Abstract
Fifty-two multidrug-resistant isolates of Mycobacterium tuberculosis representative of the currently predominant lineages in France were analyzed using repetitive-sequence-based PCR (rep-PCR) DiversiLab (DL), spoligotyping, 24-locus mycobacterial interspersed repetitive-unit–variable-number tandem-repeat typing (MIRU-VNTR), and restriction fragment length polymorphism of IS6110 (IS6110-RFLP). DL, as opposed to MIRU-VNTR and IS6110-RFLP analysis, did not allow discrimination among half of the isolates, an indication of comparatively lower resolving power.
TEXT
Multidrug-resistant (MDR) tuberculosis (TB) is a serious health threat that requires molecular procedures yielding results quickly to improve control of the diffusion of drug-resistant strains (1–4). Until recently, restriction fragment length polymorphism analysis of IS6110 (IS6110-RFLP) (5) was considered the gold standard for Mycobacterium tuberculosis strain typing (5, 6). Two PCR-based methods, spoligotyping and mycobacterial interspersed repetitive-unit–variable-number tandem-repeat typing (MIRU-VNTR), which can be performed with very small quantities of crude DNA, have gradually supplanted RFLP. The signatures revealed by spoligotyping identify strains at the clade or subclade level (7, 8, 9), but a major limitation of this method is its inferior discriminatory power compared with that of IS6110-RFLP and complete 24-locus MIRU-VNTR (10). On the other hand, 24-locus MIRU-VNTR is fairly rapid and generates numerical values that can easily be compared in interlaboratory studies. Because of particular advantages over IS6110-RFLP and spoligotyping at the technical level and in discriminatory power, MIRU-VNTR is considered to be the new reference standard for molecular epidemiological studies (11).
Repetitive-sequence-based PCR (rep-PCR) is a rapid typing procedure using primers that bind to multiple noncoding repetitive sequences interspersed throughout the bacterial genome. The strain-specific band patterns generated by rep-PCR can be used to determine the similarity of bacterial isolates at the genomic level, as the repetitive sequences throughout the genome enable discrimination of interstrain variations on the basis of amplicon size and amount (12–17). However, rep-PCR typing is notorious for its susceptibility to minor variations in experimental conditions and reagents, resulting in poor reproducibility. The DiversiLab (DL) microbial typing system (bioMérieux, Marcy l'Etoile, France) (18) consists of a semiautomated highly standardized rep-PCR (18, 19). Studies reporting on DL performance in the analysis of mycobacteria are scarce and have been done with specific aims, e.g., rapid genotyping of nontuberculosis mycobacteria (20–24), analysis of M. tuberculosis microevolution within a patient (25), monitoring of TB outbreaks (1, 3, 26), and comparison of DL to other molecular techniques for M. tuberculosis typing (1, 19, 27, 28).
In this study, we provide a comprehensive evaluation of DL by comparing its ability to discriminate among MDR TB isolates for which epidemiological data are available.
Fifty-two M. tuberculosis stricto sensu clinical isolates and the H37Rv reference strain (French National Reference Center for Mycobacteria) were analyzed. All 52 were MDR isolates previously characterized by spoligotyping, 24-locus MIRU-VNTR, and IS6110-RFLP, including three East African-Indian (EAI) strains (one EAI3-IND and two EAI2-PHL) and six Beijing, five Haarlem (three H1 and two H3), six Latin American-Mediterranean LAM9, four URAL, three Cameroon (CAM), six S, and 19 T-related (seven T1, six T2, and six T2-T3) strains. The Beijing, Haarlem, LAM, and T families represent approximately 80% of the MDR strains circulating in France (10). The available epidemiological data are summarized in Table 1. For spoligotyping, MIRU-VNTR, and rep-PCR typing, DNA was extracted from a loopful (ca. 10 μl) of colonies grown on Lowenstein-Jensen agar, using the UltraClean microbial DNA isolation kit (Mo Bio Laboratories, Solana Beach, CA) according to the manufacturer's recommendations. The procedure and interpretation of IS6110-RFLP analysis were performed according to the standardized protocol recommended by van Embden et al. (6). RFLP patterns were compared and analyzed using the computerized GelCompar software system (Applied Maths, Sint-Martens-Latem, Belgium) with the unweighted-pair group method using average linkages (UPGMA). Spoligotyping was performed as described by Abadia et al., with a Luminex microbead-based flow cytometry device (29). To determine the lineages of the 52 isolates, spoligotypes in binary format were converted to an octal code for comparison with the M. tuberculosis SpolDB4 database containing all spoligotype international types (SIT) (8). Standard 24-locus MIRU-VNTR typing was performed as previously described (11) with the MIRU-VNTR typing kit of GenoScreen. The 24 numerical values generated by MIRU-VNTR were compared with those existing in the MIRU-VNTRplus database (http://www.miru-vntrplus.org). Finally, DL was performed following the manufacturer's instructions. DNA fingerprint patterns were analyzed with the Web-based DiversiLab software, version 3.4, which uses the Pearson correlation coefficient and UPGMA for automatic comparison of the rep-PCR-based DNA fingerprints (18, 19). A percentage similarity set at ≥93% was used as the threshold in the cluster analysis (19).
TABLE 1.
Epidemiological information concerning the isolates for which links were suggested by the molecular analysis
| Lineage | Isolate no. | SIT no. | Epidemiological information | Country of birth | Yr of isolation |
|---|---|---|---|---|---|
| LAM9 | S15 | 1106 | Household contacts with S16 and S17 | France | 2007 |
| S16 | 1106 | Same family as S17 | Portugal | 2007 | |
| S17 | 1106 | Same family as S16 | France | 2007 | |
| S18 | 1106 | NLa | Portugal | 2011 | |
| T1 | S38 | 53 | Same family as S40 | Guinea | 2006 |
| S39 | 53 | NL | Guinea | 2006 | |
| S40 | 53 | Same family as S38 | Guinea | 2007 | |
| URAL | S12 | 262 | NL | Romania | 2006 |
| S13 | 262 | NL | Romania | 2008 | |
| S14 | 262 | NL | Romania | 2006 | |
| T2 | S35 | 52 | NL | DR Congob | 2006 |
| S36 | 712 | NL | DR Congo | 2006 | |
| S37 | 712 | NL | DR Congo | 2006 | |
| T1-Ghana | S41 | 53 | NL but traveled to Africa | France | 2009 |
| S42 | 53 | NL | Ivory Coast | 2006 | |
| S43 | 53 | NL | Ivory Coast | 2006 | |
| S | S29 | 466 | NL | Portugal | 2008 |
| S30 | 466 | NL | Algeria | 2007 | |
| S31 | 34 | NL | France | 2006 | |
| S32 | 34 | NL | Pakistan | 2008 | |
| S33 | 1063 | NL | Algeria | 2007 | |
| Haarlem-H3 | S22 | 50 | NL | France | 2006 |
| S23 | 50 | NL | Togo | 2006 | |
| LAM9 | S20 | 42 | NL | Armenia | 2007 |
| S21 | 42 | NL | France | 2006 | |
| Haarlem-H1 | S9 | 62 | NL | France | 2006 |
| S10 | 47 | NL | Unknown | 2006 | |
| Beijing | S1 | Beijing-like | NL | China | 2007 |
| S2 | 1 | NL | Unknown | 2008 | |
| S3 | 1 | NL | Ukraine | 2007 | |
| S4 | 1 | NL | Ukraine | 2007 | |
| S5 | 1 | NL | France | 2007 | |
| S6 | 1 | NL | Congo-Brazzaville | 2006 | |
| T2-T3 | S46 | 73 | NL | DR Congo | 2008 |
| S47 | 73 | NL | Angola | 2008 | |
| S48 | 73 | NL | DR Congo | 2007 | |
| S49 | 73 | NL | DR Congo | 2007 | |
| S50 | 73 | NL | Angola | 2006 | |
| S51 | 73 | NL | DR Congo | 2007 |
NL, not linked by contact tracing, with differences in resistance profiles and resistance gene mutations.
DR Congo, Democratic Republic of the Congo.
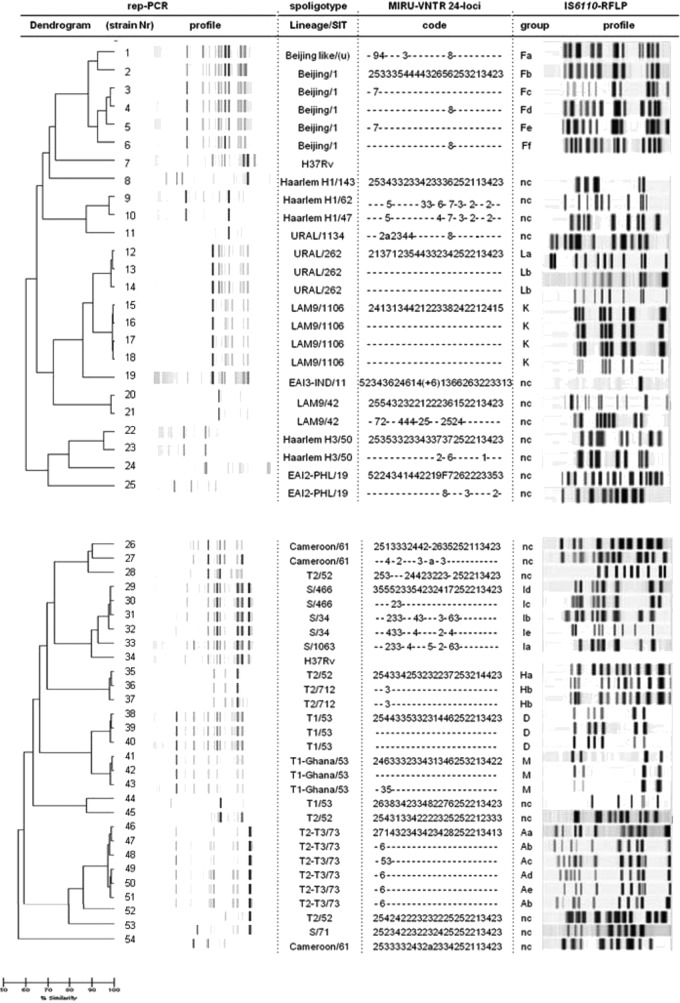
Globally, based on a similarity cutoff of 93%, the results obtained with DL were in full agreement with those generated by the other typing methods for 27 (52%) of the 52 isolates analyzed. Seven of them, belonging to two groups, with epidemiological links and sharing identical SIT numbers, MIRU codes, and RFLP profiles within each group, were correctly allocated to two subsets of high similarity levels (>98%) by DL (the four LAM9 isolates S15 to S18 and the three T1 isolates S38 to S40) (Table 1 and Fig. 1). Likewise, seven isolates with identical SIT numbers, MIRU codes, and RFLP patterns but no obvious epidemiological relationships apart from the country of origin showed similarity of >98% within each of the three corresponding DL clusters (S12 to S14 in the URAL family, S36 and S37 in the T2 family, and S41 and S42 in the T1-Ghana family) (Table 1 and Fig. 1). Finally, 13 epidemiologically unrelated isolates (not shown in Table 1) had unique SIT, MIRU-VNTR, and RFLP profiles and were clearly not linked according to the DL results, which set them well apart from the remaining isolates: S11 (URAL); S8 (H1); S19, S24, and S25 (EAI); S53 (S); S26, S27, and S54 (CAM); S44 (T1); and S28, S45, and S52 (T2) (Fig. 1).
FIG 1.
rep-PCR DiversiLab (DL), spoligotyping, 24-locus MIRU-VNTR, and IS6110-RFLP analysis of Mycobacterium tuberculosis isolates. First column, dendrogram and virtual band profiles generated using DL analysis. A scale of similarity (%) as determined using DL is shown at the bottom of the figure. Second column, spoligotyping-based lineage names and SIT numbers (u, unknown SIT). Third column, MIRU codes. Within each strain family, dashes indicate that the MIRU code does not differ from that written in full. Fourth column, RFLP cluster designation and RFLP profiles (nc, nonclustered profile).
In contrast, and comparatively to spoligotyping, 24-locus MIRU-VNTR, and IS6110-RFLP, the rep-PCR results suggested false linkages in a significant proportion (13/52; 25%) of the isolates, including five S, two H1, two Beijing, two H3, and two LAM9 strains without epidemiological links. This finding is clearly illustrated considering the five isolates of the S family (S29 to S33; Table 1), which, using DL, were all grouped into a single high-similarity cluster (97% for S29 to S32 and 93% for S33) although they displayed different SIT, MIRU, and RFLP patterns (Fig. 1). Four further isolates of the same SIT but clearly distinct MIRU and RFLP patterns were also unexpectedly grouped into two clusters, i.e., H3 isolates S22 and S23 and LAM9 isolates S20 and S21, with DL pattern similarities of 95% and 98%, respectively (Table 1 and Fig. 1). Finally, four isolates with different SIT, MIRU, and RFLP patterns appeared unexpectedly linked using DL (cluster S9 and S10 in clade H1 and cluster S1 and S2 in the Beijing family, with 97% and 93% similarity, respectively) (Table 1 and Fig. 1).
The interpretation of the DL results was problematic for 12 (23%) isolates (four Beijing, six T2-T3, one T1, one T2). With respect to the Beijing strains S3 to S6 (SIT1) (Table 1), the two epidemiologically unrelated isolates S3 and S5 were considered linked using DL (97% similarity), which is concordant with the MIRU results (identical MIRU codes) but not with the RFLP patterns, which differed from one another by three bands (Fig. 1). Conversely, isolates S4 and S6 were not linked using DL (92% similarity, i.e., below the cutoff) despite their identical MIRU codes, but in accordance with the RFLP results showing significant differences between the corresponding patterns (changes in at least three bands) (Fig. 1). More strikingly, isolates S3 and S4, which differed in their MIRU and RFLP patterns, were closely linked according to DL (98% similarity) (Fig. 1). Jang et al. (27) previously suggested that DL discriminates efficiently among Beijing family strains with near-identical IS6110-RFLP patterns, in contrast with our data which rather suggest that DL is not reliable for analysis of Beijing strains. Finally, regarding the six epidemiologically unlinked T2-T3 (SIT73) isolates S46 to S51 (Table 1), the high similarity (97%) observed within the single cluster as found with DL was not consistent with the significant differences observed in their MIRU codes and especially their RFLP patterns (Fig. 1). Similarly, using DL, isolate S43 (T1) was tightly associated with S41 and S42 despite variations in two MIRU loci, and isolate S35 (T2) was linked to S36 and S37 despite their different SIT numbers, a single-locus MIRU variation, and changes in two bands in their IS6110 fingerprints (Fig. 1). Taken together, these results suggest that DL might generate false linkages compared to MIRU-VNTR and RFLP, which have gold-standard status in molecular typing.
In conclusion, we observed here that DL did not allow discrimination in nearly half of the isolates which had been unquestionably differentiated by other techniques, indicating that rep-PCR has a lower resolving power than MIRU-VNTR and IS6110 RFLP analysis in M. tuberculosis typing. These results are in disagreement with those of Cangelosi et al. (19), who reported that the discriminatory power of rep-PCR was at least as good as that of IS6110-RFLP for M. tuberculosis, but they are in agreement with those of Masala et al. (28), who suggested that the MIRU-VNTR and IS6110-RFLP typing methods have greater discriminatory power than rep-PCR DL. In light of the clear limitations highlighted in this study, confirmation by MIRU-VNTR and/or RFLP analysis is required to substantiate MDR TB transmission when clonal relationships are suggested using DL.
ACKNOWLEDGMENTS
This work was supported by a grant from the Ministère de la Recherche (program grant UPMC ER5).
We thank Ekkehard Collatz for his critical reading of the manuscript.
We have no commercial associations that might pose a conflict of interest.
Footnotes
Published ahead of print 10 September 2014
REFERENCES
- 1.Ashworth M, Horan KL, Freeman R, Oren E, Narita M, Cangelosi GA. 2008. Use of PCR-based Mycobacterium tuberculosis genotyping to prioritize tuberculosis outbreak control activities. J. Clin. Microbiol. 46:856–862. 10.1128/JCM.01146-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brudey K, Gordon M, Moström P, Svensson L, Jonsson B, Sola C, Ridell M, Rastogi N. 2004. Molecular epidemiology of Mycobacterium tuberculosis in western Sweden. J. Clin. Microbiol. 42:3046–3051. 10.1128/JCM.42.7.3046-3051.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Freeman R, Kato-Maeda M, Hauge KA, Horan KL, Oren E, Narita M, Wallis CK, Cave D, Nolan CM, Small PM, Cangelosi GA. 2005. Use of rapid genomic deletion typing to monitor a tuberculosis outbreak within an urban homeless population. J. Clin. Microbiol. 43:5550–5554. 10.1128/JCM.43.11.5550-5554.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lari N, Rindi L, Bonanni D, Rastogi N, Sola C, Tortoli E, Garzelli C. 2007. Three-year longitudinal study of genotypes of Mycobacterium tuberculosis isolates in Tuscany, Italy. J. Clin. Microbiol. 45:1851–1857. 10.1128/JCM.00170-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McEvoy CR, Falmer AA, Gey van Pittius NC, Victor TC, van Helden PD, Warren RM. 2007. The role of IS6110 in the evolution of Mycobacterium tuberculosis. Tuberculosis 87:393–404. 10.1016/j.tube.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 6.van Embden JD, Cave MD, Crawford JT, Dale JW, Eisenach KD, Gicquel B, Hermans P, Martin C, McAdam R, Shinnick TM. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kato-Maeda M, Metcalfe JZ, Flores L. 2011. Genotyping of Mycobacterium tuberculosis: application in epidemiologic studies. Future Microbiol. 6:203–216. 10.2217/fmb.10.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brudey K, Driscoll JR, Rigouts L, Prodinger WM, Gori A, Al-Hajoj SA, Allix C, Aristimuño L, Arora J, Baumanis V, Binder L, Cafrune P, Cataldi A, Cheong S, Diel R, Ellermeier C, Evans JT, Fauville-Dufaux M, Ferdinand S, Garcia de Viedma D, Garzelli C, Gazzola L, Gomes HM, Guttierez MC, Hawkey PM, van Helden PD, Kadival GV, Kreiswirth BN, Kremer K, Kubin M, Kulkarni SP, Liens B, Lillebaek T, Ho ML, Martin C, Martin C, Mokrousov I, Narvskaïa O, Ngeow YF, Naumann L, Niemann S, Parwati I, Rahim Z, Rasolofo-Razanamparany V, Rasolonavalona T, Rossetti ML, Rüsch-Gerdes S, Sajduda A, Samper S, Shemyakin IG, Singh UB, Somoskovi A, Skuce RA, van Soolingen D, Streicher EM, Suffys PN, Tortoli E, Tracevska T, Vincent V, Victor TC, Warren RM, Yap SF, Zaman K, Portaels F, Rastogi N, Sola C. 2006. Mycobacterium tuberculosis complex genetic diversity: mining the fourth international spoligotyping database (SpolDB4) for classification, population genetics and epidemiology. BMC Microbiol. 6:23. 10.1186/1471-2180-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Embden JD, van Gorkom T, Kremer K, Jansen R, van Der Zeijst BA, Schouls LM. 2000. Genetic variation and evolutionary origin of the direct repeat locus of Mycobacterium tuberculosis complex bacteria. J. Bacteriol. 182:2393–2401. 10.1128/JB.182.9.2393-2401.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sougakoff W. 2011. Molecular epidemiology of multidrug-resistant isolates of Mycobacterium tuberculosis. Clin. Microbiol. Infect. 17:800–805. 10.1111/j.1469-0691.2011.03577.x. [DOI] [PubMed] [Google Scholar]
- 11.Supply P, Allix C, Lesjean S, Cardoso-Oelemann M, Rüsch-Gerdes S, Willery E, Savine E, de Haas P, van Deutekom H, Roring S, Bifani P, Kurepina N, Kreiswirth B, Sola C, Rastogi N, Vatin V, Gutierrez MC, Fauville M, Niemann S, Skuce R, Kremer K, Locht C, van Soolingen D. 2006. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 44:4498–4510. 10.1128/JCM.01392-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bourdon N, Lemire A, Fines-Guyon M, Auzou M, Périchon B, Courvalin P, Cattoir V, Leclercq R. 2011. Comparison of four methods, including semi-automated rep-PCR, for the typing of vancomycin-resistant Enterococcus faecium. J. Microbiol. Methods 84:74–80. 10.1016/j.mimet.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 13.Brolund A, Hæggman S, Edquist PJ, Gezelius L, Olsson-Liljequist B, Wisell KT, Giske CG. 2010. The DiversiLab system versus pulsed-field gel electrophoresis: characterisation of extended spectrum β-lactamase producing Escherichia coli and Klebsiella pneumoniae. J. Microbiol. Methods 83:224–230. 10.1016/j.mimet.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 14.Chuang YC, Wang JT, Chen ML, Chen YC. 2010. Comparison of an automated repetitive-sequence-based PCR microbial typing system with pulsed-field gel electrophoresis for molecular typing of vancomycin-resistant Enterococcus faecium. J. Clin. Microbiol. 48:2897–2901. 10.1128/JCM.00136-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fluit AC, Terlingen AM, Andriessen L, Ikawaty R, van Mansfeld R, Top J, Cohen Stuart JW, Leverstein-van Hall MA, Boel CH. 2010. Evaluation of the DiversiLab system for detection of hospital outbreaks of infections by different bacterial species. J. Clin. Microbiol. 48:3979–3989. 10.1128/JCM.01191-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ratkai C, Peixe LV, Grosso F, Freitas AR, Antunes P, Fodor E, Hajdú E, Nagy E. 2010. Successful application of the DiversiLab repetitive-sequence-based PCR typing system for confirmation of the circulation of a multiresistant Pseudomonas aeruginosa clone in different hospital wards. Diagn. Microbiol. Infect. Dis. 67:202–206. 10.1016/j.diagmicrobio.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 17.Tenover FC, Gay EA, Frye S, Eells SJ, Healy M, McGowan JE., Jr 2009. Comparison of typing results obtained for methicillin-resistant Staphylococcus aureus isolates with the DiversiLab system and pulsed-field gel electrophoresis. J. Clin. Microbiol. 47:2452–2457. 10.1128/JCM.00476-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Healy M, Huong J, Bittner T, Lising M, Frye S, Raza S, Schrock R, Manry J, Renwick A, Nieto R, Woods C, Versalovic J, Lupski JR. 2005. Microbial DNA typing by automated repetitive-sequence-based PCR. J. Clin. Microbiol. 43:199–207. 10.1128/JCM.43.1.199-207.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cangelosi GA, Freeman RJ, Lewis KN, Livingston-Rosanoff D, Shah KS, Milan SJ, Goldberg SV. 2004. Evaluation of a high-throughput repetitive-sequence-based PCR system for DNA fingerprinting of Mycobacterium tuberculosis and Mycobacterium avium complex isolates. J. Clin. Microbiol. 42:2685–2693. 10.1128/JCM.42.6.2685-2693.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Christianson S, Wolfe J, Soualhine H, Sharma MK. 2012. Comparison of repetitive-sequence-based polymerase chain reaction with random amplified polymorphic DNA analysis for rapid genotyping of nontuberculosis mycobacteria. Can. J. Microbiol. 58:953–964. 10.1139/w2012-068. [DOI] [PubMed] [Google Scholar]
- 21.Bussone G, Brossier F, Roudiere L, Bille E, Sekkal N, Charlier C, Gilquin J, Lanternier F, Lecuit M, Lortholary O, Catherinot E. 2012. Recurrent Mycobacterium avium infection after seven years of latency in a HIV-infected patient receiving efficient antiretroviral therapy. J. Infect. 64:613–617. 10.1016/j.jinf.2011.12.020. [DOI] [PubMed] [Google Scholar]
- 22.Falkinham JO., III 2010. Hospital water filters as a source of Mycobacterium avium complex. J. Med. Microbiol. 59:1198–1202. 10.1099/jmm.0.022376-0. [DOI] [PubMed] [Google Scholar]
- 23.Yanong RP, Pouder DB, Falkinham JO., III 2010. Association of mycobacteria in recirculating aquaculture systems and mycobacterial disease in fish. J. Aquat. Anim. Health 22:219–223. 10.1577/H10-009.1. [DOI] [PubMed] [Google Scholar]
- 24.Zelazny AM, Root JM, Shea YR, Colombo RE, Shamputa IC, Stock F, Conlan S, McNulty S, Brown-Elliott BA, Wallace RJ, Jr, Olivier KN, Holland SM, Sampaio EP. 2009. Cohort study of molecular identification and typing of Mycobacterium abscessus, Mycobacterium massiliense, and Mycobacterium bolletii. J. Clin. Microbiol. 47:1985–1995. 10.1128/JCM.01688-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Al-Hajoj SA, Akkerman O, Parwati I, al-Gamdi S, Rahim Z, van Soolingen D, van Ingen J, Supply P, van der Zanden AG. 2010. Microevolution of Mycobacterium tuberculosis in a tuberculosis patient. J. Clin. Microbiol. 48:3813–3816. 10.1128/JCM.00556-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tardin A, Dominicé Dao M, Ninet B, Janssens JP. 2009. Tuberculosis cluster in an immigrant community: case identification issues and a transcultural perspective. Trop. Med. Int. Health 14:995–1002. 10.1111/j.1365-3156.2009.02325.x. [DOI] [PubMed] [Google Scholar]
- 27.Jang MH, Choi GE, Shin BM, Lee SH, Kim SR, Chang CL, Kim JM. 2011. Comparison of an automated repetitive sequence-based PCR microbial typing system with IS6110-restriction fragment length polymorphism for epidemiologic investigation of clinical Mycobacterium tuberculosis isolates in Korea. Korean J. Lab. Med. 31:282–284. 10.3343/kjlm.2011.31.4.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masala S, Molicotti P, Bua A, Zumbo A, Delogu G, Sechi LA, Zanetti S. 2010. Molecular characterization of Sardinian Mycobacterium tuberculosis isolates by IS6110 restriction fragment length polymorphism, MIRU-VNTR and rep-PCR. New Microbiol. 33:155–162. [PubMed] [Google Scholar]
- 29.Abadia E, Zhang J, Ritacco V, Kremer K, Ruimy R, Rigouts L, Gomes HM, Elias AR, Fauville-Dufaux M, Stoffels K, Rasolofo-Razanamparany V, Garcia de Viedma D, Herranz M, Al-Hajoj S, Rastogi N, Garzelli C, Tortoli E, Suffys PN, van Soolingen D, Refrégier G, Sola C. 2011. The use of microbead-based spoligotyping for Mycobacterium tuberculosis complex to evaluate the quality of the conventional method: providing guidelines for quality assurance when working on membranes. BMC Infect. Dis. 11:110. 10.1186/1471-2334-11-110. [DOI] [PMC free article] [PubMed] [Google Scholar]