Abstract
For rapid results, we adapted the CarbaNP test by using 5-hour-old cultures; the sensitivity and specificity for detection of carbapenemase production were 100% for non-OXA-producing organisms. The sensitivity from early cultures was 94% for detection of carbapenemase production in Enterobacteriaceae strains. Utilizing younger cultures allows the test to be incorporated into a single day's workflow, facilitating timely patient care and antimicrobial stewardship.
TEXT
Carbapenemase-producing Enterobacteriaceae strains pose significant problems in clinical practice because of limited treatment options (1). Prompt detection of these organisms with current screening techniques, including the modified Hodge test (MHT), growth on selective media, and antimicrobial susceptibility testing, is difficult and time-consuming (2–5). Molecular methods are the gold standard for identification of carbapenemase genes (5, 6), but they are expensive, require technical experience, and are unable to detect novel genes (6). The CarbaNP test is inexpensive, rapid, and easy to perform and interpret; it utilizes imipenem hydrolysis, visualized by a color change, to identify carbapenemase production in Enterobacteriaceae and Pseudomonas species (7–10). In previous publications, isolates were taken from 1-day-old cultures (11) or the age of the culture was not mentioned (7–10). This study aims to determine whether the age of the isolates and the media used affect the ability of the CarbaNP test to detect carbapenemase production.
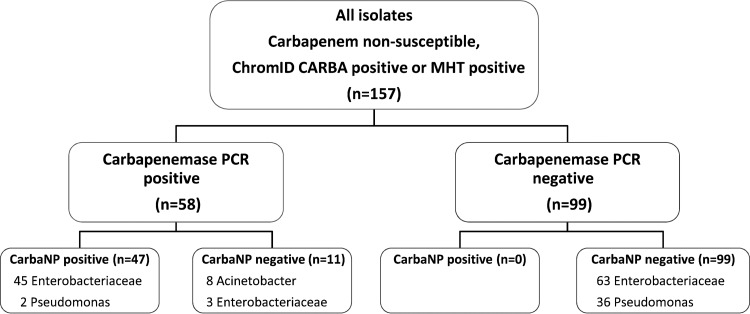
A total of 157 isolates (Enterobacteriaceae, n = 111; Pseudomonas spp., n = 38; Acinetobacter spp., n = 8) were suspected to harbor a carbapenemase because they had at least one of the following features: disc diffusion testing using CLSI guidelines (12) demonstrating decreased susceptibility to ertapenem with a zone diameter of <22 mm, meropenem MIC of ≥0.5 mg/liter determined by using a Vitek 2 system (bioMérieux Inc., Durham, NC), growth on ChromID Carba medium (bioMérieux, Inc., Durham, NC), or a positive MHT result. These isolates were recovered from clinical or environmental specimens at three tertiary hospital microbiology laboratories in Melbourne, Australia, between 2004 and 2014.
All isolates underwent PCR testing using primers designed for the carbapenemase genes blaIMP, blaKPC, blaNDM, blaOXA-23-like, blaOXA-24/40-like, blaOXA-48, blaOXA-51-like, blaOXA-58-like, blaOXA-48-like, and blaVIM (13–17). Fifty-eight isolates were confirmed to be carbapenemase producers by PCR, with typing as follows: IMP, n = 34; NDM, n = 6; KPC, n = 3; VIM, n = 2; OXA-23, n = 6; OXA-23-like/OXA-51-like, n = 1; OXA-24, n = 1; OXA-181, n = 2; OXA-24/40, n = 1; OXA-48, n = 1; OXA-51, n = 1. IMP and NDM carbapenemases were subtyped by sequencing for selected isolates.
The CarbaNP test was performed according to published methods (1, 8, 18), with modifications of culture age and inoculum. Solution A contained phenol red solution with ZnSO4, and solution B contained phenol red solution, ZnSO4 and 6 mg/ml imipenem (7, 8, 18). All isolates were plated from a single colony onto 2 sets of 5% horse blood agar (HBA), MacConkey agar, and ChromID medium. One set of plates was incubated at 35°C in air while the other set was incubated in 5% CO2. After 5 h of incubation (5-hour-old cultures), isolates underwent CarbaNP testing. Plates were reincubated to complete a total of 18 to 24 h of incubation (1-day-old cultures) before being retested with the CarbaNP test. Plates were stored at 4 to 8°C for 2 to 3 weeks and then retrieved for a final CarbaNP test. Each isolate was tested in two 1.5-ml Eppendorf tubes containing 100 μl Tris-HCl lysis buffer. One calibrated 1-μl loopful of 5-hour-old colonies was utilized for suspension, while 3 to 5 well-isolated colonies from 1-day-old cultures or stored plates were used. One hundred microliters of solution A was added to one tube, while 100 μl of solution B was added to the other tube. Tubes were then incubated at 35°C and checked for a color change from red to orange/yellow, indicating a positive reaction and carbapenemase production. All tests were performed in triplicate on separate days, and isolates were tested by an investigator who was blinded to the PCR results.
Carbapenemase PCR and CarbaNP results for all isolates are summarized in Fig. 1. Carbapenemase PCR and CarbaNP results (5-hour and 1-day) for carbapenemase-producing organisms (CPOs) are shown in Table 1. Using 1-day-old cultures, we found that 45 of the 58 carbapenemase-producing organisms (CPOs) from HBA were CarbaNP positive, with only 2 NDM Klebsiella pneumoniae and 11 OXA isolates remaining negative after 2 h (sensitivity, 78%; specificity, 100%; P < 0.0001). Using 5-hour-old cultures, the CarbaNP test was able to identify 47/58 CPOs taken from HBA (sensitivity, 81%; specificity, 100%; P < 0.0001) (Table 1 and Fig. 1 and 2). The CarbaNP test was 100% sensitive and specific (P < 0.0001) when OXA-producing organisms were excluded. The use of 5-hour-old cultures may increase the reliability of the CarbaNP results, as a greater proportion of positive results yielded a distinct yellow color than with 1-day-old cultures. When taken from 5-hour-old cultures, almost all non-OXA CarbaNP-positive isolates (43/45 isolates) were positive by 10 min (Table 1 and Fig. 1 and 2). Two isolates required 30 min to become CarbaNP positive, within a total test time of 5.5 h. An OXA-48 and OXA-181 isolate was CarbaNP positive after 1 h. The CarbaNP test with 5-hour-old cultures yielded positive results for all isolates with positive results for 1-day-old cultures. There were no false-positive CarbaNP results (i.e., all carbapenemase gene PCR-negative isolates were CarbaNP negative).
FIG 1.
CarbaNP and carbapenemase PCR results for isolates suspected to harbor a carbapenemase, from 5-h-old HBA cultures. See Table 1 for details on carbapenemase PCR-positive isolates.
TABLE 1.
Carbapenemase PCR typing and CarbaNP results from 5-hour-old and 1-day-old cultures from HBA for carbapenemase-producing organisms
| Organism | Carbapenemase | n | 5-h CarbaNPa |
1-day CarbaNP |
Meropenem MIC (mg/liter) | ||
|---|---|---|---|---|---|---|---|
| Result | Time (min) | Result | Time (min) | ||||
| Citrobacter freundii | IMP | 1 | + | 5 | + | 5 | ≥16 |
| Enterobacter aerogenes | OXA-51-like | 1 | − | NA | − | NA | ≥16 |
| Enterobacter cloacae | IMP | 7 | + | 5–10 | + | 5–15 | 1 to ≥16 |
| Escherichia coli | IMP | 2 | + | 5 | + | 5–10 | ≥16 |
| KPC | 1 | + | 5 | + | 15 | ≥16 | |
| NDM | 1 | + | 5 | + | 5 | ≥16 | |
| NDM-7 | 1 | + | 5 | + | 5 | ≥16 | |
| OXA-48 | 1 | + | 60 | + | 60 | ≥16 | |
| OXA-24/40 | 1 | − | NA | − | NA | 0.25 | |
| K. pneumoniae | IMP | 6 | + | 5–10 | + | 5 | 1 to ≥16 |
| IMP-14 | 1 | + | 5 | + | 5 | ≥16 | |
| KPC | 2 | + | 5 | + | 5 | ≥16 | |
| NDM-1 | 2 | + | 5 | + | 10 | ≥16 | |
| NDM | 2 | + | 5 | − | NA | ≥16 | |
| OXA-181 | 1 | + | 60 | + | 60 | ≥16 | |
| OXA-181 | 1 | − | NA | − | NA | 4 | |
| Serratia marcescens | IMP | 9 | + | 5–10 | + | 5–10 | 4 to ≥16 |
| IMP-4 | 8 | + | 5–10 | + | 5–15 | 8 to ≥16 | |
| A. baumannii | OXA-23, OXA-24, OXA-23/51 | 8 | − | NA | − | NA | ≥16 |
| Pseudomonas aeruginosa | VIM | 2 | + | 30 | + | 120 | ≥16 |
+, positive result; −, negative result; NA, not applicable.
FIG 2.
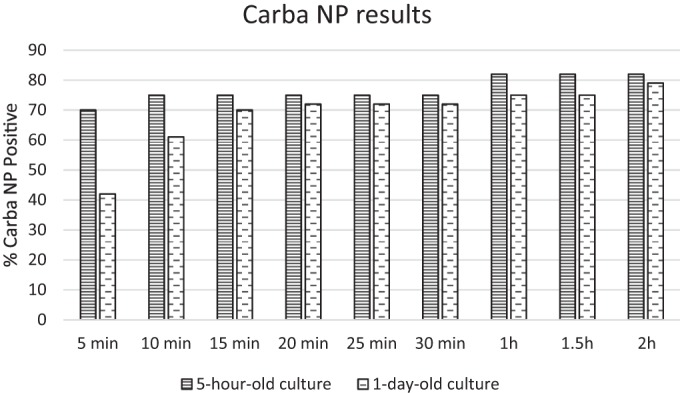
CarbaNP results for carbapenemase PCR-positive organisms from 5-hour-old and 1-day-old cultures using horse blood agar.
We also found that the sensitivity of the CarbaNP test varied when isolates were taken from different media, confirming previous reports (19). From 1-day-old cultures, only 34/58 isolates were CarbaNP positive from MacConkey agar (sensitivity, 59%; specificity, 100%; P < 0.0001), 45/58 were positive from HBA, and 47/58 were positive from ChromID medium (sensitivity, 81%; specificity, 100%; P < 0.0001). From 5-hour-old cultures, 47/58 isolates were CarbaNP positive from both ChromID medium and HBA. No differences in the CarbaNP test results were observed for plates that had been incubated at 35°C in air versus 5% CO2. Final CarbaNP results after 2 h for the plates stored for 2 to 3 weeks were the same as the results for 1-day-old colonies from the respective media. It should be noted that testing from MacConkey agar is less reliable and should be avoided. We also found that halving the amount of Tris-HCl lysis buffer and reagent solutions to 50 μl did not affect the results of the CarbaNP test when we tested PCR-positive (n = 58) and PCR-negative (n = 99) isolates. Given that Tris-HCl lysis buffer is the most expensive component of the reagents, this is an important finding that is particularly relevant for microbiology laboratories in developing countries, where the CarbaNP test would be especially useful.
This study suggests that the CarbaNP test performs well during the growth phase of Enterobacteriaceae strains. A possible explanation for this finding involves altered carbapenemase gene expression during this time. Differences in inocula represent another possible explanation for differences in the performance of the CarbaNP test with 5-hour-old cultures versus 1-day-old cultures, as suggested in a recent report (20). We note that the original authors have improved the CarbaNP test by using a smaller amount of bacteria (one-fourth to one-third of a 10-μl loop) (20). A limitation of this study is that differences in both culture age and methods of inoculum preparation make it difficult to explain instances in which results differed for the two CarbaNP test methods.
We demonstrated the high sensitivity of the 5-hour CarbaNP test across a range of Enterobacteriaceae species producing a number of different carbapenemases. The low CPO prevalence in Australia results in a limited representation of certain CPO types. Additional studies are required to confirm our findings for important CPO types prevalent in other countries.
A strength of our study is that it includes many carbapenemase-negative but non-carbapenem-susceptible isolates, confirming the high specificity of the CarbaNP test (100%) in a low-prevalence setting. In contrast, for our isolates the specificity of the MHT was 49% and specificity of ChromID medium was 39%, in comparison with PCR testing.
Our study confirms the findings of previous studies that showed that OXA producers are more difficult to detect with the CarbaNP test (9). Other studies did not test OXA-23-like/OXA-51-like, OXA 23, and OXA 24 Acinetobacter baumannii complex organisms, all of which failed to yield positive results in this study. Recently, the CarbAcineto NP test, utilizing sodium chloride, has been developed to better identify carbapenemases in these organisms (21).
In this study, adapting the CarbaNP test for use with 5-hour-old cultures improved the turnaround time for results, in comparison with traditional 1-day-old cultures, and maintained high test sensitivity. Utilizing younger cultures allows incorporation of the test into a single day's workflow, facilitating timely patient care (by guiding antimicrobial therapy), antimicrobial stewardship, and infection control. Rapid identification is possible using matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI–TOF MS), and future efforts may involve reevaluating existing susceptibility methods to determine whether they are reliable when performed with younger cultures, to further improve turnaround time.
ACKNOWLEDGMENTS
We thank Gillian Wood, Austin Hospital Microbiology Department, and Chris Lee and Adam Jenney, Alfred Hospital Microbiology Department, for additional isolates. We also thank the Melbourne Diagnostic Unit, University of Melbourne, and Jan Bell at SA Pathology for molecular diagnostics.
We declare no conflicts of interest.
Footnotes
Published ahead of print 20 August 2014
REFERENCES
- 1.Nordmann P, Naas T, Poirel L. 2011. Global spread of carbapenemase producing Enterobacteriaceae. Emerg. Infect. Dis. 17:1791–1798. 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Australian Group on Antimicrobial Resistance. 2011. Gram-negative survey: 2011 antimicrobial susceptibility report. Australian Group on Antimicrobial Resistance, Perth, Australia: http://www.agargroup.org/files/AGAR%20GNB11%20Report%20FINAL.pdf. [Google Scholar]
- 3.Australian Group on Antimicrobial Resistance. 2012. Gram-negative survey: 2012 antimicrobial susceptibility report. Australian Group on Antimicrobial Resistance, Perth, Australia: http://www.agargroup.org/files/AGAR%20GNB12%20Report%20FINAL.pdf. [Google Scholar]
- 4.Espedido B, Partridge S, Iredell J. 2008. blaIMP-4 in different genetic contexts in Enterobacteriaceae isolates from Australia. Antimicrob. Agents Chemother. 52:2984–2987. 10.1128/AAC.01634-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nordmann P, Poirel L. 2013. Strategies for identification of carbapenemase-producing Enterobacteriaceae. J. Antimicrob. Chemother. 68:487–489. 10.1093/jac/dks426. [DOI] [PubMed] [Google Scholar]
- 6.Kruse E, Aurbach U, Wisplinghoff H. 2013. Carbapenem-resistant Enterobacteriaceae: laboratory detection and infection control practices. Curr. Infect. Dis. Rep. 15:549–558. 10.1007/s11908-013-0373-x. [DOI] [PubMed] [Google Scholar]
- 7.Nordmann P, Poirel L, Dortet L. 2012. Rapid detection of carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 18:1503–1507. 10.3201/eid1809.120355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dortet L, Poirel L, Nordmann P. 2012. Rapid detection of carbapenemase-producing Pseudomonas spp. J. Clin. Microbiol. 50:3773–3776. 10.1128/JCM.01597-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tijet N, Boyd D, Patela S, Mulveyb M, Melanoa R. 2013. Evaluation of the CarbaNP test for rapid detection of carbapenemase-producing Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 57:4578–4580. 10.1128/AAC.00878-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang T, Berhin C, Bogaerts P, Glupczynski Y. 21 May 2014. Comparative evaluation of two chromogenic tests for rapid detection of carbapenemase in Enterobacteriaceae and in Pseudomonas aeruginosa isolates. J. Clin. Microbiol. 10.1128/JCM.00643-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vasoo S, Cunningham S, Kohner P, Simner P, Mandrekar J, Lolans K, Hayden M, Patel R. 2013. Comparison of a novel, rapid chromogenic biochemical assay, the CarbaNP test, with the modified Hodge test for detection of carbapenemase-producing Gram-negative bacilli. J. Clin. Microbiol. 51:3097–3101. 10.1128/JCM.00965-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clinical and Laboratory Standards Institute. 2013. Performance standards for antimicrobial susceptibility testing; 23rd informational supplement. Document M100-S23. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 13.Endimiani A, Carias LL, Hujer AM, Bethel CR, Hujer KM, Perez F, Hutton RA, Fox WR, Hall GS, Jacobs MR, Paterson DL, Rice LB, Jenkins SG, Tenover FC, Bonomo RA. 2008. Presence of plasmid-mediated quinolone resistance in Klebsiella pneumoniae isolates possessing blaKPC in the United States. Antimicrob. Agents Chemother. 52:2680–2682. 10.1128/AAC.00158-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poirel L, Héritier C, Tolün V, Nordmann P. 2004. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 48:15–22. 10.1128/AAC.48.1.15-22.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pitout JD, Gregson DB, Poirel L, McClure J-A, Le P, Church DL. 2005. Detection of Pseudomonas aeruginosa producing metallo-β-lactamases in a large centralized laboratory. J. Clin. Microbiol. 43:3129–3135. 10.1128/JCM.43.7.3129-3135.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mushtaq S, Irfan S, Sarma JB, Doumith M, Pike R, Pitout J, Livermore DM, Woodford N. 2011. Phylogenetic diversity of Escherichia coli strains producing NDM-type carbapenemases. J. Antimicrob. Chemother. 66:2002–2005. 10.1093/jac/dkr226. [DOI] [PubMed] [Google Scholar]
- 17.Woodford N, Ellington MJ, Coelho JM, Turton JF, Ward ME, Brown S, Amyes SG, Livermore DM. 2006. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int. J. Antimicrob. Agents 27:351–353. 10.1016/j.ijantimicag.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 18.Dortet L, Poirel L, Nordmann P. 2012. Rapid identification of carbapenemase types in Enterobacteriaceae and Pseudomonas spp. by using a biochemical test. Antimicrob. Agents Chemother. 56:6437–6440. 10.1128/AAC.01395-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dortet L, Brechard L, Poirel L, Nordmann P. 2014. Impact of the isolation medium for detection of carbapenemase-producing Enterobacteriaceae using an updated version of the CarbaNP test. J. Med. Microbiol. 63:772–776. 10.1099/jmm.0.071340-0. [DOI] [PubMed] [Google Scholar]
- 20.Dortet L, Poirel L, Nordmann P. 2014. Further proof of concept for the CarbaNP test. Antimicrob. Agents Chemother. 58:1269. 10.1128/AAC.01825-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dortet L, Poirel L, Nordmann P. 2013. Rapid detection of carbapenemase-producing Acinetobacter spp., poster D-599. Abstr. Intersci. Conf. Antimicrob. Agents Chemother. 2013. http://www.icaaconline.com/php/icaac2013abstracts/data/papers/2013/D/2013_D-599.htm. [Google Scholar]