Abstract
Cytomegalovirus resistance to antivirals is a major problem in transplant recipients. We evaluated the impact of five mutations (A594V, L595F, and E655K in the UL97 gene and V526L and E756K in the UL54 gene), detected in a blood sample from a stem cell transplant recipient, on drug susceptibilities and replicative capacities of recombinant viruses.
TEXT
Cytomegalovirus (CMV) infection remains a leading cause of morbidity and mortality in hematopoietic stem cell transplant (HSCT) recipients (1). Three antiviral agents (ganciclovir [GCV], foscarnet [FOS], and cidofovir [CDV]) are approved for the treatment of CMV diseases (2). Prophylaxis and preemptive treatment with GCV may result in the emergence of drug resistance (3). Genotypic testing, which can be achieved directly in blood samples, is increasingly being used for the diagnosis of drug resistance mutations in UL97 phosphotransferase and UL54 DNA polymerase genes. However, the detection of newly emerging mutations not previously linked to drug resistance requires their transfer into recombinant viruses to assess drug phenotypes (4). Resistance mutations are detected most often in the UL97 gene and confer resistance to GCV alone (5). In general, UL54 mutations emerge after prolonged GCV exposure and thereby increase the level of GCV resistance conferred by mutations already present in the UL97 gene (6–9). Infections with multidrug-resistant CMV isolates that contain several mutations in the UL97 and/or UL54 genes were reported in immunocompromised patients (10–12). Such CMV infections may be associated with a single isolate that harbors multiple mutations in the UL97 and/or UL54 genes or may result from mixed virus subpopulations that harbor different mutations (6). Three mutations in the UL97 gene (A594V, L595F, and E655K) and two mutations in the UL54 gene (V526L and E756K) were detected in a blood sample from an HSCT recipient who was not responding to GCV therapy. In this report, we characterize the role of the unknown E655K and V526L mutations and analyze the impact of the five mutations introduced alone or in different combinations on the drug resistance phenotypes and replicative capacities of recombinant viruses.
A 4-year-old child was diagnosed with acute lymphoblastic leukemia (ALL) for which she received chemotherapy in August 2001. She experienced a relapse of ALL in December 2004 and later received an allogeneic cord blood transplant (CMV-seropositive recipient, CMV-seronegative donor) on 27 February 2005. The child did not receive anti-CMV prophylaxis other than intravenous (i.v.) acyclovir from 3 days before to 20 days after the transplant. CMV reactivation in her blood was first evidenced by a qualitative nucleic acid sequence-based amplification (NASBA) assay (Organon Teknika) on 7 April 2005. Intravenous GCV was administered at a dose of 5 mg/kg of body weight twice daily (BID) from 12 April to 17 June 2005, at which time the CMV viral load (as measured by the Cobas Amplicor Monitor test [Roche Diagnostics]) was negative (i.e., <600 copies/ml). The NASBA and Amplicor CMV Monitor assays rapidly became positive in July 2005, and i.v. GCV was resumed from 20 July to 15 September 2005. The patient's viral load remained positive on therapy, which prompted a switch to FOS therapy along with CMV hyperimmunoglobulins (Cytogam; CSL Behring) on 16 September 2005 and a request for a genotypic assay on 19 September 2005 (after a total of 120 days of GCV and 3 days of FOS). A total of five mutations, three (A594V, L595F, and E655K) in the UL97 gene and two (V526L and E756K) in the UL54 gene, were detected by Sanger sequencing on a single clinical sample. In the absence of a viral isolate and further cloning analysis, we were unable to determine whether these mutations were present in a single virus population or in a number of distinct virus subpopulations. The patient's viral load subsequently became undetectable after 4 October 2005. During that period, the patient had suspected CMV pneumonitis based on diffuse interstitial lung infiltrates in a computed tomography (CT) scan performed on 21 April 2005. However, no bronchoalveolar lavage was performed to confirm this diagnosis. The patient was still alive as of August 2014.
Human lung fibroblasts (MRC-5; ATCC CCL-171 cell line) and human foreskin fibroblasts (HFFs) were maintained in minimal essential medium (MEM) (Gibco/Invitrogen) supplemented with 10% fetal bovine serum (FBS) (Gibco/Invitrogen). We used a bacterial artificial chromosome (BAC) plasmid containing the wild-type CMV genome derived from the laboratory strain AD169, i.e., pHB5 (13), in which we integrated the Gaussia luciferase (GLuc) reporter gene under the control of the CMV major immediate early promoter, i.e., pHB5-GLuc (14). The different mutations were introduced alone or sequentially in the UL97 and/or UL54 gene of pHB5-GLuc by en passant mutagenesis, as previously described (14, 15). MRC-5 cells were transfected with wild-type and mutant pHB5-GLuc by electroporation, and the reconstituted recombinant viruses were collected after 2 to 3 weeks. The introduction of the desired mutation(s) was confirmed by sequencing the entire UL97 and UL54 genes of mutated pHB5-GLuc and reconstituted viruses.
Testing of the wild-type and mutant recombinant virus susceptibilities to antiviral agents was performed in HFFs by the GLuc yield reduction assay (14). Briefly, newly confluent cells were infected with different recombinant viruses at a multiplicity of infection (MOI) of 0.002 to 0.004 in MEM plus 2% FBS. Infected cells were incubated with increasing concentrations of GCV (0 to 800 μM), FOS (0 to 2,000 μM), or CDV (0 to 40 μM) (all from Sigma-Aldrich) in MEM plus 2% FBS. The 50% effective concentration (EC50) that reduced GLuc activity by 50% was determined in each cell culture supernatant by using the coelenterazine substrate (14). Drug resistance was defined by an EC50 that was ≥3-fold higher than that for the wild type, whereas a ratio between 2 and 3 was considered to indicate a borderline level of resistance.
The replicative capacities of wild-type and mutant recombinant viruses were determined in HFFs by the GLuc reporter-based assay (14). Newly confluent HFFs were infected with the different recombinant viruses at an MOI of 0.002, and the GLuc activities were assayed in cell culture supernatants that were collected daily for 7 days.
We analyzed the impact on drug susceptibility phenotypes of the five mutations detected in this patient after they were introduced alone or in different combinations in the UL97 and/or UL54 genes of recombinant viruses. As previously reported (16), the A594V and L595F mutations in the UL97 gene conferred 6.4-fold and 11.4-fold increases, respectively, in the GCV EC50s over that for the wild type (Table 1). The unknown E655K mutation, localized outside UL97 regions usually associated with GCV resistance (4), corresponded to polymorphism (1.7-fold increase in the EC50). The E756K mutation in the UL54 gene conferred resistance to GCV, FOS, and CDV (8.1-, 8.0-, and 2.7-fold increases in the EC50s, respectively), as previously reported (6). The unknown V526L mutation in the UL54 gene conferred resistance to GCV (5.5-fold increase) and a borderline level of resistance to CDV (2.5-fold increase) but did not alter the susceptibility to FOS (1.8-fold increase). Mutations localized in the δ-region C/ExoIII conserved region of the DNA polymerase typically confer various levels of resistance to GCV and CDV but not to FOS (4). The combinations of A594V with E756K or V526L were additive for resistance to GCV (12.7- or 13.2-fold increases, respectively), whereas the levels of resistance to FOS and CDV remained almost similar to that conferred by E756K or V526L mutation alone. Similarly, the GCV EC50 was additive (18.9-fold increase) for the L595F and V526L double-mutant recombinant virus compared to single mutants, without marked changes for susceptibilities to FOS and CDV. A combination of the V526L and E756K mutations resulted in a significant increase of the GCV EC50 (>25.0-fold increase; P < 0.001), without notable effects for FOS and CDV susceptibilities. The sequential introduction of the three UL97 mutations (E655K, L595F, and A594V) to the V526L-E756K UL54 double mutant resulted in viable recombinant viruses with high-level GCV resistance.
TABLE 1.
Susceptibilities of wild-type and mutant recombinant viruses to antivirals by the Gaussia luciferase (GLuc) yield reduction assay
| Mutation(s) (gene) | Susceptibility data |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ganciclovir |
Foscarnet |
Cidofovir |
|||||||
| EC50a (mean ± SD) (μM) | Fold increaseb | No. of determinations | EC50 (mean ± SD) (μM) | Fold increase | No. of determinations | EC50 (mean ± SD) (μM) | Fold increase | No. of determinations | |
| Wild type | 4.09 ± 1.89 | 1.0 | 11 | 42.63 ± 14.44 | 1.0 | 8 | 0.38 ± 0.17 | 1.0 | 8 |
| A594V (UL97) | 26.19 ± 10.59 | 6.4 | 3 | NDc | ND | ||||
| L595F (UL97) | 46.51 ± 5.28 | 11.4 | 3 | ND | ND | ||||
| E655K (UL97) | 7.02 ± 2.29 | 1.7 | 5 | ND | ND | ||||
| E756K (UL54) | 33.23 ± 8.36 | 8.1 | 3 | 340.20 ± 81.25 | 8.0 | 3 | 1.03 ± 0.34 | 2.7 | 3 |
| V526L (UL54) | 22.41 ± 11.34 | 5.5 | 4 | 77.38 ± 29.22 | 1.8 | 5 | 0.97 ± 0.25 | 2.5 | 3 |
| E756K (UL54) and A594V (UL97) | 51.70 ± 22.38 | 12.7 | 4 | 227.30 ± 51.65 | 5.3 | 3 | 1.10 ± 0.43 | 2.9 | 3 |
| V526L (UL54) and A594V (UL97) | 54.44 ± 13.07 | 13.2 | 4 | 70.52 ± 7.39 | 1.7 | 3 | 1.02 ± 0.42 | 2.7 | 4 |
| V526L (UL54) and L595F (UL97) | 77.97 ± 13.51 | 18.9 | 3 | 81.27 ± 4.88 | 1.9 | 3 | 1.35 ± 0.32 | 3.5 | 3 |
| V526L-E756K (UL54) | >100.00d | >25.0 | 3 | 215.80 ± 67.12 | 5.1 | 3 | 1.24 ± 0.23 | 3.2 | 3 |
| V526L-E756K (UL54) and E655K (UL97) | >100.00d | >25.0 | 3 | 266.70 ± 59.31 | 6.3 | 3 | 1.61 ± 0.15 | 4.2 | 4 |
| V526L-E756K (UL54) and E655K-L595F (UL97) | >100.00d | >25.0 | 3 | 202.40 ± 25.69 | 4.8 | 3 | 1.68 ± 0.58 | 4.4 | 4 |
| V526L-E756K (UL54) and E655K-L595F-A594V (UL97) | >100.00d | >25.0 | 3 | 227.53 ± 137.62 | 5.3 | 3 | 1.01 ± 0.31 | 2.7 | 3 |
EC50 is the effective concentration of antiviral drug that reduces GLuc activity by 50%. Results are the mean ± SD of n determinations.
Fold increase over that for the wild-type recombinant virus.
ND, not determined.
P < 0.001 compared with each single mutant recombinant virus by using a one-way analysis of variance (ANOVA) (GraphPad Prism version 5.00).
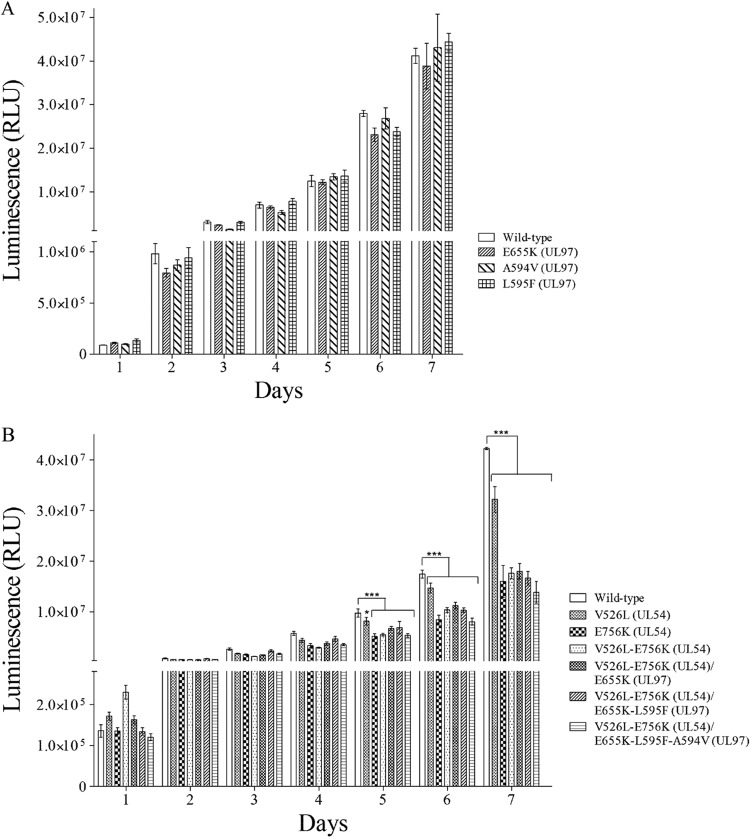
The viral growth of UL97 mutant recombinant viruses was similar to that of the wild-type virus (Fig. 1A). As previously reported (6, 17), the E756K mutation in the UL54 gene caused a significant decrease in the replicative capacity compared with that of the wild type (0.25-log reduction on day 7 postinfection; P < 0.001) (Fig. 1B). The unknown V526L mutation conferred a slight replicative defect that was more evident on day 7 (0.13-log reduction; P < 0.001) postinfection. The replicative capacity of the V526L-E756K double mutant was affected to an extent similar to that of the E756K single mutant, whereas the sequential addition of UL97 mutations had no effect.
FIG 1.
Replicative capacities of wild-type and recombinant viruses harboring mutations in the UL97 (A) and UL54 and/or UL97 (B) genes determined by the Gaussia luciferase (GLuc) reporter-based assay over time. Human foreskin fibroblasts were infected with the different recombinant viruses at a multiplicity of infection of 0.002, and the GLuc activities were measured in cell culture supernatants sampled daily for 7 days. Results represent the means ± standard deviation of sextuplicate determinations of one representative experiment. RLU, relative light unit. *, P < 0.05, and ***, P < 0.001, compared with wild type by using a two-way analysis of variance (ANOVA) test (GraphPad Prism version 5.00).
In conclusion, the emergence of multiple UL97 and/or UL54 mutations might result in a multidrug-resistant CMV variant with an extremely high GCV resistance level associated with a slightly reduced in vitro replicative capacity. Our results highlight the need to develop new antiviral strategies that target other viral proteins (18).
ACKNOWLEDGMENTS
This study was supported by Canadian Institutes of Health Research grant MOP-86583 (to G.B.).
G.B. is the holder of the Canada research chair on emerging viruses and antiviral resistance.
Footnotes
Published ahead of print 20 August 2014
REFERENCES
- 1.Sedky M, Mekki Y, Mialou V, Bleyzac N, Girard S, Salama E, Abdel Rahman H, Bertrand Y. 5 December 2013. Cytomegalovirus infection in pediatric allogenic hematopoietic stem cell transplantation. A single center experience. Pediatr. Hematol. Oncol. 10.3109/08880018.2013.859188. [DOI] [PubMed] [Google Scholar]
- 2.Andrei G, De Clercq E, Snoeck R. 2009. Drug targets in cytomegalovirus infection. Infect. Disord. Drug Targets 9:201–222. 10.2174/187152609787847758. [DOI] [PubMed] [Google Scholar]
- 3.Boeckh M. 2011. Complications, diagnosis, management, and prevention of CMV infections: current and future. Hematology 2011:305–309. 10.1182/asheducation-2011.1.305. [DOI] [PubMed] [Google Scholar]
- 4.Lurain NS, Chou S. 2010. Antiviral drug resistance of human cytomegalovirus. Clin. Microbiol. Rev. 23:689–712. 10.1128/CMR.00009-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Erice A. 1999. Resistance of human cytomegalovirus to antiviral drugs. Clin. Microbiol. Rev. 12:286–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chou S, Lurain NS, Thompson KD, Miner RC, Drew WL. 2003. Viral DNA polymerase mutations associated with drug resistance in human cytomegalovirus. J. Infect. Dis. 188:32–39. 10.1086/375743. [DOI] [PubMed] [Google Scholar]
- 7.Chou S, Van Wechel LC, Lichy HM, Marousek GI. 2005. Phenotyping of cytomegalovirus drug resistance mutations by using recombinant viruses incorporating a reporter gene. Antimicrob. Agents Chemother. 49:2710–2715. 10.1128/AAC.49.7.2710-2715.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chou S, Marousek GI, Van Wechel LC, Li S, Weinberg A. 2007. Growth and drug resistance phenotypes resulting from cytomegalovirus DNA polymerase region III mutations observed in clinical specimens. Antimicrob. Agents Chemother. 51:4160–4162. 10.1128/AAC.00736-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scott GM, Weinberg A, Rawlinson WD, Chou S. 2007. Multidrug resistance conferred by novel DNA polymerase mutations in human cytomegalovirus isolates. Antimicrob. Agents Chemother. 51:89–94. 10.1128/AAC.00633-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blackman SC, Lurain NS, Witte DP, Filipovich AH, Groen P, Schleiss MR. 2004. Emergence and compartmentalization of fatal multi-drug-resistant cytomegalovirus infection in a patient with autosomal-recessive severe combined immune deficiency. J. Pediatr. Hematol. Oncol. 26:601–605. 10.1097/01.mph.0000135283.77668.6a. [DOI] [PubMed] [Google Scholar]
- 11.Levi ME, Mandava N, Chan LK, Weinberg A, Olson JL. 2006. Treatment of multidrug-resistant cytomegalovirus retinitis with systemically administered leflunomide. Transpl. Infect. Dis. 8:38–43. 10.1111/j.1399-3062.2006.00128.x. [DOI] [PubMed] [Google Scholar]
- 12.Germi R, Mariette C, Alain S, Lupo J, Thiebaut A, Brion JP, Epaulard O, Saint Raymond C, Malvezzi P, Morand P. 2014. Success and failure of artesunate treatment in five transplant recipients with disease caused by drug-resistant cytomegalovirus. Antiviral Res. 101:57–61. 10.1016/j.antiviral.2013.10.014. [DOI] [PubMed] [Google Scholar]
- 13.Borst EM, Hahn G, Koszinowski UH, Messerle M. 1999. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. J. Virol. 73:8320–8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drouot E, Piret J, Boivin G. 2013. Novel method based on “en passant” mutagenesis coupled with a Gaussia luciferase reporter assay for studying the combined effects of human cytomegalovirus mutations. J. Clin. Microbiol. 51:3216–3224. 10.1128/JCM.01275-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tischer BK, Smith GA, Osterrieder N. 2010. En passant mutagenesis: a two step markerless red recombination system. Methods Mol. Biol. 634:421–430. 10.1007/978-1-60761-652-8_30 20677001. [DOI] [PubMed] [Google Scholar]
- 16.Chou S, Waldemer RH, Senters AE, Michels KS, Kemble GW, Miner RC, Drew WL. 2002. Cytomegalovirus UL97 phosphotransferase mutations that affect susceptibility to ganciclovir. J. Infect. Dis. 185:162–169. 10.1086/338362. [DOI] [PubMed] [Google Scholar]
- 17.Chevillotte M, Schubert A, Mertens T, von Einem J. 2009. Fluorescence-based assay for phenotypic characterization of human cytomegalovirus polymerase mutations regarding drug susceptibility and viral replicative fitness. Antimicrob. Agents Chemother. 53:3752–3761. 10.1128/AAC.00165-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chemaly RF, Ullmann AJ, Stoelben S, Richard MP, Bornhauser M, Groth C, Einsele H, Silverman M, Mullane KM, Brown J, Nowak H, Kolling K, Stobernack HP, Lischka P, Zimmermann H, Rubsamen-Schaeff H, Champlin RE, Ehninger G, AIC246 Study Team 2014. Letermovir for cytomegalovirus prophylaxis in hematopoietic-cell transplantation. N. Engl. J. Med. 370:1781–1789. 10.1056/NEJMoa1309533. [DOI] [PubMed] [Google Scholar]