Abstract
Briefly incubated agar cultures from positive blood cultures were used for antimicrobial susceptibility testing (AST) by Vitek 2. The cultivation time until inoculation was 3.8 h for Gram-positive cocci and 2.4 h for Gram-negative rods. The error rates were low, providing early and reliable AST without additional time or cost expenditure.
TEXT
Since the outcome of sepsis is dependent on appropriate and timely treatment (1), the identification of pathogens and availability of antimicrobial susceptibility testing (AST) should occur as soon as possible (2). Routinely, positive blood culture (BC) broth is plated onto agar and incubated for 18 to 24 h before inoculation of Vitek 2 cards for combined identification and AST (3, 4). Recently, the rapid identification of bacteria from positive BCs by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) has been reported from cultures very briefly incubated on solid medium (5). Here, we evaluated the feasibility and accuracy of accelerated AST using the inoculation of Vitek 2 cards with biomass very briefly incubated on agar after subcultivation of positive BC broth.
(This work was presented in part at the 53rd Interscience Conference on Antimicrobial Agents and Chemotherapy, 10 to 13 September 2013, Denver, CO [6].)
A total of 138 blood samples flagged as positive by an automated BC system (BD Bactec 9240; BD Diagnostics, Heidelberg, Germany) and Gram stained as Gram-positive cocci (GPC) or Gram-negative rods (GNR) were processed as follows: two drops of BC broth were subcultured on Columbia blood agar. Starting from 2 h of incubation at 36°C in air with 5% CO2, the plates were inspected hourly, and a 0.5 to 0.63 McFarland standard inoculum was prepared at the first time point at which the growth appeared sufficient. Subsequently, Vitek 2 (bioMérieux SA, Marcy l'Étoile, France) testing was performed. For comparison, a 0.5 to 0.63 McFarland standard inoculum from 24-h cultures was tested by Vitek 2, as recommended by the manufacturer (3). The choice of a definite Vitek 2 AST card was based on the Gram stain from BC broth or/and early MALDI-TOF identification, performed as previously described (5). AST was done in triplicate, i.e., three AST cards were used for each inoculum to minimize the impact of reproducibility errors on the comparison between rapid and standard testing. The median values were used for analysis. Additionally, the detection of particular resistance mechanisms was evaluated: cefoxitin screening for methicillin resistance in staphylococci, inducible clindamycin resistance in staphylococci and Streptococcus agalactiae, and the production of extended-spectrum β-lactamases (ESBL) in Enterobacteriaceae.
Of 138 BCs, 13 ambiguously identified samples, 8 samples with technical errors, 5 isolates that could not be tested by Vitek 2, and 8 mixed cultures were excluded. Thus, 104 isolates (68 GPC and 36 GNR) were available for analysis by rapid AST (Table 1).
TABLE 1.
Microorganisms available for AST results comparisona
| Organism | No. (%) available for AST |
|---|---|
| Gram-positive cocci | 68 (100) |
| Staphylococcus aureus | 8 (11.8) |
| Coagulase-negative staphylococci | 40 (58.8) |
| Enterococcus spp. | 17 (25.0) |
| Streptococcus pneumoniae | 2 (2.9) |
| Streptococcus agalactiae | 1 (1.5) |
| Gram-negative rods | 36 (100) |
| Escherichia coli | 25 (69.4) |
| Klebsiella pneumoniae | 5 (13.9) |
| Enterobacter cloacae | 4 (11.1) |
| Serratia marcescens | 1 (2.8) |
| Pseudomonas aeruginosa | 1 (2.8) |
n = 104.
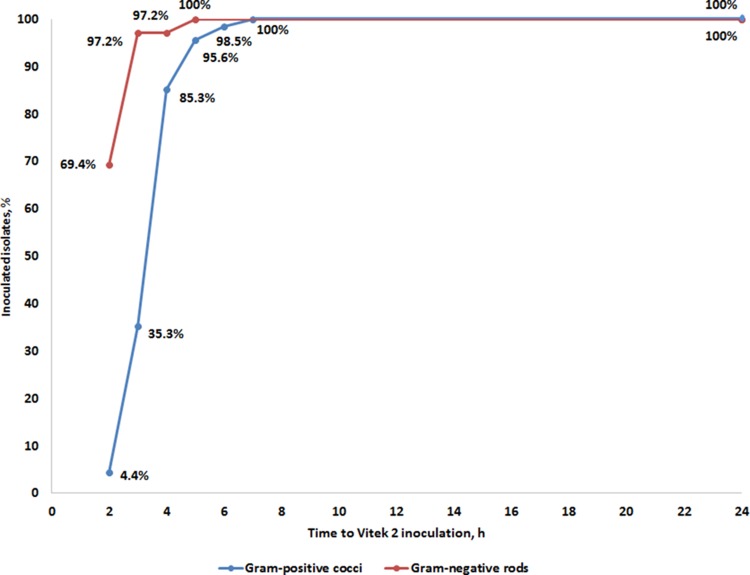
The mean time necessary for cultivating plated positive BC broth on agar until successful inoculation of Vitek 2 cards was as short as 3.8 h for GPC and 2.4 h for GNR (Table 2). For GPC, inoculation was possible at 4 h in 85.3% of the isolates, with all specimens being inoculated within 7 h (Fig. 1). At 3 h, inoculation was possible with all GNR isolates, with the exception of a Pseudomonas aeruginosa isolate, which was inoculated at 5 h (Fig. 1). The mean cultivation times to inoculation of Vitek 2 cards and to an AST result for different bacterial species or groups are shown in Fig. S1 in the supplemental material. An AST result was available 20.2 h and 21.8 h earlier than with the standard method for GPC and GNR, respectively (Table 2).
TABLE 2.
Time to successful inoculation and AST result for Gram-positive cocci and Gram-negative rods in Vitek 2
| Time period | Time (mean ± SD) (h) for: |
|||||
|---|---|---|---|---|---|---|
| Gram-positive cocci (n = 68) |
Gram-negative rods (n = 36) |
|||||
| Brief culture | Control culture | P valuea | Brief culture | Control culture | P valuea | |
| Cultivation time until Vitek 2 inoculation | 3.8 ± 0.9 | 24.0 | <0.0001 | 2.4 ± 0.6 | 24.0 | <0.0001 |
| Duration of Vitek 2 AST | 9.8 ± 1.1 | 9.8 ± 1.8 | NSb | 8.8 ± 1.4 | 9.0 ± 2.8 | NS |
| Total time from positive BC subculture to AST result | 13.6 ± 1.4 | 33.8 ± 1.8 | <0.0001 | 11.2 ± 1.7 | 33.0 ± 2.8 | <0.0001 |
By Wilcoxon signed-rank test.
NS, not significant.
FIG 1.
Cultivation time until Vitek 2 inoculation required for successful AST for Gram-positive cocci and Gram-negative rods.
Very major error (VME) (false-susceptible result of rapid AST), major error (ME) (false-resistant result of rapid AST), and minor error (mE) (false categorization involving intermediate result) rates, and categorical agreement (CA) (results within the same category) and essential agreement (EA) (MIC difference ≤ 1 double dilution step) were 1.6%, 0.3%, 0.1%, 99.2%, and 99.1% for GPC (Table 3) and 0%, 0.5%, 0.8%, 99.2%, and 99.3% for GNR (Table 4), respectively. In general, the performance of rapid AST in our study was better or similar to that reported in studies using differential centrifugation for the direct inoculation of Vitek 2 cards from positive BCs (7–15). We report error rates here, as required by both the International Organization for Standardization and the U.S. Food and Drug Administration (16, 17), which recommend that the rate of VME should be calculated using the number of isolates which have been determined as resistant by the reference method as the denominator. To obtain the rate of ME, number of ME should be divided by the number of susceptible isolates determined by the reference method. The mE rate is calculated based on the total number of isolates. Of note, only a minority of studies (7, 10, 15) investigating direct inoculation of Vitek 2 cards for AST presented their results in that way, while the most of studies on rapid Vitek 2 AST reported VME and ME error rates based on the total number of isolates (8, 9, 12–14, 18). If calculated based on the number of total isolates, the VME and ME rates are inherently lower and would amount in our study to 0.5% and 0.2% for GPC and 0 and 0.3% for GNR, respectively. The results of all three AST tests within the triplicate experiments were the same for 99.4% and 99.0% of the isolate-antibiotic combinations for rapid and standard testing, respectively (P = 0.246), demonstrating the high reproducibility of the rapid method.
TABLE 3.
Performance of rapid antimicrobial susceptibility testing of Gram-positive cocci (n = 68)
| Antimicrobial agent (n) | No. of isolate-antibiotic combinations | No. of isolates that werea: |
No. (%) of errors |
Categorical agreement (%) | Essential agreement (%) | |||
|---|---|---|---|---|---|---|---|---|
| Resistant | Susceptible | Very major | Major | Minor | ||||
| Staphylococcus spp. (48) | 872 | 275 | 586 | 4 (1.5) | 2 (0.3) | 0 | 99.3 | 99.0 |
| Penicillin G | 48 | 42 | 6 | 100 | 100 | |||
| Oxacillin | 48 | 30 | 18 | 100 | 97.9 | |||
| Clindamycin | 48 | 22 | 26 | 100 | 100 | |||
| Erythromycin | 48 | 30 | 18 | 100 | 100 | |||
| Levofloxacin | 48 | 25 | 23 | 100 | 100 | |||
| Moxifloxacin | 48 | 15 | 23 | 100 | 100 | |||
| Gentamicin | 48 | 19 | 29 | 100 | 100 | |||
| Tobramycin | 48 | 23 | 25 | 100 | 100 | |||
| Vancomycin | 48 | 0 | 48 | 100 | 100 | |||
| Teicoplanin | 48 | 7 | 41 | 2 | 1 | 93.8 | 89.6 | |
| Linezolid | 48 | 0 | 48 | 100 | 97.9 | |||
| Tetracycline | 48 | 18 | 29 | 1 | 97.9 | 97.9 | ||
| Tigecycline | 48 | 0 | 48 | 100 | 100 | |||
| Trimethoprim-sulfamethoxazole | 48 | 17 | 31 | 100 | 100 | |||
| Fusidic acid | 48 | 9 | 39 | 1 | 1 | 95.8 | 97.9 | |
| Fosfomycin | 48 | 14 | 34 | 100 | 100 | |||
| Rifampin | 48 | 4 | 44 | 100 | 100 | |||
| Nitrofurantoin | 48 | 0 | 48 | 100 | 100 | |||
| Mupirocin | 8 | 0 | 8 | 100 | 100 | |||
| Enterococcus spp. (17) | 238 | 91 | 144 | 1 (1.1) | 0 | 1 (0.4) | 99.2 | 98.7 |
| Penicillin G | 17 | 13 | 4 | 100 | 94.1 | |||
| Ampicillin | 17 | 10 | 7 | 100 | 100 | |||
| Ampicillin-sulbactam | 17 | 13 | 4 | 100 | 100 | |||
| Imipenem | 17 | 10 | 7 | 100 | 100 | |||
| Erythromycin | 17 | 14 | 0 | 1 | 94.1 | 94.1 | ||
| Levofloxacin | 17 | 10 | 7 | 100 | 100 | |||
| Vancomycin | 17 | 1 | 16 | 100 | 100 | |||
| Teicoplanin | 17 | 1 | 16 | 100 | 100 | |||
| Linezolid | 17 | 1 | 16 | 100 | 100 | |||
| Tetracycline | 17 | 8 | 9 | 100 | 100 | |||
| Tigecycline | 17 | 0 | 17 | 100 | 100 | |||
| Trimethoprim-sulfamethoxazole | 17 | 0 | 17 | 100 | 100 | |||
| Quinupristin-dalfopristin | 17 | 6 | 11 | 100 | 100 | |||
| Nitrofurantoin | 17 | 4 | 13 | 1 | 94.1 | 100 | ||
| Total Gram-positive cocci (68) | 1,163 | 377 | 767 | 6 (1.6)b,c | 2 (0.3)b | 1 (0.1)b | 99.2b | 99.1b,c |
The number of results within the intermediate category can be calculated by subtracting the resistant and susceptible results from the number of isolate-antibiotic combinations tested.
Streptococci (S. pneumoniae, n = 2 and S. agalactiae, n = 1) are included in the total number of Gram-positive cocci and in the calculation of the error and agreement rates for all Gram-positive cocci. No separate calculation of error and agreement rates was performed for streptococci because of the small number of isolates.
One VME (tetracycline in a S. pneumoniae isolate) and no other errors were observed in streptococci. Essential agreement (EA) was observed for all streptococcal isolate-antibiotic combinations.
TABLE 4.
Performance of rapid antimicrobial susceptibility testing of Gram-negative rods (n = 36)
| Antimicrobial agent | No. of isolate-antibiotic combinationsb | No. of isolates that werea: |
No. (%) of errors |
Categorical agreement (%) | Essential agreement (%) | |||
|---|---|---|---|---|---|---|---|---|
| Resistant | Susceptible | Very major | Major | Minor | ||||
| Ampicillin | 35 | 29 | 6 | 100 | 100 | |||
| Ampicillin-sulbactam | 35 | 20 | 15 | 1 | 97.1 | 97.1 | ||
| Piperacillin | 1 | 0 | 0 | 100 | 100 | |||
| Piperacillin-tazobactam | 36 | 9 | 18 | 2 | 94.4 | 97.2 | ||
| Cefuroxime | 35 | 16 | 19 | 100 | 100 | |||
| Cefuroxime axetil | 35 | 15 | 20 | 100 | 100 | |||
| Cefotaxime | 36 | 10 | 26 | 1 | 97.2 | 97.2 | ||
| Cefpodoxime | 35 | 14 | 21 | 100 | 100 | |||
| Ceftazidime | 36 | 4 | 25 | 100 | 100 | |||
| Cefepime | 1 | 0 | 1 | 100 | 100 | |||
| Ertapenem | 35 | 0 | 35 | 1 | 97.1 | 97.1 | ||
| Imipenem | 36 | 0 | 35 | 100 | 100 | |||
| Meropenem | 36 | 0 | 35 | 100 | 100 | |||
| Aztreonam | 1 | 0 | 0 | 100 | 100 | |||
| Ciprofloxacin | 36 | 10 | 26 | 100 | 100 | |||
| Moxifloxacin | 35 | 10 | 25 | 100 | 100 | |||
| Gentamicin | 36 | 1 | 34 | 100 | 100 | |||
| Amikacin | 1 | 0 | 1 | 100 | 100 | |||
| Tobramycin | 1 | 0 | 1 | 100 | 100 | |||
| Tetracycline | 35 | 16 | 19 | 1 | 97.1 | 100 | ||
| Tigecycline | 35 | 1 | 29 | 1 | 97.1 | 100 | ||
| Trimethoprim-sulfamethoxazole | 36 | 14 | 22 | 100 | 100 | |||
| Colistin | 1 | 0 | 1 | 100 | 100 | |||
| Total | 609 | 169 | 414 | 0 | 2 (0.5) | 5 (0.8) | 99.2 | 99.3 |
The number of results in the intermediate category can be calculated by subtracting the resistant and susceptible results from the number of isolate-antibiotic combinations tested.
One as a number of isolate-antibiotic combinations for some antibiotics is due to the fact that those antibiotics are analyzed only in the card AST-N248, which was used for testing of the only isolate of P. aeruginosa.
Among 48 staphylococci tested, 30 were methicillin resistant as detected by positive cefoxitin screening (12.5% [1/8] of Staphylococcus aureus isolates and 72.5% [29/40] of coagulase-negative staphylococci). Thereby, 100% agreement was observed between the rapid and standard methods. Inducible clindamycin resistance was identified in three Staphylococcus epidermidis isolates. Again, this mechanism was detected concordantly by the two methods. Among 30 Escherichia coli and Klebsiella pneumoniae isolates, ESBL production was reported in eight isolates (7 E. coli and 1 K. pneumoniae) when short-term cultures were used. The standard method identified one additional ESBL producer (E. coli); however, this ESBL phenotype was not confirmed by an additional test.
The microbial counts of 0.5 McFarland standard suspensions from the briefly incubated cultures and 24-h cultures were 3.2 × 107 and 9.4 × 107 CFU/ml, respectively (P < 0.0001; for a comparison for a subgroup of isolates, see Table S1 in the supplemental material). This difference is not high, which might explain the accuracy of the rapid results.
In conclusion, the inoculation of brief solid medium cultures into Vitek 2 provided early and reliable AST without additional time or cost expenditure. However, further studies are necessary to confirm the reliability of this method with more pathogens with different resistance phenotypes. The thorough control of mature cultures is essential, as mixed cultures can occur. This method can be combined with the rapid MALDI-TOF identification from short-term agar cultures (5).
Supplementary Material
Footnotes
Published ahead of print 27 August 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.02400-14.
REFERENCES
- 1.Kumar A, Roberts D, Wood KE, Light B, Parrillo JE, Sharma S, Suppes R, Feinstein D, Zanotti S, Taiberg L, Gurka D, Kumar A, Cheang M. 2006. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit. Care Med. 34:1589–1596. 10.1097/01.CCM.0000217961.75225.E9. [DOI] [PubMed] [Google Scholar]
- 2.Kerremans JJ, Verboom P, Stijnen T, Hakkaart-van Roijen L, Goessens W, Verbrugh HA, Vos MC. 2008. Rapid identification and antimicrobial susceptibility testing reduce antibiotic use and accelerate pathogen-directed antibiotic use. J. Antimicrob. Chemother. 61:428–435. 10.1093/jac/dkm497. [DOI] [PubMed] [Google Scholar]
- 3.bioMérieux SA. 2010. Vitek 2 systems product information. 410791, Marcy-l'Étoile, France. [Google Scholar]
- 4.Petti CA, Weinstein MP, Carroll KC. 2011. Systems for detection and identification of bacteria and yeasts, p 15–26 In Versalovic J, Carroll KC, Funke G, Jorgensen JH, Landry ML, Warnock DW. (ed), Manual of clinical microbiology 10th ed. ASM Press, Washington, DC. [Google Scholar]
- 5.Idelevich EA, Schüle I, Grünastel B, Wüllenweber J, Peters G, Becker K. 2014. Rapid identification of microorganisms from positive blood cultures by MALDI-TOF mass spectrometry subsequent to very short-term incubation on solid medium. Clin. Microbiol. Infect. 10.1111/1469-0691.12640. [DOI] [PubMed] [Google Scholar]
- 6.Idelevich EA, Schüle I, Grünastel B, Wüllenweber J, Peters G, Becker K. 2013. Acceleration of antimicrobial susceptibility testing from positive blood cultures by inoculation of short incubated solid medium cultures into Vitek-2, presentation D-604. Abstr. 53rd Intersci. Conf. Antimicrob. Agents Chermother., 10 to 13 September 2013, Denver, CO. [Google Scholar]
- 7.Bruins MJ, Bloembergen P, Ruijs GJ, Wolfhagen MJ. 2004. Identification and susceptibility testing of Enterobacteriaceae and Pseudomonas aeruginosa by direct inoculation from positive Bactec blood culture bottles into Vitek 2. J. Clin. Microbiol. 42:7–11. 10.1128/JCM.42.1.7-11.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Cueto M, Ceballos E, Martinez-Martinez L, Perea EJ, Pascual A. 2004. Use of positive blood cultures for direct identification and susceptibility testing with the Vitek 2 system. J. Clin. Microbiol. 42:3734–3738. 10.1128/JCM.42.8.3734-3738.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gherardi G, Angeletti S, Panitti M, Pompilio A, Di Bonaventura G, Crea F, Avola A, Fico L, Palazzo C, Sapia GF, Visaggio D, Dicuonzo G. 2012. Comparative evaluation of the Vitek-2 Compact and Phoenix systems for rapid identification and antibiotic susceptibility testing directly from blood cultures of Gram-negative and Gram-positive isolates. Diagn. Microbiol. Infect. Dis. 72:20–31. 10.1016/j.diagmicrobio.2011.09.015. [DOI] [PubMed] [Google Scholar]
- 10.Kerremans JJ, Goessens WH, Verbrugh HA, Vos MC. 2004. Accuracy of identification and susceptibility results by direct inoculation of Vitek 2 cards from positive Bactec cultures. Eur. J. Clin. Microbiol. Infect. Dis. 23:892–898. 10.1007/s10096-004-1247-9. [DOI] [PubMed] [Google Scholar]
- 11.Ling TK, Liu ZK, Cheng AF. 2003. Evaluation of the Vitek 2 system for rapid direct identification and susceptibility testing of Gram-negative bacilli from positive blood cultures. J. Clin. Microbiol. 41:4705–4707. 10.1128/JCM.41.10.4705-4707.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lupetti A, Barnini S, Morici P, Ghelardi E, Nibbering PH, Campa M. 2013. Saponin promotes rapid identification and antimicrobial susceptibility profiling of Gram-positive and Gram-negative bacteria in blood cultures with the Vitek 2 system. Eur. J. Clin. Microbiol. Infect. Dis. 32:493–502. 10.1007/s10096-012-1762-z. [DOI] [PubMed] [Google Scholar]
- 13.Munoz-Dávila MJ, Yagüe G, Albert M, García-Lucas T. 2012. Comparative evaluation of Vitek 2 identification and susceptibility testing of Gram-negative rods directly and isolated from BacT/Alert-positive blood culture bottles. Eur. J. Clin. Microbiol. Infect. Dis. 31:663–669. 10.1007/s10096-011-1356-1. [DOI] [PubMed] [Google Scholar]
- 14.Prod'hom G, Durussel C, Greub G. 2013. A simple blood-culture bacterial pellet preparation for faster accurate direct bacterial identification and antibiotic susceptibility testing with the VITEK 2 system. J. Med. Microbiol. 62:773–777. 10.1099/jmm.0.049361-0. [DOI] [PubMed] [Google Scholar]
- 15.Quesada MD, Gimenez M, Molinos S, Fernandez G, Sanchez MD, Rivelo R, Ramirez A, Banque G, Ausina V. 2010. Performance of Vitek-2 Compact and overnight MicroScan panels for direct identification and susceptibility testing of Gram-negative bacilli from positive FAN BacT/Alert blood culture bottles. Clin. Microbiol. Infect. 16:137–140. 10.1111/j.1469-0691.2009.02907.x. [DOI] [PubMed] [Google Scholar]
- 16.ISO. 2007. ISO 20776-2:2007. Clinical laboratory testing and in vitro diagnostic test systems–susceptibility testing of infectious agents and evaluation of performance of antimicrobial susceptibility test devices–part 2: evaluation of performance of antimicrobial susceptibility test devices. International Organization for Standardization; Geneva, Switzerland: http://www.iso.org/iso/catalogue_detail.htm?csnumber=41631. [Google Scholar]
- 17.U.S. FDA. 2009. Guidance for industry and FDA: Class II special controls guidance document: antimicrobial susceptibility test (AST) systems. U.S. Food and Drug Administration, Silver Spring, MD: http://www.fda.gov/downloads/MedicalDevices/DeviceRegulationandGuidance/GuidanceDocuments/UCM388961.pdf. [Google Scholar]
- 18.Romero-Gómez MP, Gómez-Gil R, Paño-Pardo JR, Mingorance J. 2012. Identification and susceptibility testing of microorganism by direct inoculation from positive blood culture bottles by combining MALDI-TOF and Vitek-2 Compact is rapid and effective. J. Infect. 65:513–520. 10.1016/j.jinf.2012.08.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.