasive isolates. The arrays and typing scheme described here proved to be a convenient tool for genotyping large numbers of clinical/veterinary isolates and thus might help obtain insight into the epidemiology of S. agalactiae.
INTRODUCTION
Streptococcus agalactiae, also commonly referred to as group B streptococcus (GBS), belongs to the gastrointestinal and genitourinary flora in humans and can cause, e.g., urinary tract infections. More importantly, it can be found in the vagina, and up to 20 to 40% of pregnant women are colonized (1–4). S. agalactiae is a major cause of septicemia, meningitis, and pneumonia in newborns (5). Mother-to-child transmission may lead to neonatal infection in 1 to 2 infants per 1,000 live births, with mortality rates ranging from 10 to 15% for early onset disease and 2 to 6% for late-onset disease (6–8). For prevention, it is strongly recommended to collect vaginal and rectal swabs between 35 and 37 weeks after gestation for screening and to eradicate GBS if present. The recommended antibiotics for decolonization and therapy include penicillin G, ampicillin, or first-/second-generation cephalosporins. Resistance to the β-lactams has rarely been observed (9). Other options, e.g., in case of allergy, are erythromycin or clindamycin, although there is a considerable risk of resistance (10–14). Tetracyclines can be given to nonpregnant adults only, and under some conditions, other compounds, like vancomycin or linezolid, may be considered.
S. agalactiae is also an important pathogen in animals, especially in cattle, in which it causes bovine mastitis. It may also cause meningoencephalitis and septicemia in fish (15).
Several typing techniques have been described. These include serotyping (16–20), restriction fragment length polymorphism analysis (21), ribotyping (22), pulsed-field gel electrophoresis (23), multilocus enzyme electrophoresis (24, 25), analysis of randomly amplified polymorphic DNA (26), amplified cps restriction polymorphism analysis (27) and multilocus sequence typing (MLST) (28).
For the present work, a microarray for the typing of GBS was designed and validated with reference strains and clinical isolates.
MATERIALS AND METHODS
Bacterial isolates.
Eight fully sequenced strains, i.e., S. agalactiae NEM316 (GenBank accession no. AL732656.1), A909 (CP000114.1), 2603V/R (AE009948), COH1 (AAJR00000000.1), strain 515 (AAJP00000000.1), H36B (AAJS00000000.1), 18RS21 (AAJO00000000.1), and CJB111 (AAJQ00000000.1) were tested and used for quality control purposes and protocol optimization.
One hundred thirty-two isolates from the Dresden University Hospital were collected between 2008 and mid-2010. This mainly included routine vaginal and rectal swabs but also other clinical samples, like swabs from the ears, eyes, and throats of newborns (n = 8), as well as swabs from (chronic) wounds (n = 18), blood cultures (n = 9), urine (n = 14), breast milk samples (n = 4), and swabs from the male genital tract (n = 4).
Additionally, 97 isolates (courtesy of R. Berner, Freiburg/Dresden) from invasive infections of newborns, i.e., septicemia, meningitis, and pneumonia, were collected across Germany as part of the ESPED (i.e., the Surveillance Unit for Rare Pediatric Diseases in Germany) study and were genotyped. Furthermore, surveillance isolates from different parts of Germany (courtesy of M. van der Linden, National Reference Centre for Streptococci, Aachen, Germany [n = 119], and S. Weber, Bioscientia MVZ, Saarbrücken, Germany [n = 36]) and from the hospital of the University of West Indies, St. Augustine, Trinidad and Tobago (courtesy of P. Akpaka; n = 34), were processed. One isolate originated from a laboratory proficiency test (supplied by Instand e.V. [http://www.instandev.de/en.html]). Finally, 21 isolates from bovine mastitis cases were included (courtesy of K. Kadlec, Friedrich-Loeffler-Institute, Neustadt-Mariensee, Germany).
Ethics statement.
The isolates were obtained during routine diagnostics and were analyzed anonymously. No patient data beyond classification as infant or adult and sample type were stored. Ethics approval and informed consent were thus not required.
Preparation of genomic DNA.
The isolates were incubated for 12 to 24 h at 37°C on Columbia blood agar. One inoculation loop of colony material was harvested and placed in 100 μl of lysis buffer comprising 10 μl of achromopeptidase (catalog no. A3547-100KU; Sigma-Aldrich; 100 units dissolved in 5 ml of phosphate-buffered saline [PBS] and stored frozen in small aliquots), 2 mg of lysozyme (Sigma-Aldrich), 2 mg of RNase A (Sigma-Aldrich), 2 μl of 20 mM Tris-HCl (pH 8.0), 2 μl of 2 mM EDTA, and 1 μl of Triton X-100. After incubation (at 37°C and 550 rpm on a thermomixer), 10 μl of proteinase K and 100 μl of AL buffer (Qiagen, Hilden, Germany) were added, and another incubation step (55°C without mixing) followed. After adding 100 μl of ethanol, DNA was purified using Qiagen spin columns. Finally, DNA was rediluted in 50 μl of PCR-grade distilled water and heated (for 10 min at 70°C) to evaporate any traces of ethanol from the washing buffers.
MLST.
Representative isolates were characterized by MLST using protocols and software provided at http://pubmlst.org/sagalactiae/.
Microarray design and protocol optimization.
The microarray covered typing markers, such as capsule- and pilus-associated genes and alp genes. Additionally, macrolide/lincosamide, tetracycline, and heavy metal resistance genes, genes associated with phages and gene motility, and various genes coding for virulence factors, digestive enzymes, and certain metabolic functions were included, with variability in the presence or sequence being the main inclusion criterion (Tables 1 to 4; see also Tables S1 and S2 in the supplemental material).
TABLE 1.
Key markers for array-based strain classification, their average positions in the S. agalactiae genome sequence, and known alleles
| Median position based on NEM316, A909, and 2603V/R | Gene(s) | Presence/allele (GenBank accession no. or strain) | Representative GenBank accession no. (positions) | Code no. |
|---|---|---|---|---|
| 145000 | bac | Negative | B-0 | |
| Positive | CP000114.1 (145690–149184) | B-1 | ||
| 459000 | alp | Negative | A-0 | |
| alp-2 | AL766845.1 (114012–117392) | A-2 | ||
| alp-3 | AF245663.1 (365–2962) | A-3 | ||
| alp-4 | DQ629924.1 (1–1095) | A-4 | ||
| alp-5 | DQ629925.1 (1–1347) | A-5 | ||
| alp-bca | CP000114.1 (459256–462318) | A-B | ||
| alp-rib2 | AM050558.1 (1–735; partial) | A-R2 | ||
| alp-rib (R4) | AE009948.1 (447507–451676) | A-R4 | ||
| 647000 | pil1 locus | Negative | P1-0 | |
| pil1A/C | P1–2 | |||
| pil1A/B/C | AE009948.1 (A: 636431–639103)/(B: 631950–633614)/(C: 633703–634626) | P1–3 | ||
| 700000 | pepS8 | Negative | E-0 | |
| Positive | CP000114.1 (740435–743092) | E-1 | ||
| 877000 | fbsB | fbsB (AM050623) | AM050623 (1–1293) | F-3 |
| fbsB (AM050624) | AM050624 (1–1992) | F-4 | ||
| fbsB (515) | AAJP01000006.1 (11614–13521) | F-5 | ||
| fbsB (AY326423) | AY326423 (167–2320) | F-6 | ||
| fbsB (A909) | CP000114.1 (910378–912639) | F-9 | ||
| 1171000–1235000 | Capsule | No capsule | C-0 | |
| locus | Capsule type IA | For capsH Ia, CP000114.1 (1226719–1227858) | C-1A | |
| Capsule type IB | For capsH Ib, AAJS01000021.1 (16021–17160) | C-1B | ||
| Capsule type II | For capsH II, AAJO01000077.1 (1918–3024) | C-2 | ||
| Capsule type III | For capsH III, AL732656.1 (1279631–1280776) | C-3 | ||
| Capsule type IV | For capsH IV, AF355776.1 (6895–7992) | C-4 | ||
| Capsule type V | For capsH V, AE009948.1 (1172623–1173717) | C-5 | ||
| Capsule type VI | For capsH VI, AF337958.1 (6907–8064) | C-6 | ||
| Capsule type VII | For capsH VII, AY376403.1 (3881–4975) | C-7 | ||
| Capsule type VIII | For capsH VIII, AY375363.1 (4219–5367) | C-8 | ||
| Capsule type IX | For capsH IX, EF157290.1 (1–330; partial) | C-9 | ||
| 1256000 | hylB | Lacking hylB | H-0 | |
| hylB (NEM316) | AL732656.1 (1308821–1311775) | H-3 | ||
| hylB (A909) | CP000114.1 (1256012–1258993) | H-9 | ||
| 1353000 | abiG-I/-II, | Lacking abiG-I/-II, Q8DZ34 | Q-0 | |
| Q8DZ34 | abiG-I/-II, Q8DZ34 (NEM316) | AL732656.1 (1408678–1409268)/(1409265–1410113)/(1410217–1413012) | Q-3 | |
| abiG-I/-II, Q8DZ34 (H36B) | AAJS01000027.1 (10052–10642)/(10639–11483)/(11569–14370) | Q-6 | ||
| 1419000–1422000 | pil2 locus | Lacking pil2 | P2-0 | |
| pil2 A/B/C | pil2a (18RS21) | AAJO01000002.1 (A: 21274–23979)/(B: 19027–21144)/(C: 16227–17153) | P2-A1 | |
| pil2a (NEM316) | AL732656.1 (A: 1528058–1530763)/(B: 1525911–1527935)/(C: 1523114–1524040) | P2-A3 | ||
| pil2a (ABC020013424) | A: EU929280.1 (12346–15051)/B: EU929899.1 (1–2082) | P2-A4 | ||
| pil2a (515) | AAJP01000012.1 (A: 12346–15051)/(B: 15181–17208)/(C: 19079–20005) | P2-A5 | ||
| pil2a (H36B) | AAJS01000017.1 (A: 21508–24198)/(B: 19308-21389)/(C: 16511–17437) | P2-A6 | ||
| pil2a (ABC020016680) | A: EU929258.1 (1–2691)/B: EU929876.1 (1–2115) | P2-A8 | ||
| pil2a (ABC020005369) | A: EU929282.1 (1–2691)/B: EU929901.1 (1–2049) | P2-A9 | ||
| pil2b (COH1) | AAJR01000022.1 (B: 7660–11964)/(C: 14416–15087) | P2-B1 | ||
| pil2b (527.25) | A: AM051289.1 (1–1365) | P2-B5 | ||
| pil2b (A909) | CP000114.1 (A: 1420792–1422300)/(B: 1422341–1426641)/C:(1419218–1419889) | P2-B9 | ||
| 1482000–1487000 | nss, srr | Lacking nss/srr/rogB2 | N-0 | |
| rogB2 | nss (COH1) + srr (COH1) | AAJR01000012.1 (16899–17828)/(28428-3168) | N-1 | |
| nss + srr + rogB2 (NEM316) | AL732656.1 (1587233–1588240)/(1588571–1592503)/(1531047–1532576) | N-3 | ||
| nss + srr + rogB2 (A909) | CP000114.1 (1482334–1483341)/(1483672–1487448)/(1487831–1489323) | N-9 | ||
| 1944000 | rgfC/A/D/B | Lacking rgf | R-0 | |
| rgf (COH1) | AAJR01000004.1 (A: 44577 −45329)/(B: 45545–46360)/(C: 43246–44580)/(D: 45304–45432) | R-1 | ||
| rgf (515) | AAJP01000002.1 (A: 18747 −19499)/(B: 19776–20591)/(C: 18379–18739) | R-5 | ||
| rgf (A909) | CP000114.1 (A: 1920710–1921462)/(B: 1921678–1922493)/(C: 1919379–1920713)/(D: 1921437–1921565) | R-9 |
TABLE 4.
Prevalence of resistance genes in clinical isolates
| Resistance gene(s) | Prevalences for isolates by type: |
|||||||
|---|---|---|---|---|---|---|---|---|
| Total |
Clinical, Germany |
Clinical, Trinidad and Tobago |
Veterinary, Germany |
|||||
| No. resistant | % | No. resistant | % | No. resistant | % | No. resistant | % | |
| erm(A) | 26 | 5.8 | 26 | 6.8 | 0 | 0.0 | 0 | 0.0 |
| erm(B) | 47 | 10.5 | 44 | 11.5 | 0 | 0.0 | 3 | 14.3 |
| erm(C) | 8 | 1.8 | 8 | 2.1 | 0 | 0.0 | 0 | 0.0 |
| tet(M) | 345 | 77.0 | 306 | 79.7 | 24 | 70.6 | 10 | 47.6 |
| emrB/qacA | 448 | 100.0 | 384 | 100.0 | 34 | 100.0 | 21 | 100.0 |
| cadC | 95 | 21.2 | 86 | 22.4 | 6 | 17.6 | 0 | 0.0 |
| cadD | 400 | 89.3 | 352 | 91.7 | 32 | 94.1 | 10 | 47.6 |
| merR + merA | 213 | 47.5 | 176 | 45.8 | 30 | 88.2 | 1 | 4.8 |
For each target, all relevant entries were retrieved from GenBank. One entry was selected as the reference, and its coding sequence was excised. All BLAST hits for that sequence were downloaded into a local database excising and aligning all coding sequences related to each target gene. These alignments were inspected visually. Based on sequence similarities, the sequences were classified into paralogues and allelic variants. The consensus regions in the alignments of all available sequences of each target were chosen for the probe design. The probe sequences were selected for specificity and, in order to yield similar signal intensities, for having similar G+C contents, lengths, and melting temperatures. The resulting probe sequences were again BLASTed against all available sequences to theoretically exclude false-positive reactions due to cross-reactivity or false-negative reactions due to sequence variations.
Two hundred thirteen probes were spotted on arrays that were mounted into ArrayStrips (Alere Technologies, Jena, Germany). The probe lengths ranged from 25 to 36 bases, with a median of 30 bases. The probes were designed to have approximately identical binding temperatures. There were 208 primers with lengths ranging from 18 to 28 bases; their median length was 23 bases. The primers were designed to have similar annealing temperatures in order to be used in a multiplex reaction.
Probes and primers (see Table S1 in the supplemental material) were synthesized and purified by Metabion (Martinsried, Germany). The DNA microarrays were based on the ArrayStrip platform (Alere Technologies), with arrays being mounted in standard 8-well strips. The buffers and reagents were taken from the HybPlus kit (Alere Technologies), unless stated otherwise.
For validation and protocol optimization, completely sequenced strains (S. agalactiae COH1, A909, 515, H36B, 18RS21, CJB 111, 2603VR, and CIP 8245; for GenBank accession numbers, see above) were used. First, hybridization experiments under high-, medium-, and low-stringency conditions were simulated by mapping the probe sequences against known genome sequences. Next, real experiments were performed, and the results were compared to those from the simulations. The hybridization temperature was kept at 55°C, but washing temperatures varied between 25°C and 55°C until the experiments yielded results matching the high-stringency prediction (Table 2; see Table S2 in the supplemental material).
TABLE 2.
Array-based typing results
| Hybridization pattern | MLST result |
Reference strain (GenBank accession no.) | Identical genome sequences, GenBank accession no., and predicted ST (in silico analysis only) | No. of isolates (type) from: |
Code no. of 11 markers used for array typing |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | ST | Germany (clinical) | Trinidad and Tobago (clinical) | Germany (cattle) | bac | alp | pil1 | pepS8 | fbsB | Capsule | hylB | abiG/Q8DZ34 | pil 2 | nss/srr/rogB2 | rgf | |||
| HP-01 | CC19/01 | ST1 | CJB111 (AAJQ00000000.1) | CCUG 44140, ANDL01, ST1; CCUG 49072, ANDP01, ST524; CCUG 49100, ANDR01, ST1; FSL S3-023, ANCP01, ST1; GB00013, ALSO01, ST1; Gottschalk 13227, ANEZ01, ST1; Gottschalk 2864, ANFA01, ST1 | 56 | 2 | 2 | B-0 | A-03 | P1-3 | E-1 | F-9 | C-05 | H-3 | Q-0 | P2-A3 | N-9 | R-9 |
| HP-02 | 09mas018883, HF952104.1, ST1 | 1 | 0 | 0 | B-0 | A-03 | P1-3 | E-1 | F-9 | C-05 | H-3 | Q-6 | P2-A3 | N-9 | R-9 | |||
| HP-03 | 0 | 0 | 1 | B-0 | A-03 | P1-0 | E-1 | F-9 | C-05 | H-3 | Q-0 | P2-A3 | N-9 | R-9 | ||||
| HP-04 | CC19/01 | ST1 | 1 | 0 | 0 | B-0 | A-03 | P1-3 | E-1 | F-9 | C-07 | H-3 | Q-0 | P2-B9 | N-9 | R-9 | ||
| HP-05 | CC19/01 | ST1 | 1 | 0 | 0 | B-0 | A-03 | P1-3 | E-1 | F-9 | C-1B | H-9 | Q-0 | P2-A3 | N-9 | R-9 | ||
| HP-06 | CC19/01 | ST14 | 1 | 0 | 0 | B-0 | A-05 | P1-3 | E-1 | F-9 | C-X | H-3 | Q-0 | P2-B9 | N-9 | R-9 | ||
| HP-07 | CC19/01 | ST297 | 0 | 0 | 2 | B-0 | A-05 | P1-2 | E-1 | F-9 | C-04 | H-3 | Q-0 | P2-B9 | N-9 | R-1 | ||
| HP-08 | CC19/01 | ST425 | 0 | 6 | 0 | B-0 | A-BC | P1-0 | E-1 | F-5 | C-04 | H-3 | Q-0 | P2-B1 | N-1 | R-5 | ||
| HP-09 | 0 | 1 | 0 | B-0 | A-BC | P1-0 | E-1 | F-5 | C-00 | H-3 | Q-0 | P2-B1 | N-1 | R-5 | ||||
| HP-10 | CC19/02 | ST2 | 1 | 0 | 0 | B-0 | A-05 | P1-3 | E-0 | F-9 | C-04 | H-3 | Q-3 | P2-A3 | N-9 | R-X | ||
| HP-11 | CCUG 28551, ANDA01, ST2 | 1 | 0 | 1 | B-0 | A-05 | P1-3 | E-0 | F-9 | C-04 | H-3 | Q-0 | P2-A3 | N-9 | R-9 | |||
| HP-12 | CC19/02 | GB00901, ALUK01, ST459 | 3 | 0 | 0 | B-0 | A-05 | P1-3 | E-1 | F-9 | C-04 | H-3 | Q-6 | P2-A4 | N-9 | R-9 | ||
| HP-13 | CC19/04 | ST5 | 1 | 0 | 0 | B-0 | A-05 | P1-2 | E-1 | F-9 | C-1A | H-3 | Q-0 | P2-B9 | N-9 | R-9 | ||
| HP-14 | CC19/10 | ST6 | 1 | 0 | 0 | B-1 | A-03 | P1-3 | E-1 | F-9 | C-02 | H-3 | Q-3 | P2-A5 | N-3 | R-9 | ||
| HP-15 | CC19/10 | ST6 | H36B (AAJS00000000.1) | 0 | 0 | 0 | B-1 | A-BC | P1-0 | E-1 | F-9 | C-1B | H-9 | Q-6 | P2-A6 | N-3 | R-9 | |
| HP-16 | CC19/10 | ST7 | A909 (CP000114.1) | STIR-CD-28, ALRT01, ST500 CF01173, CAQB01, ST7 | 1 | 0 | 0 | B-1 | A-BC | P1-3 | E-1 | F-9 | C-1A | H-9 | Q-0 | P2-B9 | N-9 | R-9 |
| HP-17 | CC19/10 | ST8 | FSL S3-014, ANCR01, ST8; Gottschalk 999B, ANFD01, ST8 | 19 | 0 | 2 | B-1 | A-BC | P1-3 | E-1 | F-9 | C-1B | H-9 | Q-0 | P2-A3 | N-3 | R-9 | |
| HP-18 | 1 | 0 | 0 | B-1 | A-BC | P1-3 | E-1 | F-9 | C-1B | H-9 | Q-0 | P2-A5 | N-3 | R-9 | ||||
| HP-19 | 1 | 0 | 0 | B-1 | A-BC | P1-3 | E-1 | F-9 | C-1B | H-9 | Q-0 | P2-A9 | N-3 | R-9 | ||||
| HP-20 | CC19/10 | ST12 | 10 | 0 | 0 | B-1 | A-BC | P1-3 | E-1 | F-9 | C-02 | H-9 | Q-0 | P2-A3 | N-3 | R-9 | ||
| HP-21 | CC19/10 | ST10 | 9 | 0 | 0 | B-1 | A-BC | P1-3 | E-1 | F-9 | C-05 | H-3 | Q-0 | P2-A3 | N-3 | R-9 | ||
| HP-22 | 0 | 0 | 1 | B-1 | A-BC | P1-3 | E-1 | F-9 | C-05 | H-3 | Q-0 | P2-A9 | N-3 | R-9 | ||||
| HP-23 | CC19/10 | ST12 | 2 | 0 | 0 | B-1 | A-BC | P1-3 | E-1 | F-9 | C-02 | H-3 | Q-3 | P2-A4 | N-9 | R-9 | ||
| HP-24 | CC19/10 | ST12 | 1 | 0 | 0 | B-1 | A-BC | P1-0 | E-1 | F-9 | C-02 | H-3 | Q-3 | P2-A4 | N-9 | R-9 | ||
| HP-25 | 1 | 0 | 0 | B-1 | A-BC | P1-3 | E-1 | F-9 | C-02 | H-3 | Q-3 | P2-A4 | N-9 | R-X | ||||
| HP-26 | CC19/10 | ST569 | 3 | 0 | 1 | B-1 | A-BC | P1-3 | E-1 | F-9 | C-02 | H-3 | Q-0 | P2-A5 | N-3 | R-9 | ||
| HP-27 | CC19/10 | ST579slv | 1 | 0 | 0 | B-1 | A-BC | P1-3 | E-1 | F-9 | C-04 | H-3 | Q-0 | P2-A5 | N-3 | R-9 | ||
| HP-28 | 1 | 0 | 0 | B-1 | A-BC | P1-3 | E-1 | F-9 | C-02 | H-3 | Q-0 | P2-A8 | N-3 | R-9 | ||||
| HP-29 | 1 | 0 | 0 | B-1 | A-BC | P1-3 | E-1 | F-9 | C-02 | H-3 | Q-0 | P2-X | N-3 | R-9 | ||||
| HP-30 | CC19/17 | ST17 | COH1 (AAJR00000000.1) | FSL S3-102, ANCS01, ST31; GB00112, AKXO01, ST17; M732, ALQW01, ST17; Melin 455511, ALRA01, ST17 | 62a | 2 | 0 | B-0 | A-R4 | P1–3 | E-1 | F-5 | C-03 | H-3 | Q-0 | P2-B1 | N-1 | R-1 |
| HP-31 | 1 | 0 | 0 | B-0 | A-R4 | P1-3 | E-1 | F-5 | C-03 | H-3 | Q-0 | P2-X | N-1 | R-1 | ||||
| HP-32 | CC19/17 | ST291 | 5 | 0 | 0 | B-0 | A-R4 | P1-3 | E-1 | F-5 | C-04 | H-3 | Q-0 | P2-B1 | N-1 | R-1 | ||
| HP-33 | 1 | 0 | 0 | B-0 | A-R4 | P1-0 | E-1 | F-5 | C-03 | H-3 | Q-0 | P2-B1 | N-1 | R-1 | ||||
| HP-34 | CC19/17 | ST17 | 1 | 0 | 0 | B-0 | A-R4 | P1-0 | E-1 | F-5 | C-03 | H-3 | Q-6 | P2-B1 | N-1 | R-1 | ||
| HP-35 | CC19/19 | ST44 or 178 | Gottschalk 1003A, ALSI01, ST19 | 42 | 1 | 0 | B-0 | A-R4 | P1-3 | E-1 | F-9 | C-03 | H-3 | Q-6 | P2-A1 | N-3 | R-5 | |
| HP-36 | CC19/19 | ST19 | 18RS21 (AAJO00000000.1) | CCUG 91, ANCY01, ST28; GB00588, ALTQ01, ST447 | 20 | 2 | 0 | B-0 | A-R4 | P1-3 | E-1 | F-9 | C-02 | H-3 | Q-6 | P2-A1 | N-3 | R-5 |
| HP-37 | 2 | 0 | 0 | B-0 | A-R4 | P1-3 | E-1 | F-9 | C-02 | H-3 | Q-0 | P2-A1 | N-3 | R-5 | ||||
| HP-38 | CC19/19 | ST28 | 4 | 1 | 0 | B-0 | A-R4 | P1-3 | E-1 | F-9 | C-02 | H-3 | Q-6 | P2-A1 | N-0 | R-5 | ||
| HP-39 | 0 | 1 | 0 | B-0 | A-R4 | P1-3 | E-1 | F-9 | C-02 | H-3 | Q-6 | P2-X | N-0 | R-5 | ||||
| HP-40 | CC19/19 | ST193 | FSL S3-003, ALQJ01, ST19 | 12 | 0 | 0 | B-0 | A-R4 | P1-3 | E-1 | F-9 | C-03 | H-3 | Q-0 | P2-A1 | N-3 | R-5 | |
| HP-41 | 1 | 0 | 0 | B-0 | A-R4 | P1-x | E-1 | F-9 | C-03 | H-3 | Q-0 | P2-A1 | N-3 | R-5 | ||||
| HP-42 | CC19/19 | ST110 | 2603V/R (AE009948) | 4 | 1 | 0 | B-0 | A-R4 | P1-3 | E-1 | F-9 | C-05 | H-3 | Q-6 | P2-A1 | N-3 | R-5 | |
| HP-43 | 2 | 0 | 0 | B-0 | A-R4 | P1-0 | E-1 | F-9 | C-05 | H-3 | Q-6 | P2-A1 | N-3 | R-5 | ||||
| HP-44 | CC19/19 | ST19 | 4 | 0 | 0 | B-0 | A-05 | P1-3 | E-1 | F-9 | C-05 | H-3 | Q-6 | P2-A1 | N-3 | R-5 | ||
| HP-45 | 1 | 0 | 0 | B-0 | A-R4 | P1-0 | E-1 | F-9 | C-03 | H-3 | Q-6 | P2-A1 | N-3 | R-5 | ||||
| HP-46 | CC19/22 | ST22 | 6 | 0 | 0 | B-0 | A-BC | P1-0 | E-0 | F-6 | C-02 | H-3 | Q-0 | P2-A9 | N-3 | R-5 | ||
| HP-47 | 1 | 0 | 0 | B-0 | A-BC | P1-0 | E-0 | F-6 | C-02 | H-3 | Q-3 | P2-A9 | N-3 | R-5 | ||||
| HP-48 | CC23 | ST23 | Strain 515 (AAJP00000000.1), “RV2”b | FSL S3-090, ANCQ01, ST23; Gottschalk 31825, ANFB01, ST23 | 53c | 3 | 1 | B-0 | A-05 | P1-0 | E-0 | F-5 | C-1A | H-3 | Q-0 | P2-A5 | N-3 | R-5 |
| HP-49 | CC23 | ST24 | 3 | 9 | 0 | B-0 | A-BC | P1-0 | E-0 | F-5 | C-1A | H-3 | Q-0 | P2-A5 | N-3 | R-5 | ||
| HP-50 | CC23 | ST144 | 1 | 5 | 0 | B-0 | A-R4 | P1-0 | E-0 | F-5 | C-1A | H-3 | Q-0 | P2-A5 | N-3 | R-5 | ||
| HP-51 | 1 | 0 | 0 | B-0 | A-05 | P1-0 | E-0 | F-5 | C-1A | H-3 | Q-3 | P2-A5 | N-3 | R-5 | ||||
| HP-52 | 1 | 0 | 0 | B-0 | A-05 | P1-0 | E-0 | F-5 | C-05 | H-3 | Q-0 | P2-A5 | N-3 | R-5 | ||||
| HP-53 | 2 | 0 | 0 | B-0 | A-05 | P1-x | E-0 | F-5 | C-1A | H-3 | Q-0 | P2-A5 | N-3 | R-5 | ||||
| HP-54 | CC23 | ST23 | NEM316 (AL732656.1) | 5 | 0 | 0 | B-0 | A-02 | P1-3 | E-0 | F-5 | C-03 | H-3 | Q-3 | P2-A3 | N-3 | R-5 | |
| HP-55 | CC23 | ST23 | 2 | 0 | 0 | B-0 | A-02 | P1-3 | E-0 | F-5 | C-03 | H-3 | Q-3 | P2-A5 | N-3 | R-5 | ||
| HP-56 | 0 | 0 | 1 | B-0 | A-02 | P1-3 | E-0 | F-5 | C-03 | H-3 | Q-3 | P2-A8 | N-3 | R-5 | ||||
| HP-57 | CC23 | ST23 | 14 | 0 | 0 | B-0 | A-02 | P1-3 | E-0 | F-5 | C-1A | H-3 | Q-3 | P2-A8 | N-3 | R-5 | ||
| HP-58 | 1 | 0 | 0 | B-0 | A-02 | P1-0 | E-0 | F-5 | C-1A | H-3 | Q-3 | P2-A8 | N-3 | R-5 | ||||
| HP-59 | 1 | 0 | 0 | B-0 | A-02 | P1-0 | E-0 | F-5 | C-1A | H-3 | Q-X | P2-A8 | N-3 | R-5 | ||||
| HP-60 | CC26 | ST26 | 1 | 0 | 0 | B-0 | A-00 | P1-0 | E-0 | F-4 | C-05 | H-3 | Q-6 | P2-A9 | N-3 | R-5 | ||
| HP-61 | 1 | 0 | 0 | B-0 | A-00 | P1-0 | E-0 | F-4 | C-05 | H-3 | Q-0 | P2-A9 | N-3 | R-5 | ||||
| HP-62 | CC103 | ST314 | 0 | 0 | 2 | B-0 | A-05 | P1-0 | E-1 | F-3 | C-1A | H-9 | Q-6 | P2-B5 | N-9 | R-5 | ||
| HP-63 | 0 | 0 | 1 | B-0 | A-00 | P1-0 | E-1 | F-3 | C-03 | H-9 | Q-0 | P2-B5 | N-9 | R-5 | ||||
| HP-64 | CC103 | ST314 | 1 | 0 | 2 | B-0 | A-05 | P1-0 | E-1 | F-3 | C-1A | H-9 | Q-0 | P2-B5 | N-9 | R-5 | ||
| HP-65 | 0 | 0 | 1 | B-0 | A-05 | P1-0 | E-1 | F-3 | C-03 | H-9 | Q-0 | P2-B5 | N-9 | R-5 | ||||
| HP-66 | 1 | 0 | 0 | B-0 | A-X | P1-0 | E-1 | F-3 | C-1A | H-9 | Q-6 | P2-B5 | N-9 | R-5 | ||||
| HP-67 | 0 | 0 | 1 | B-0 | A-05 | P1-0 | E-1 | F-3 | C-1A | H-9 | Q-6 | P2-X | N-9 | R-5 | ||||
| HP-68 | CC130 | ST130 | GB00300, ALTJ01, ST130 | 3 | 0 | 0 | B-1 | A-BC | P1-0 | E-1 | F-3 | C-09 | H-3 | Q-6 | P2-A1 | N-3 | R-9 | |
| HP-69 | 1 | 0 | 0 | B-0 | A-03 | P1-3 | E-1 | F-9 | C-02 | H-3 | Q-3 | P2-A3 | N-9 | R-9 | ||||
| HP-70 | 1 | 0 | 0 | B-0 | A-05 | P1-0 | E-1 | F-9 | C-02 | H-3 | Q-3 | P2-X | N-9 | R-9 | ||||
| HP-71 | 0 | 0 | 1 | B-0 | A-05 | P1-X | E-1 | F-9 | C-1A | H-3 | Q-6 | P2-B9 | N-9 | R-5 | ||||
| HP-72 | 1 | 0 | 0 | B-0 | A-R4 | P1-2 | E-1 | F-9 | C-02 | H-3 | Q-3 | P2-B9 | N-9 | R-9 | ||||
| HP-73 | 1 | 0 | 0 | B-0 | A-R4 | P1-3 | E-1 | F-5 | C-03 | H-3 | Q-X | P2-A1 | N-3 | R-5 | ||||
| HP-74 | 1 | 0 | 0 | B-1 | A-BC | P1-0 | E-1 | F-9 | C-1B | H-9 | Q-0 | P2-X | N-3 | R-9 | ||||
| HP-75 | 0 | 0 | 1 | B-1 | A-BC | P1-0 | E-1 | F-9 | C-02 | H-3 | Q-6 | P2-A1 | N-3 | R-9 | ||||
| HP-76 | 1 | 0 | 0 | B-1 | A-BC | P1-3 | E-1 | F-9 | C-02 | H-3 | Q-3 | P2-A3 | N-3 | R-9 | ||||
Includes two isolates from clinically/epidemiologically established transmission.
A strain from the proficiency testing program for German diagnostic microbiology laboratories, 2010/Febr./RV4, obtained from INSTAND eV.
Includes two isolates from another clinically/epidemiologically established transmission event.
Microarray procedures.
A linear and thermally synchronized primer elongation reaction was used. For one labeling reaction, 1.5 μl of a primer mixture containing one primer per target sequence (0.135 μmol/liter each), an oligonucleotide mixture, and biotin-16-dUTP, as well as 1 to 2 μg of unfragmented RNA-free target DNA, were utilized. Amplification was performed using a standard thermocycler with initial denaturation at 96°C for 5 min, followed by 55 cycles (60 s at 96°C, 20 s at 50°C, and 40 s at 72°C) and final storage at 4°C.
First, the arrays were prewashed with 200 μl of double-distilled water and 200 μl of hybridization buffer 1 (both steps were 5 min at 55°C and 550 rpm on the thermomixer). Labeled DNA samples were diluted with 90 μl of hybridization buffer 1 and then transferred into the ArrayStrip well. After that, the samples were incubated at 55°C and 550 rpm for 60 min and then removed completely. The arrays were washed twice with 200 μl of washing buffer C2 (for 2 min and for 5 min, at 35°C and 550 rpm). Next, the microarrays were incubated with 100 μl of horseradish peroxidase-streptavidin (15 min at 30°C and 550 rpm). After removal of this solution, the arrays were washed twice with 200 μl of washing buffer C5 at 30°C and 550 rpm for 2 min and 5 min, respectively. Finally, 100 μl of precipitating substrate was added. After 5 min of incubation at room temperature (without shaking), the substrate was removed and the arrays were scanned and analyzed using the ArrayMate reader (Alere Technologies). The signal intensities were calculated as previously described (29). For breakpoint determination, the median values of the intensities of those probes of species markers [fbsB (consensus), femH (locus 2), gtfB (variant 1), gtfB (variant 2), nss1, and nss2] that yielded a raw value of >0.2 and the biotin control were calculated. Each probe that yielded a signal of >2/3 of this median value was considered positive, and signals between 1/3 and 2/3 of this median value were considered ambiguous. If the median was <0.5, the entire experiment was considered invalid. If mutually exclusive alleles of species markers [gtfB (variant 1) and gtfB (variant 2) or nss1 and nss2] were detected together, the experiment was considered invalid, assuming contamination.
eBURST.
Analysis of clonal complexes was performed using the software provided on eBURST (see http://eburst.mlst.net/v3/enter_data/single/). The MLST profiles were downloaded from http://pubmlst.org/perl/bigsdb/bigsdb.pl?db=pubmlst_sagalactiae_seqdef&page=downloadProfiles&scheme_id=1 (see Table S3 in the supplemental material). For an eBURST analysis of the array hybridization profiles, 11 markers (see below and Table 2) were selected, and their presence or their alleles were coded numerically.
RESULTS
A full list of the data sets for the tested strains and isolates can be found in Table S2 in the supplemental material, while the abridged results are provided in Tables 2 to 4.
Typing GBS by MLST.
Thirty-six isolates that were representative of the common array hybridization patterns were subjected to MLST, and the MLST types for the sequenced reference strains were deduced from published genome sequences (Table 2). For further analysis, clonal complexes (CC) were introduced in analogy to Staphylococcus aureus terminology using the eBURST algorithm (http://eburst.mlst.net/). CC19 was the most common and diverse CC. It was quite inhomogeneous with regard to both the MLST- and array-based (see below) typing approaches. CC19 included sequence type 1 (ST1), ST2, ST5, ST6, ST7, ST8, ST10, ST12, ST14, ST17, ST19, ST22, ST44/178, ST110, ST193, ST291, ST297, ST425, and ST569, as well as a single-locus variant (SLV) of ST579. eBURST shows that CC19 comprises several distinct clusters that were recognized as independent clonal complexes (CC01, CC17, CC19, and CC22) before linking STs were identified. These clusters were designated CC19/01, CC19/17, etc. (Table 2). Additional CC19/02 and CC19/05 clusters were designated because their MLST profiles link CC19/01, or CC19/10, respectively, to CC19/19. CC23 was the second most common CC, with ST23, ST24, and ST144. To CC26 and CC130, only one ST each (ST26 and ST130) was assigned. For CC103, two ST314 isolates were identified by MLST.
Genotyping GBS by microarray hybridization.
Putative typing markers (e.g., alp alleles and capsule types) vary independently of each other and independently of their ST/CC affiliation (Table 2). Thus, it is not possible to assign an isolate to an ST/CC based on a single marker.
For array-based typing, 11 markers (bac, alp, pil1 locus, pepS8, fbsB, capsule locus, hylB, abiG-I/-II plus Q8DZ34, pil2 locus, nss plus srr plus rogB2, and rgfC/A/D/B; Table 2) were selected that provide a framework for splitting GBS organisms into strains. These markers were selected based on variability affecting either sequences, allowing assignment to clear-cut alleles or their presence/absence. In general, the typing markers are distributed rather evenly across the genome sequence. For each marker locus, the presence or absence or a small number of different alleles was interrogated. This allowed, in analogy to MLST, a conversion of the hybridization results for these markers into 11 numerical codes (Table 2). Ubiquitous genes, such as cylD/cylE (β-hemolysin locus), were not suitable as typing markers. Highly mobile plasmid-borne (such as resistance genes) or phage-borne genes were also not considered, although they can be detected by the array. Their use for further discerning the variants within strains still needs to be evaluated.
Details on the 11 markers will be discussed below in the order of their localizations in the S. agalactiae genome that are conserved among known genomes (30). The gene positions in the following paragraphs are provided as medians of the starting positions from the fully and conventionally sequenced genomes of NEM316, A909, and 2603V/R.
The bac gene encodes a surface protein binding the Fc part of IgA (31). Because of an extremely variable repeat region (32), only its presence or absence, rather than specific alleles, was interrogated. Its presence apparently correlated with alleles 4, 9, and 10 of the MLST gene adhP, or roughly with affiliation to CC19/10 and CC130.
Genes encoding alpha, alpha-like (33), Bca, and Rib proteins are all regarded as alleles of the alp gene (for GenBank accession numbers, see Table 1). The alleles alp-2, alp-3, alp-4 (= alp-6), and alp-5 (= alp-7, alp-epsilon), alp-bca (= alpha C or C-alpha), alp-rib(R4) (= R4, rib), and alp-rib2 were covered. The alleles alp-rib(R4) and alp-bca were the most common alp alleles in human isolates; in the cattle group, alp-5 was most prevalent. Three isolates, including both CC26 isolates, were alp negative (or carried an alternative undetectable allele). There was no correlation between alp and alleles of the neighboring MLST markers tkt, glck, and atr.
S. agalactiae pili (34) are composed of PilB and the accessory pilus proteins PilA and PilC (35, 36). There are two loci. The first one (pilA/B/C-1) is situated around position 650000 in the genome sequence. In a vast majority of isolates, either the three pil1 genes together or no pil1 genes at all were detected. Four isolates yielded only signals for pilA/C-1. For the second pilus locus, there are two mutually exclusive genomic islands (pil2a and pil2b) at identical positions in the genome sequence. Eighteen probes recognized seven pil2a alleles, while another eight probes yielded three pil2b patterns, one of which matches only an incompletely known sequence (GenBank accession no. AM051289.1). In the human isolates, the pil2a genes were generally more common (Table 2). In the cattle isolates, pil2b was as common as pil2a. Three isolates yielded unassignable patterns. There were no isolates lacking pil2. The loci adjacent to pil2 are gtfA/B (encoding glycosyltransferase, around position 1466000) and rogB1 (encoding the transcriptional regulator RogB, locus 1, position 1475000). Two alleles of gtfA/B were discerned to correlate with pil2a or pil2b, respectively. The presence of rogB1 is associated with pil2a. Thus, gtfA/B and rogB1 yield no additional typing information.
Another variable genomic island is detected by probes for pepS8 (peptidase S8). All CC19/22, CC23, and CC26 isolates, as well as a part of CC19/02, did not contain this island.
The gene fbsB encodes a fibrinogen binding protein (37, 38). Five hybridization patterns were generated by eight probes; isolates lacking fbsB were not observed.
A cluster of 16 genes encodes the capsule, of which there are 10 types (Ia, Ib, II, III, IV, V, VI, VII, VIII, and IX). The different prevalences of the German, Caribbean, cattle, and human isolates are shown in Table 3. Capsule type III was overrepresented among the invasive isolates. Types VI, VII, and IX were very rare, and type VIII was not found (although the array can identify it; therefore, a type VIII reference isolate was kindly provided by K. Zürn and F. Lander, Dresden University Medical Centre). One Caribbean isolate yielded signals with a part of the capsule IV probes (cpsJ-IV, cpsM-IV, and cpsM-IV) only.
TABLE 3.
Prevalences of marker alleles
| Presence/allele (accession no. or strain) | Code no. | Prevalences for isolates by type: |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total |
Human, Germany |
Human, Trinidad and Tobago |
Veterinary, Germany |
Human, invasive infection |
Human, localized infection |
Human, carriage |
|||||||||
| No. | % | No. | % | No. | % | No. | % | No. | % | No. | % | No. | % | ||
| bac negative | B-0 | 384 | 85,7 | 327 | 85.2 | 34 | 100.0 | 16 | 76.2 | 100 | 87.0 | 42 | 87.5 | 219 | 85.9 |
| bac positive | B-1 | 64 | 14.3 | 57 | 14.8 | 0 | 0.0 | 5 | 23.8 | 15 | 13.0 | 6 | 12.5 | 36 | 14.1 |
| alp negative | A-0 | 3 | 0.7 | 2 | 0.5 | 0 | 0.0 | 1 | 4.8 | 1 | 0.9 | 1 | 2.1 | 0 | 0.0 |
| alp-2 | A-2 | 25 | 5.6 | 23 | 6.0 | 0 | 0.0 | 1 | 4.8 | 4 | 3.5 | 9 | 18.8 | 10 | 3.9 |
| alp-3 | A-3 | 67 | 15.0 | 61 | 15.9 | 2 | 5.9 | 3 | 14.3 | 19 | 16.5 | 9 | 18.8 | 35 | 13.7 |
| alp-4 | A-4 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
| alp-5 | A-5 | 86 | 19.2 | 70 | 18.2 | 3 | 8.8 | 11 | 52.4 | 20 | 17.4 | 8 | 16.7 | 45 | 17.6 |
| alp-bca | A-B | 89 | 19.9 | 66 | 17.2 | 16 | 47.1 | 5 | 23.8 | 18 | 15.7 | 6 | 12.5 | 58 | 22.7 |
| alp-R2 | A-R2 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
| alp-rib (R4) | A-R4 | 177 | 39.5 | 161 | 41.9 | 13 | 38.2 | 0 | 0.0 | 53 | 46.1 | 15 | 31.3 | 106 | 41.6 |
| alp atypical/unidentified | A-X | 1 | 0.2 | 1 | 0.3 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 1 | 0.4 |
| pil1 negative | P1-0 | 120 | 26.8 | 83 | 21.6 | 24 | 70.6 | 10 | 47.6 | 24 | 20.9 | 9 | 18.8 | 74 | 29.0 |
| pil1A/C | P1–2 | 4 | 0.9 | 2 | 0.5 | 0 | 0.0 | 2 | 9.5 | 0 | 0.0 | 0 | 0.0 | 2 | 0.8 |
| pil1A/B/C | P1–3 | 320 | 71.4 | 296 | 77.1 | 10 | 29.4 | 8 | 38.1 | 89 | 77.4 | 39 | 81.3 | 178 | 69.8 |
| pil1 atypical/unidentified | P1-X | 4 | 0.9 | 3 | 0.8 | 0 | 0.0 | 1 | 4.8 | 2 | 1.7 | 0 | 0.0 | 1 | 0.4 |
| pepS8 negative | E-0 | 118 | 26.3 | 95 | 24.7 | 17 | 50.0 | 3 | 14.3 | 29 | 25.2 | 16 | 33.3 | 67 | 26.3 |
| pepS8 positive | E-1 | 330 | 73.7 | 289 | 75.3 | 17 | 50.0 | 18 | 85.7 | 86 | 74.8 | 32 | 66.7 | 188 | 73.7 |
| fbsB (AM050623) | F-3 | 12 | 2.7 | 5 | 1.3 | 0 | 0.0 | 7 | 33.3 | 0 | 0.0 | 1 | 2.1 | 4 | 1.6 |
| fbsB (AM050624) | F-4 | 2 | 0.4 | 2 | 0.5 | 0 | 0.0 | 0 | 0.0 | 1 | 0.9 | 1 | 2.1 | 0 | 0.0 |
| fbsB (515) | F-5 | 187 | 41.7 | 155 | 40.4 | 26 | 76.5 | 2 | 9.5 | 61 | 53.0 | 22 | 45.8 | 98 | 38.4 |
| fbsB (AY326423) | F-6 | 7 | 1.6 | 7 | 1.8 | 0 | 0.0 | 0 | 0.0 | 1 | 0.9 | 0 | 0.0 | 6 | 2.4 |
| fbsB (A909) | F-9 | 240 | 53.6 | 215 | 56.0 | 8 | 23.5 | 12 | 57.1 | 52 | 45.2 | 24 | 50.0 | 147 | 57.6 |
| Capsule type: | |||||||||||||||
| Ia | C-1A | 107 | 23.9 | 80 | 20.8 | 17 | 50.0 | 7 | 33.3 | 25 | 21.7 | 14 | 29.2 | 58 | 22.7 |
| Ib | C-1B | 26 | 5.8 | 23 | 6.0 | 0 | 0.0 | 2 | 9.5 | 10 | 8.7 | 0 | 0.0 | 13 | 5.1 |
| II | C-2 | 64 | 14.3 | 57 | 14.8 | 4 | 11.8 | 2 | 9.5 | 11 | 9.6 | 4 | 8.3 | 46 | 18.0 |
| III | C-3 | 137 | 30.6 | 129 | 33.6 | 3 | 8.8 | 3 | 14.3 | 47 | 40.9 | 14 | 29.2 | 71 | 27.8 |
| IV | C-4 | 20 | 4.5 | 11 | 2.9 | 6 | 17.6 | 3 | 14.3 | 1 | 0.9 | 4 | 8.3 | 12 | 4.7 |
| V | C-5 | 88 | 19.6 | 79 | 20.6 | 3 | 8.8 | 4 | 19.0 | 21 | 18.3 | 11 | 22.9 | 50 | 19.6 |
| VI | C-6 | 1 | 0.2 | 1 | 0.3 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 1 | 0.4 |
| VII | C-7 | 1 | 0.2 | 1 | 0.3 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 1 | 0.4 |
| VIII | C-8 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
| IX | C-9 | 3 | 0.7 | 3 | 0.8 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 1 | 2.1 | 2 | 0.8 |
| No capsule/truncated cap locus | C-0 | 1 | 0.2 | 0 | 0.0 | 1 | 2.9 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 1 | 0.4 |
| hylB negative | H-0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
| hylB (NEM316) | H-3 | 401 | 89.5 | 348 | 90.6 | 34 | 100.0 | 12 | 57.1 | 102 | 88.7 | 46 | 95.8 | 234 | 91.8 |
| hylB (A909) | H-9 | 47 | 10.5 | 36 | 9.4 | 0 | 0.0 | 9 | 42.9 | 13 | 11.3 | 2 | 4.2 | 21 | 8.2 |
| abiG/Q8DZ34 negative | Q-0 | 309 | 69.0 | 261 | 68.0 | 28 | 82.4 | 15 | 71.4 | 92 | 80.0 | 28 | 58.3 | 169 | 66.3 |
| abiG/Q8DZ34 (NEM316) | Q-3 | 36 | 8.0 | 34 | 8.9 | 0 | 0.0 | 1 | 4.8 | 4 | 3.5 | 10 | 20.8 | 20 | 7.8 |
| abiG/Q8DZ34 (H36B) | Q-6 | 101 | 22.5 | 87 | 22.7 | 6 | 17.6 | 5 | 23.8 | 17 | 14.8 | 10 | 20.8 | 66 | 25.9 |
| abiG/Q8DZ34 (unidentified) | Q-X | 2 | 0.4 | 2 | 0.5 | 0 | 0.0 | 0 | 0.0 | 2 | 1.7 | 0 | 0.0 | 0 | 0.0 |
| pil2 negative | P2-0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
| pil2a (18RS21) | P2-A1 | 104 | 23.2 | 96 | 25.0 | 5 | 14.7 | 1 | 4.8 | 19 | 16.5 | 9 | 18.8 | 73 | 28.6 |
| pil2a (NEM316) | P2-A3 | 115 | 25.7 | 105 | 27.3 | 2 | 5.9 | 6 | 28.6 | 32 | 27.8 | 13 | 27.1 | 62 | 24.3 |
| pil2a (ABC020013424) | P2-A4 | 7 | 1.6 | 7 | 1.8 | 0 | 0.0 | 0 | 0.0 | 1 | 0.9 | 3 | 6.3 | 3 | 1.2 |
| pil2a (515) | P2-A5 | 88 | 19.6 | 67 | 17.4 | 17 | 50.0 | 2 | 9.5 | 24 | 20.9 | 7 | 14.6 | 53 | 20.8 |
| pil2a (H36B) | P2-A6 | 1 | 0.2 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
| pil2a (ABC020016680) | P2-A8 | 18 | 4.0 | 17 | 4.4 | 0 | 0.0 | 1 | 4.8 | 2 | 1.7 | 8 | 16.7 | 7 | 2.7 |
| pil2a (ABC020005369) | P2-A9 | 11 | 2.5 | 10 | 2.6 | 0 | 0.0 | 1 | 4.8 | 2 | 1.7 | 1 | 2.1 | 7 | 2.7 |
| pil2a (unidentified) | P2-AX | 6 | 1.3 | 5 | 1.3 | 1 | 2.9 | 0 | 0.0 | 2 | 1.7 | 0 | 0.0 | 4 | 1.6 |
| pil2b (COH1) | P2-B1 | 79 | 17.6 | 69 | 18.0 | 9 | 26.5 | 0 | 0.0 | 33 | 28.7 | 7 | 14.6 | 38 | 14.9 |
| pil2b (527.25) | P2-B5 | 8 | 1.8 | 2 | 0.5 | 0 | 0.0 | 6 | 28.6 | 0 | 0.0 | 0 | 0.0 | 2 | 0.8 |
| pil2b (A909) | P2-B9 | 9 | 2.0 | 5 | 1.3 | 0 | 0.0 | 3 | 14.3 | 0 | 0.0 | 0 | 0.0 | 5 | 2.0 |
| pil2b (unidentified) | P2-BX | 2 | 0.4 | 1 | 0.3 | 0 | 0.0 | 1 | 4.8 | 0 | 0.0 | 0 | 0.0 | 1 | 0.4 |
| nss/srr/rogB2 negative | N-0 | 6 | 1.3 | 4 | 1.0 | 2 | 5.9 | 0 | 0.0 | 1 | 0.9 | 0 | 0.0 | 5 | 2.0 |
| nss/srr (COH1) | N-1 | 80 | 17.9 | 70 | 18.2 | 9 | 26.5 | 0 | 0.0 | 33 | 28.7 | 7 | 14.6 | 39 | 15.3 |
| nss/srr/rogB2 (NEM316) | N-3 | 268 | 59.8 | 234 | 60.9 | 21 | 61.8 | 7 | 33.3 | 62 | 53.9 | 29 | 60.4 | 164 | 64.3 |
| nss/srr/rogB2 (A909) | N-9 | 94 | 21.0 | 76 | 19.8 | 2 | 5.9 | 14 | 66.7 | 19 | 16.5 | 12 | 25.0 | 47 | 18.4 |
| rgf negative | R-0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
| rgf (COH1) | R-1 | 75 | 16.7 | 70 | 18.2 | 2 | 5.9 | 2 | 9.5 | 33 | 28.7 | 7 | 14.6 | 32 | 12.5 |
| rgf (515) | R-5 | 233 | 52.0 | 188 | 49.0 | 30 | 88.2 | 10 | 47.6 | 48 | 41.7 | 24 | 50.0 | 146 | 57.3 |
| rgf (A909) | R-9 | 138 | 30.8 | 124 | 32.3 | 2 | 5.9 | 9 | 42.9 | 34 | 29.6 | 17 | 35.4 | 75 | 29.4 |
| rgf atypical/unidentified | R-X | 2 | 0.4 | 2 | 0.5 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 2 | 0.8 |
Another typing factor was hylB (hyaluronate lyase), with two distinct alleles.
Two variants of a gene cluster comprising abiG-I, abiG-II (encoding the abortive infection proteins I and II, respectively), and Q8DZ34 (CHAP domain containing protein/N-acetylmuramoyl-l-alanine amidase) were discerned. An NEM316-like variant was more common than an H36B-like variant; two isolates were unassignable. Most isolates did not contain these variants, and there were no major differences between the different populations tested.
Another variable gene cluster comprises nss, srr, and rogB2, encoding a putative nucleotide sugar synthetase, a serine-rich repeat 1 glycoprotein mediating the penetration of the blood-brain barrier (39), and a second locus of the transcriptional regulator, respectively. Three variants of this genomic island were discerned.
The GBS genome sequence comprises several regulatory loci, including the rgf quorum-sensing locus (55). Three different variants were identified, with rgfB being conserved, rgfA and rgfC having two or three alleles, respectively, and rgfD being absent from several lineages and from the genome sequence of S. agalactiae strain 515 (GenBank accession no. AAJP01000002.1).
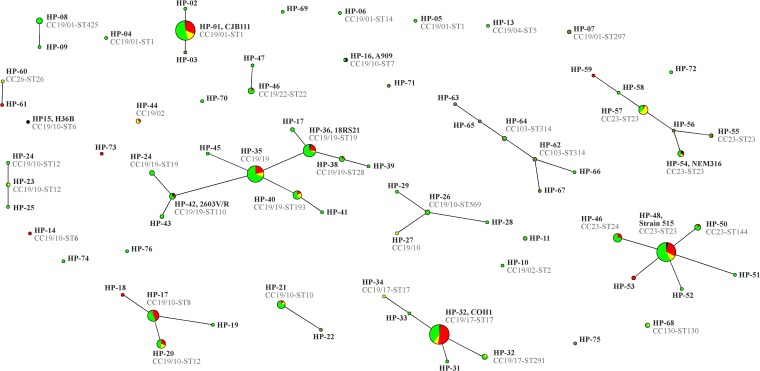
Analyzing the hybridization results for these 11 markers, 76 distinct strain profiles were distinguished (notated as hybridization profiles [HP], followed by sequential numbers). When applying eBURST on these markers, some of the HPs clustered. Generally, the HPs showed some correlation with the MLST-defined CCs/STs (Table 2 and Fig. 1). The isolates that showed that identical or related HPs belonged to the same CC, but isolates that are related by MLST sometimes show dissimilar hybridization patterns.
FIG 1.
eBURST population snapshot graph based on hybridization patterns for 11 variable marker genes. The sizes of the circles correlate with the numbers of isolates. The color codes indicate provenance: red are invasive isolates, yellow are isolates from local infections, and green are from carriage. Black indicates reference strains, and brown are isolates from local infections (mastitis) in cattle.
The highest diversity (46 HPs) was observed in CC19. Many isolates displaying similar HPs belonged to the same cluster within CC19 (Fig. 1) having the same STs or single locus variants thereof.
Nine HPs were not assigned to CCs/STs, as they yielded patterns that were dissimilar to any isolates with known STs.
Population structure.
CC19 was the largest CC, accounting for 75% of the human isolates from Germany, 50% of the human isolates from Trinidad and Tobago, and 32% of the bovine isolates from Germany. For CC23, the corresponding figures were 22%, 50%, and 6%, respectively. HP-08 and HP-09 (CC19/01-ST425) and HP-49 and HP-50 (CC23-ST24 and -144, respectively) appeared to predominate in Trinidad and Tobago. The cattle isolates belonged to diverse lineages, but CC103 was more common in this group than in the other groups, with 23% of the bovine isolates being associated with this CC. With regard to clinical background (Fig. 1), the highest proportion of invasive isolates was observed for CC19/17, followed by CC19/01, CC19/10, and CC23.
Detection of antibiotic resistance markers.
Table 4 shows the data for the antibiotic resistance genes. The isolates were screened for erm(A), erm(B), and erm(C), which code for methylases conferring macrolide/clindamycin resistance by methylation of the binding site at the ribosome. No erm genes were detected in the Caribbean isolates. In Germany, the most common erm gene was erm(B). erm(A) and erm(C) were less common, and they were not found among the cattle isolates. The tetracycline resistance marker tet(M) was common among human isolates but less common among the veterinary isolates.
All isolates harbored emrB/qacA (multidrug resistance transporter).
Detection of heavy metal resistance markers.
Table 4 shows the data for the heavy metal resistance markers. Two genes associated with cadmium resistance (cadC, encoding a cadmium efflux system accessory protein, and cadD, encoding cadmium resistance protein D) were interrogated. These genes are not associated with each other, and cadD was much more common. The mercury resistance genes merA (encoding a mercuric reductase) and merR (encoding a mercuric resistance operon regulatory protein) were always found together. Nearly half of the human isolates from Germany and 88% of the isolates from Trinidad and Tobago but only a single veterinary isolate were found positive. While merA/R were common in CC19 and CC23, they were rare in other CCs.
DISCUSSION
S. agalactiae is a common cause of illness in infants and is transferred vertically. Besides establishing such a transmission in individual cases, molecular typing of this organism should also address whether there are hypervirulent clones. It would be relevant to know the geographic distribution of such clones, so as to identify regions where a risk for infants might be above average. Moreover, it would be interesting to know if there is a possible zoonotic risk or if, conversely, livestock can be infected with GBS from humans.
With regard to population structure, we observed that CC19 predominated among the human isolates from Germany, whereas in Trinidad and Tobago, CC19 and CC23 were equally common. In cattle, CC19 was also common, but another prevalent lineage is CC103 (40), which is rare among human isolates. These observations might indicate both geographic- and host-specific differences in population structure. With regard to host specificity, one virulence factor, scpB (encoding a complement-inactivating C5a peptidase), might be involved (see Table S2 in the supplemental material). It was present in 412 of 418 human isolates (98.6%) but in only 10 of 21 cattle isolates (48%). All isolates belonging to the bovine-associated CC103 (including two human isolates) lacked scpB. It was also absent in some bovine isolates belonging to human lineages (one of two HP-07, CC19/01-ST297 isolates, one HP-56, and a CC23 isolate).
When studying the population structure, a major disadvantage of MLST/eBURST became visible. Some previously described CCs (CC1, CC17, and CC22) are not recognized as CCs anymore but lumped together into CC19. Similar observations were made for S. aureus, as nowadays, e.g., distinct CC5 and CC8 are merged together by eBURST (41). Strains linking two distinct CCs can theoretically be common ancestors of both, but they also might be recent hybrids or recombinants. MLST/eBURST is not able to visualize the difference so that distinct CCs are merged together when linking strains are found. In S. aureus, for instance, the hybrid strain ST239 links CC8 to the unrelated CC30 (42). Both lineages became linked to a third unrelated CC (CC45), with the identification of a recombinant strain (ST2249) that emerged from ST239 and CC45 parents (43). Thus, with a broadening database, the current concept of CCs will obscure phylogenetic patterns and, eventually, an entire species might be regarded as a single CC, especially if it is a species in which horizontal gene transfer is as common as it is in S. agalactiae (44). Hence, clusters within CC19 (such as CC19/01 and CC19/17) were distinguished (Table 2) to visualize the relationships within the large CC19.
With DNA microarrays, it is possible to genotype high numbers of isolates, with the method being less cumbersome than MLST and providing additional information on specific virulence and resistance determinants.
Similarly to what has been shown for S. aureus, hybridization patterns appear to correspond to STs and CCs (29). Compared to S. aureus, this correlation in GBS is poor. It is possible to predict an ST/CC affiliation from a hybridization pattern if it is identical or similar to a previously MLS-typed strain. However, isolates belonging to the same CC/ST might yield different hybridization patterns, and it not possible to predict a hybridization pattern from a known ST. Otherwise, we frequently observed (Table 2) that similar strains differed in single features, such as capsule genes, alp, or pilus genes, and that these genes vary independently of each other. Some ubiquitous genes or gene clusters that apparently always reside at the same position in the genome sequence (30) show a number of distinct alleles, and the identity of an actual allele at a given position is of a random nature. This might be explained by a high number of past recombination events (30, 44) or, theoretically, also by convergent evolution.
In both cases, typing by array and by MLST would not reflect a true phylogeny in terms of providing information on the evolutionary history and provenance of a given strain but just create a random pattern to be compared with other random patterns. The identities of two isolates might then indicate, e.g., a transmission event, but a mere similarity between two isolates would not necessarily mean they were related, i.e., have a recent common ancestor. The resolution of a typing method might be increased with a greater number of interrogated markers. The markers described here could be combined with the MLST scheme, creating an 18-marker framework. This approach could be used experimentally or, in the near future, purely in silico by extracting handy information out of the abundance of data generated by next-generation sequencing. Besides, erm, cad, mer, or tet genes could also be used to prove the identity or nonidentity of isolates, provided that data on the frequency of their acquisition or loss becomes available.
In order to understand the possible differences in virulence within S. agalactiae, future studies should focus not only on ST/CC affiliations but also on the alleles of relevant genes or gene clusters, because these might vary within an ST/CC. No single factor was identified that distinguishes between invasive and carriage isolates. However, it was observed that invasive isolates are less likely to carry abiG-I/II and Q8DZ34, but they are more likely to show the presence of the pil1 locus, fbsB (515), and capsule type III (13, 45), as well as COH1-like alleles of pil2b, nss/srr, and rgf (Table 3). This does not necessarily mean that these factors are involved in invasiveness. They also just might be surrogate markers being linked to invasive strains. Several of these markers appear together in CC19/17 isolates. Previously, CC19 (including ST01, ST17, ST19, and ST28) was found in neonatal sepsis cases from very diverse geographic origins (30, 46–48), and the CC19/17 cluster has also frequently been observed in invasive isolates (49, 50).
Other virulence factors include speM (encoding exotoxin M) and the cyl operon (see Table S2 in the supplemental material). The speM gene was found in only seven isolates. The cyl operon was always found, and thus, its detection cannot be helpful for predicting virulence. However, in vivo expression might be a very different issue, and possibly, the key to understanding the difference between invasive and carriage isolates is not the mere presence or absence of specific genes or alleles but their expression.
Generally, it is not possible to safely distinguish invasive from carriage isolates and to predict an individual fate based on available typing data. Even if some strains are associated with an increased risk of infection, other strains cannot be considered harmless. For practical purposes, this warrants screening for and eradication of any S. agalactiae colonization prior to delivery, regardless of the genotyping data. Vaccination might become an additional tool for preventing infection, and a vaccine covering capsule types Ia, Ib, and III is currently being tested (51). Capsule types are geographically unevenly distributed, as the comparison between the German and Caribbean isolates and other work (50, 52, 53) indicate. Thus, the efficacy of a capsule-based vaccination might vary in different countries and also might change over time (54), emphasizing a need for typing GBS.
For the sake of statistical significance, future studies should target more isolates from defined clinical conditions and healthy controls and aim to compare S. agalactiae populations from different regions and host species. The arrays described here provide a convenient tool for typing large numbers of clinical/veterinary isolates and thus might help to obtain insights in GBS epidemiology.
Supplementary Material
ACKNOWLEDGMENTS
We thank R. Berner (Dresden University Hospital), M. van der Linden (National Reference Centre for Streptococci, Aachen, Germany), S. Weber (Sheikh Khalifa Medical City, Abu Dhabi, UAE), P. Akpaka (University of West Indies, St. Augustine, Trinidad and Tobago), K. Kadlec (Friedrich Loeffler Institute, Neustadt-Mariensee, Germany), M. Rench (Baylor College of Medicine, Houston, TX), the Institut Pasteur, K. Hochauf, and F. Gunzer (Institute for Medical Microbiology and Hygiene, IMMH, Dresden, Germany), and the colleagues of Dresden University Hospital for collecting and providing the strains and isolates, W. Rudolph (IMMH Dresden) for MLST sequencing, and S. Hildebrandt (Dresden) for advice on clinical issues. We thank A. Ruppelt (IMMH Dresden), I. Engelmann, J. Sachtschal, and G. Rößler (Alere Jena) for excellent technical assistance, and we also thank E. Jacobs, K. Hochauf, and F. Gunzer (IMMH Dresden), E. Ermantraut (Alere Jena), and the Infectognostics Research Campus Consortium Jena for their support.
The study was funded by the Faculty of Medicine Carl Gustav Carus, Technische Universität Dresden (MeDDrive program).
S.M., P.S., E.M., and R.E. are employees of Alere Technologies, the company that manufactured the assays described herein. (S.M. was employed by Alere Technologies after the experiments were finished but before the paper was written.) This had no influence on the study design, on the decision to publish, or on the preparation of the manuscript.
Footnotes
Published ahead of print 27 August 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.02411-14.
REFERENCES
- 1.Kowalska B, Niemiec KT, Drejewicz H, Polak K, Kubik P, Elmidaoui A, Gierowska-Bogusz B, Jaczynska R. 2003. Prevalence of group B streptococcal colonization in pregnant women and their newborns based on the results of examination of patients in the Obstetric and Gynecology Department of the National Research Institute of Mother and Child–a pilot study. Ginekol. Pol. 74:1223–1227 (In Polish.) [PubMed] [Google Scholar]
- 2.Kunze M, Ziegler A, Fluegge K, Hentschel R, Proempeler H, Berner R. 2011. Colonization, serotypes and transmission rates of group B streptococci in pregnant women and their infants born at a single University Center in Germany. J. Perinat. Med. 39:417–422. 10.1515/JPM.2011.037. [DOI] [PubMed] [Google Scholar]
- 3.Marconi C, Rocchetti TT, Rall VL, Carvalho LR, Borges VT, Silva MG. 2010. Detection of Streptococcus agalactiae colonization in pregnant women by using combined swab cultures: cross-sectional prevalence study. Sao Paulo Med. J. 128:60–62. 10.1590/S1516-31802010000200003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Müller-Vranjes A, Puntarić D, Curzik D, Sijanović S, Topolovec Z, Kasac Z, Miskulin M. 2011. Prevalence and significance of vaginal group B streptococcus colonization in pregnant women from Osijek, Croatia. Coll. Antropol. 35:21–26. [PubMed] [Google Scholar]
- 5.Schrag SJ, Zywicki S, Farley MM, Reingold AL, Harrison LH, Lefkowitz LB, Hadler JL, Danila R, Cieslak PR, Schuchat A. 2000. Group B streptococcal disease in the era of intrapartum antibiotic prophylaxis. N. Engl. J. Med. 342:15–20. 10.1056/NEJM200001063420103. [DOI] [PubMed] [Google Scholar]
- 6.Baker CJ, Edwards MS. 1995. Group B streptococcal infections, 3rd ed. WB Saunders, Philadelphia, PA. [Google Scholar]
- 7.Glantz JC, Kedley KE. 1998. Concepts and controversies in the management of group B streptococcus during pregnancy. Birth 25:45–53. 10.1046/j.1523-536x.1998.00045.x. [DOI] [PubMed] [Google Scholar]
- 8.Puopolo KM, Madoff LC, Eichenwald EC. 2005. Early-onset group B streptococcal disease in the era of maternal screening. Pediatrics 115:1240–1246. 10.1542/peds.2004-2275. [DOI] [PubMed] [Google Scholar]
- 9.Betriu C, Gomez M, Sanchez A, Cruceyra A, Romero J, Picazo JJ. 1994. Antibiotic resistance and penicillin tolerance in clinical isolates of group B streptococci. Antimicrob. Agents Chemother. 38:2183–2186. 10.1128/AAC.38.9.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aracil B, Miñambres M, Oteo J, De La Rosa M, Gómez-Garcés JL, Alos AJ. 2002. Susceptibility of strains of Streptococcus agalactiae to macrolides and lincosamides, phenotype patterns and resistance genes. Clin. Microbiol. Infect. 8:745–748. 10.1046/j.1469-0691.2002.00450.x. [DOI] [PubMed] [Google Scholar]
- 11.Betriu C, Culebras E, Gómez M, Rodríguez-Avial I, Sánchez BA, Ágreda MC, Picazo JJ. 2003. Erythromycin and clindamycin resistance and telithromycin susceptibility in Streptococcus agalactiae. Antimicrob. Agents Chemother. 47:1112–1114. 10.1128/AAC.47.3.1112-1114.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Azavedo JC, McGavin M, Duncan C, Low DE, McGeer A. 2001. Prevalence and mechanisms of macrolide resistance in invasive and noninvasive group B streptococcus isolates from Ontario, Canada. Antimicrob. Agents Chemother. 45:3504–3508. 10.1128/AAC.45.12.3504-3508.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schuchat A. 1998. Epidemiology of group B streptococcal disease in the United States: shifting paradigms. Clin. Microbiol. Rev. 11:497–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uh Y, Jang IH, Hwang GY, Yoon KJ, Song W. 2001. Emerging erythromycin resistance among group B streptococci in Korea. Eur. J. Clin. Microbiol. Infect. Dis. 20:52–54. 10.1007/s100960000414. [DOI] [PubMed] [Google Scholar]
- 15.Pereira Ude P, Rodrigues Dos Santos A, Hassan SS, Aburjaile FF, Soares Sde C, Ramos RT, Carneiro AR, Guimarães LC, Silva de Almeida S, Diniz CA, Barbosa MS, Gomes de Sá P, Ali A, Bakhtiar SM, Dorella FA, Zerlotini A, Araújo FM, Leite LR, Oliveira G, Miyoshi A, Silva A, Azevedo V, Figueiredo HC. 2013. Complete genome sequence of Streptococcus agalactiae strain SA20-06, a fish pathogen associated to meningoencephalitis outbreaks. Stand. Genomic Sci. 8:188–197. 10.4056/sigs.3687314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kogan G, Uhrín D, Brisson JR, Paoletti LC, Blodgett AE, Kasper DL, Jennings HJ. 1996. Structural and immunochemical characterization of the type VIII group B streptococcus capsular polysaccharide. J. Biol. Chem. 271:8786–8790. 10.1074/jbc.271.15.8786. [DOI] [PubMed] [Google Scholar]
- 17.Kong F, Lambertsen LM, Slotved HC, Ko D, Wang H, Gilbert GL. 2008. Use of phenotypic and molecular serotype identification methods to characterize previously nonserotypeable group B streptococci. J. Clin. Microbiol. 46:2745–2750. 10.1128/JCM.00189-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paoletti LJ, Bradford J, Paoletti LC. 1999. A serotype VIII strain among colonizing group B streptococcal isolates in Boston, Massachusetts. J. Clin. Microbiol. 37:3759–3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.von Hunolstein C, Parisi L, Tissi L, Recchia S, Alfarone G, Nicolini L, Volpe C, Wagner B, Motlovà J, Orefici G. 1999. Virulence properties of type VII Streptococcus agalactiae (group B streptococci) and immunochemical analysis of capsular type polysaccharide. J. Med. Microbiol. 48:983–990. 10.1099/00222615-48-11-983. [DOI] [PubMed] [Google Scholar]
- 20.Wessels MR. 1997. Biology of streptococcal capsular polysaccharides. Soc. Appl. Bacteriol. Symp. Ser. 26:20S–31S. [PubMed] [Google Scholar]
- 21.Blumberg HM, Stephens DS, Licitra C, Pigott N, Facklam R, Swaminathan B, Wachsmuth IK. 1992. Molecular epidemiology of group B streptococcal infections: use of restriction endonuclease analysis of chromosomal DNA and DNA restriction fragment length polymorphisms of ribosomal RNA genes (ribotyping). J. Infect. Dis. 166:574–579. 10.1093/infdis/166.3.574. [DOI] [PubMed] [Google Scholar]
- 22.Huet H, Martin C, Geslin P, Grimont F, Quentin R. 1993. Ribotyping of Streptococcus agalactiae strains isolated from vaginas of asymptomatic women. Res. Microbiol. 144:457–465. 10.1016/0923-2508(93)90053-5. [DOI] [PubMed] [Google Scholar]
- 23.Benson JA, Ferrieri P. 2001. Rapid pulsed-field gel electrophoresis method for group B streptococcus isolates. J. Clin. Microbiol. 39:3006–3008. 10.1128/JCM.39.8.3006-3008.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quentin R, Huet H, Wang FS, Geslin P, Goudeau A, Selander RK. 1995. Characterization of Streptococcus agalactiae strains by multilocus enzyme genotype and serotype: identification of multiple virulent clone families that cause invasive neonatal disease. J. Clin. Microbiol. 33:2576–2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Selander RK, Caugant DA, Ochman H, Musser JM, Gilmour MN, Whittam TS. 1986. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl. Environ. Microbiol. 51:873–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang GW, Kotiw M, Daggard G. 2002. A RAPD-PCR genotyping assay which correlates with serotypes of group B streptococci. Lett. Appl. Microbiol. 35:247–250. 10.1046/j.1472-765X.2002.01177.x. [DOI] [PubMed] [Google Scholar]
- 27.Manning SD, Lacher DW, Davies HD, Foxman B, Whittam TS. 2005. DNA polymorphism and molecular subtyping of the capsular gene cluster of group B streptococcus. J. Clin. Microbiol. 43:6113–6116. 10.1128/JCM.43.12.6113-6116.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones N, Bohnsack JF, Takahashi S, Oliver KA, Chan MS, Kunst F, Glaser P, Rusniok C, Crook DW, Harding RM, Bisharat N, Spratt BG. 2003. Multilocus sequence typing system for group B streptococcus. J. Clin. Microbiol. 41:2530–2536. 10.1128/JCM.41.6.2530-2536.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Monecke S, Slickers P, Ehricht R. 2008. Assignment of Staphylococcus aureus isolates to clonal complexes based on microarray analysis and pattern recognition. FEMS Immunol. Med. Microbiol. 53:237–251. 10.1111/j.1574-695X.2008.00426.x. [DOI] [PubMed] [Google Scholar]
- 30.Sørensen UBS, Poulsen K, Ghezzo C, Margarit I, Kilian M. 2010. Emergence and global dissemination of host-specific Streptococcus agalactiae clones. mBio 1(3):e00178-10. 10.1128/mBio.00178-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heden LO, Frithz E, Lindahl G. 1991. Molecular characterization of an IgA receptor from group B streptococci: sequence of the gene, identification of a proline-rich region with unique structure and isolation of N-terminal fragments with IgA-binding capacity. Eur. J. Immunol. 21:1481–1490. 10.1002/eji.1830210623. [DOI] [PubMed] [Google Scholar]
- 32.Kong F, Gidding HF, Berner R, Gilbert GL. 2006. Streptococcus agalactiae Cβ protein gene (bac) sequence types, based on the repeated region of the cell-wall-spanning domain: relationship to virulence and a proposed standardized nomenclature. J. Med. Microbiol. 55:829–837. 10.1099/jmm.0.46307-0. [DOI] [PubMed] [Google Scholar]
- 33.Creti R, Fabretti F, Orefici G, von Hunolstein C. 2004. Multiplex PCR assay for direct identification of group B streptococcal alpha-protein-like protein genes. J. Clin. Microbiol. 42:1326–1329. 10.1128/JCM.42.3.1326-1329.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lauer P, Rinaudo CD, Soriani M, Margarit I, Maione D, Rosini R, Taddei AR, Mora M, Rappuoli R, Grandi G, Telford JL. 2005. Genome analysis reveals pili in group B streptococcus. Science 309:105. 10.1126/science.1111563. [DOI] [PubMed] [Google Scholar]
- 35.Dramsi S, Caliot E, Bonne I, Guadagnini S, Prévost MC, Kojadinovic M, Lalioui L, Poyart C, Trieu-Cuot P. 2006. Assembly and role of pili in group B streptococci. Mol. Microbiol. 60:1401–1413. 10.1111/j.1365-2958.2006.05190.x. [DOI] [PubMed] [Google Scholar]
- 36.Rosini R, Rinaudo CD, Soriani M, Lauer P, Mora M, Maione D, Taddei A, Santi I, Ghezzo C, Brettoni C, Buccato S, Margarit I, Grandi G, Telford JL. 2006. Identification of novel genomic islands coding for antigenic pilus-like structures in Streptococcus agalactiae. Mol. Microbiol. 61:126–141. 10.1111/j.1365-2958.2006.05225.x. [DOI] [PubMed] [Google Scholar]
- 37.Gutekunst H, Eikmanns BJ, Reinscheid DJ. 2004. The novel fibrinogen-binding protein FbsB promotes Streptococcus agalactiae invasion into epithelial cells. Infect. Immun. 72:3495–3504. 10.1128/IAI.72.6.3495-3504.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schubert A, Zakikhany K, Pietrocola G, Meinke A, Speziale P, Eikmanns BJ, Reinscheid DJ. 2004. The fibrinogen receptor FbsA promotes adherence of Streptococcus agalactiae to human epithelial cells. Infect. Immun. 72:6197–6205. 10.1128/IAI.72.11.6197-6205.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Sorge NM, Quach D, Gurney MA, Sullam PM, Nizet V, Doran KS. 2009. The group B streptococcal serine-rich repeat 1 glycoprotein mediates penetration of the blood-brain barrier. J. Infect. Dis. 199:1479–1487. 10.1086/598217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang Y, Liu Y, Ding Y, Yi L, Ma Z, Fan H, Lu C. 2013. Molecular characterization of Streptococcus agalactiae isolated from bovine mastitis in eastern China. PLoS One 8:e67755. 10.1371/journal.pone.0067755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dabul ANG, Camargo ILBC. 2014. Clonal complexes of Staphylococcus aureus: all mixed and together. FEMS Microbiol. Lett. 351:7–8. 10.1111/1574-6968.12358. [DOI] [PubMed] [Google Scholar]
- 42.Robinson DA, Enright MC. 2004. Evolution of Staphylococcus aureus by large chromosomal replacements. J. Bacteriol. 186:1060–1064. 10.1128/JB.186.4.1060-1064.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nimmo G, Steen J, Monecke S, Ehricht R, Slickers P, Thomas JC, Grimmond SM, Robinson DA, Coombs GW. 2014. ST2249-MRSA-III: a second major Staphylococcus aureus recombinant causing healthcare infections in the 1970s. Eur. Cong. Clin. Microbiol. Infect. Dis. (ECCMID), 10 to 13 May 2014, Barcelona, Spain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brochet M, Rusniok C, Couvé E, Dramsi S, Poyart C, Trieu-Cuot P, Kunst F, Glaser P. 2008. Shaping a bacterial genome by large chromosomal replacements, the evolutionary history of Streptococcus agalactiae. Proc. Natl. Acad. Sci. U. S. A. 105:15961–15966. 10.1073/pnas.0803654105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davies HD, Jones N, Whittam TS, Elsayed S, Bisharat N, Baker CJ. 2004. Multilocus sequence typing of serotype III group B streptococcus and correlation with pathogenic potential. J. Infect. Dis. 189:1097–1102. 10.1086/382087. [DOI] [PubMed] [Google Scholar]
- 46.Brzychczy-Wloch M, Gosiewski T, Pawlik D, Szumala-Kakol A, Samead A, Heczko PB. 2012. Occurrence of the hypervirulent ST-17 clone of Streptococcus agalactiae in pregnant women and newborns. Przegl. Epidemiol. 66:395–401 (In Polish.) [PubMed] [Google Scholar]
- 47.Ryu H, Park YJ, Kim YK, Chang J, Yu JK. 2014. Dominance of clonal complex 10 among the levofloxacin-resistant Streptococcus agalactiae isolated from bacteremic patients in a Korean hospital. J. Infect. Chemother. 20:509–511. 10.1016/j.jiac.2014.03.005. [DOI] [PubMed] [Google Scholar]
- 48.Usein CR, Militaru M, Cristea V, Străut M. 2014. Genetic diversity and antimicrobial resistance in Streptococcus agalactiae strains recovered from female carriers in the Bucharest area. Mem. Inst. Oswaldo Cruz 109:189–196. 10.1590/0074-0276140431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Straková L, Musílek M, Motlová J. 2010. Multilocus sequence types in Czech neonatal GBS strains from 2004 to 2008. Epidemiol. Mikrobiol. Imunol. 59:45–47. [PubMed] [Google Scholar]
- 50.Tien N, Ho CM, Lin HJ, Shih MC, Ho MW, Lin HC, Lin HS, Chang CC, Lu JJ. 2011. Multilocus sequence typing of invasive group B streptococcus in central area of Taiwan. J. Microbiol. Immunol. Infect. 44:430–434. 10.1016/j.jmii.2011.04.013. [DOI] [PubMed] [Google Scholar]
- 51.Madhi SA, Dangor Z, Heath PT, Schrag S, Izu A, Sobanjo-Ter Meulen A, Dull PM. 2013. Considerations for a phase-III trial to evaluate a group B streptococcus polysaccharide-protein conjugate vaccine in pregnant women for the prevention of early- and late-onset invasive disease in young-infants. Vaccine 31(Suppl 4):D52–D57. 10.1016/j.vaccine.2013.02.029. [DOI] [PubMed] [Google Scholar]
- 52.Brimil N, Barthell E, Heindrichs U, Kuhn M, Lütticken R, Spellerberg B. 2006. Epidemiology of Streptococcus agalactiae colonization in Germany. Int. J. Med. Microbiol. 296:39–44. 10.1016/j.ijmm.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 53.Dutra VG, Alves VM, Olendzki AN, Dias CA, de Bastos AF, Santos GO, de Amorin EL, Sousa MA, Santos R, Ribeiro PC, Fontes CF, Andrey M, Magalhães K, Araujo AA, Paffadore LF, Marconi C, Murta EF, Fernandes PC, Jr, Raddi MS, Marinho PS, Bornia RB, Palmeiro JK, Dalla-Costa LM, Pinto TC, Botelho AC, Teixeira LM, Fracalanzza SE. 2012. Streptococcus agalactiae in Brazil: serotype distribution, virulence determinants and antimicrobial susceptibility. BMC Infect. Dis. 14:323. 10.1186/1471-2334-14-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Harrison LH, Elliott JA, Dwyer DM, Libonati JP, Ferrieri P, Billmann L, Schuchat A. 1998. Serotype distribution of invasive group B streptococcal isolates in Maryland: implications for vaccine formulation. Maryland Emerging Infections Program. J. Infect. Dis. 177:998–1002. [DOI] [PubMed] [Google Scholar]
- 55.Spellerberg B, Rozdzinski E, Martin S, Weber-Heynemann J, Lütticken R. 2002. rgf encodes a novel two-component signal transduction system of Streptococcus agalactiae. Infect. Immun. 70:2434–2440. 10.1128/IAI.70.5.2434-2440.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.