Abstract
We evaluated a multiplex real-time PCR and melting curve analysis assay (Real-Q NTM-ID kit; Biosewoom, Seoul, South Korea) for the identification of eight common nontuberculous mycobacterial species, using 30 type strains and 230 consecutive clinical isolates. The concordance rate of this assay with multigene sequence-based typing was 97.0% (223/230 isolates).
TEXT
Nontuberculous mycobacteria (NTM) are increasingly significant causes of many clinical infections (1, 2). Since patterns of antibiotic susceptibilities vary among different NTM species, the goal of achieving accurate and rapid identification of multiple NTM species is one of great interest (2, 3).
Conventional biochemical tests for mycobacterial identification have mostly been replaced by molecular methods such as DNA sequencing, line probe hybridization, PCR restriction fragment length polymorphism analysis (PRA), and real-time PCR. However, DNA sequencing remains a labor-intensive approach requiring expensive instrumentation (4, 5). Moreover, line probe hybridization and PRA are prone to carryover amplicon contamination resulting from postamplification procedures (6–8). In contrast to line probe hybridization or PRA, real-time PCR and melting curve analysis can be performed in closed systems without the risk of carryover amplicon contamination and can also be easily adapted to a high-throughput format (9–11).
In the present study, we evaluated the performance of the Real-Q NTM-ID kit (Real-Q assay; BioSewoom, Seoul, South Korea). The Real-Q assay is a multiplex real-time PCR method that incorporates melting curve analysis and is designed to target the internal transcribed spacer (ITS) region to detect and distinguish eight NTM species: Mycobacterium abscessus, M. avium, M. intracellulare, M. chelonae, M. fortuitum, M. gordonae, M. kansasii, and M. massiliense. We compared the performance of this assay to that of multigene sequence-based typing as a reference method.
This study was conducted at a tertiary-care hospital in Seoul, South Korea, and was approved by the Institutional Review Board of Samsung Medical Center. In total, 30 reference strains (25 mycobacterial and 5 other strains) and 230 consecutive clinical NTM isolates were genotyped (Table 1). Decontaminated samples were placed into a mycobacterial growth indicator tube (MGIT 960 system; Becton Dickinson, Sparks, MD) and also into 3% Ogawa agar (Shinyang, Seoul, South Korea) and cultured for 6 weeks.
TABLE 1.
Reference strains tested
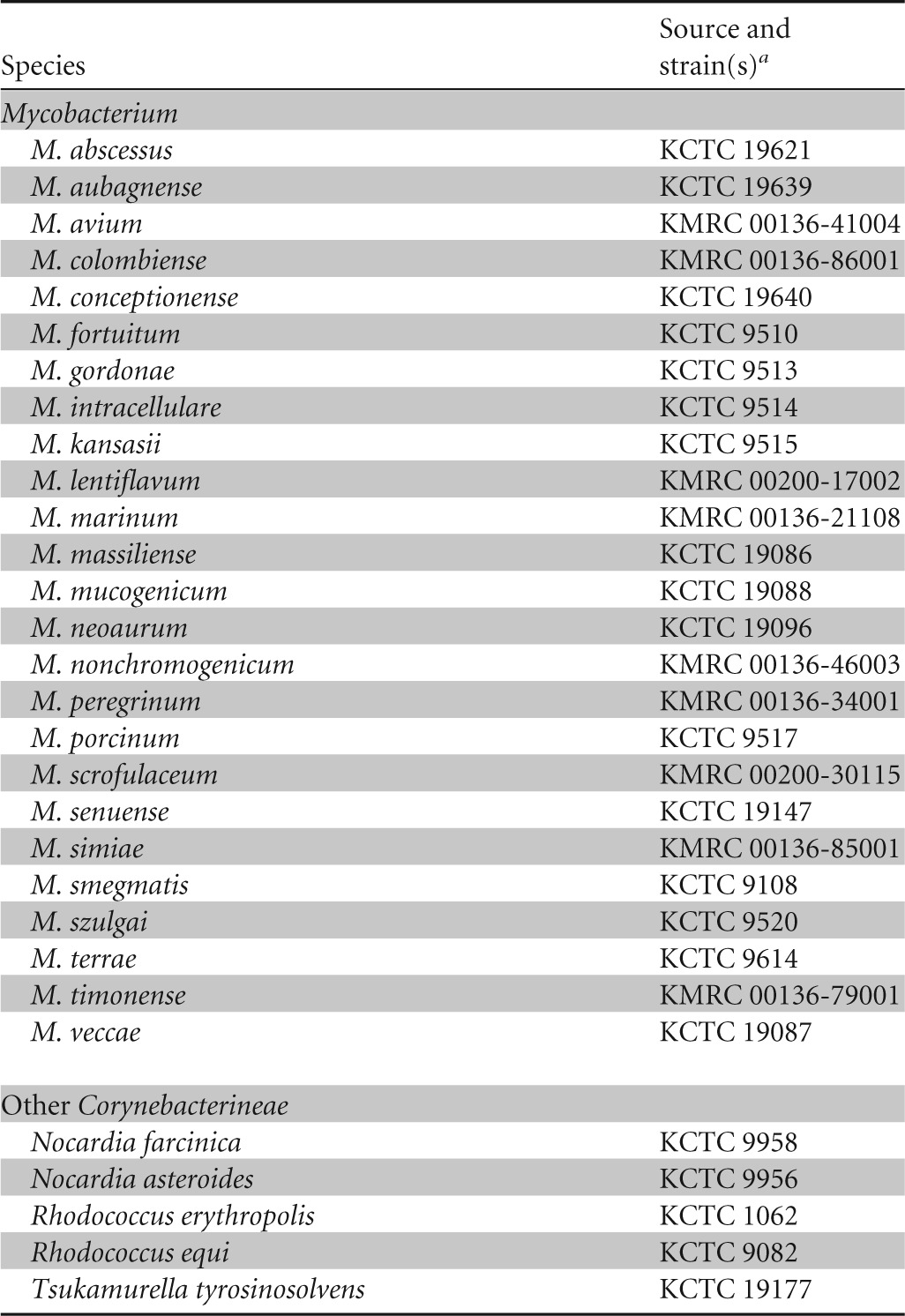
KCTC, Korean Collection for Type Cultures; KMRC, Korean Mycobacteria Resource Center.
For cultures grown on solid medium, DNA was prepared by suspending a loopful of bacteria in 100 μl DNA extraction buffer. For cultures grown in liquid MGIT culture medium (1 ml), the cultures were centrifuged at 13,000 × g for 5 min. The pellets were resuspended in 50 μl DNA extraction buffer. Both sample types were heated in a boiling water bath for 10 min. After centrifugation, the supernatants were used in the analyses.
Real-time PCRs were performed on a 7500 Fast real-time PCR system (Applied Biosystems, Foster City, CA, USA), using the Real-Q NTM-ID kit, which comprises four reaction tubes, each containing specific primer mixtures for two NTM species. Melting curve analysis using EvaGreen dye was performed after amplification (see Table S1 and Fig. S1 in the supplemental material). Each PCR was performed in a total volume of 20 μl (8 μl of the PCR mixture, 6 μl of the primer mixture, 2 μl of distilled water, 1 μl of the internal control, and 3 μl of template DNA). Thermocycling conditions included a step at 50°C for 2 min and a step at 95°C for 10 min, followed by 35 cycles of 15 s at 94°C, 30 s at 62°C, and 15 s at 72°C. The four primer sets and their melting temperatures are shown in Table S1 in the supplemental material. The melting curve was generated by heating the reaction mixtures from 60°C to 95°C after the last cycle.
The ITS region, the hsp65 gene, and the rpoB gene were sequenced according to the protocol outlined in the Clinical and Laboratory Standards Institute guideline MM18-A (12). The PCR primer sets used to amplify the ITS region, the hsp65 gene, and the rpoB gene have been previously published (8, 13, 14). The amplified sequences were analyzed using GenBank. The final sequencing result was taken as the definitive identification.
The specificities of the Real-Q assay for detecting each species were evaluated using the 30 reference strains. Only the expected PCR products were amplified, except in the case of Mycobacterium conceptionense, which produced a peak that corresponded to that of M. fortuitum.
Among the 230 isolates tested, the organism most commonly identified using the multigene sequence-based typing method was M. avium (35.2%), followed by M. intracellulare (23.9%), M. abscessus (19.1%), M. massiliense (7.8%), M. gordonae (2.2%), and other NTM species (Table 2). Approximately 92% (211/230) of the clinical isolates were mycobacterial species that the Real-Q assay could detect. Out of the 211 isolates that could be genotyped by the assay, 207 (98.1%) were correctly identified at the species level; however, two cases of M. intracellulare and two cases of M. fortuitum were incorrectly identified as out of the detection range of the assay. One strain of Mycobacterium marseillense and two strains of M. conceptionense were misidentified as M. intracellulare and M. fortuitum, respectively. In total, 223/230 strains (97.0%) yielded the expected results (Table 2). All discrepant cases were identified as a related species, defined as a species in the same complex or group. The Real-Q assay, which targets the ITS region, could not distinguish between closely related species in the M. avium complex and the M. fortuitum complex, which might be due to sequence similarities between these two complexes.
TABLE 2.
Comparison of Real-Q NTM-ID assay results with multigene sequence-based typing results
| Mycobacterial species | Total no. of isolates | No. of correct or compatiblea results obtained with Real-Q assay | Real-Q assay result for incorrectly identified isolate(s) (no. of isolates) |
|---|---|---|---|
| Detectable by Real-Q assay | |||
| M. avium complex | |||
| M. avium | 81 | 81 | |
| M. intracellulare | 55 | 53 | Out of detection range (2) |
| M. abscessus-M. chelonae complex | |||
| M. abscessus | 44 | 44 | |
| M. massiliense | 18 | 18 | |
| M. chelonae | 2 | 2 | |
| M. fortuitum complex | |||
| M. fortuitum | 3 | 1 | Out of detection range (2) |
| M. gordonae | 5 | 5 | |
| M. kansasii | 1 | 1 | |
| M. abscessus + M. intracellulare | 1 | 1 | |
| M. avium + M. massiliense | 1 | 1 | |
| Not detectable by Real-Q assay | |||
| M. avium complex | |||
| M. marseillense | 1 | 0 | M. intracellulare (1) |
| M. colombiense | 1 | 1 | |
| M. fortuitum complex | |||
| M. peregrinum | 3 | 3 | |
| M. conceptionense | 3 | 1 | M. fortuitum (2) |
| M. porcinum | 1 | 1 | |
| M. lentiflavum | 4 | 4 | |
| M. terrae complex | |||
| M. algericum | 2 | 2 | |
| M. senuense | 1 | 1 | |
| M. mucogenicum | 2 | 2 | |
| M. nebraskense | 1 | 1 | |
| Total (%) | 230 (100) | 223 (97) |
Compatible results are defined as “out of detection range” results from the Real-Q NTM-ID assay for mycobacterial species that the assay cannot detect.
The Real-Q assay correctly identified all 63 isolates in the M. abscessus-M. chelonae complex at the species level. The M. abscessus complex is frequently isolated from patients with respiratory rapidly growing mycobacteria around the world but especially in South Korea (15, 16). Koh et al. reported that the clinical impact of this species, which is classified within the M. abscessus complex, was different. The treatment response rates for clarithromycin-based antibiotic therapies were higher in patients with M. massiliense lung disease than in those with M. abscessus lung disease (16). Thus, species-level identification is important, especially in the M. abscessus complex. Differentiation between M. abscessus and M. massiliense is one advantage of the Real-Q assay compared with most other commercially available kits. The Real-Q assay could reduce the turnaround time of species identification compared with standard probe hybridization-based methods. The average turnaround time for the real-time PCR assay was less than 2.5 h. Furthermore, the Real-Q assay requires less hands-on time than do line probe assays.
However, the main limitation of this assay is its inability to identify rare isolates at the species level. Thus, additional procedures will sometimes be needed for the species-level identification of clinically important isolates. Nevertheless, the Real-Q assay can rapidly detect most clinically important NTM species with good specificity; thus, this assay could be useful as a routine method in clinical settings.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by BioSewoom (Seoul, South Korea). The sponsor had no involvement in the study design, data interpretation, or writing of the manuscript.
Footnotes
Published ahead of print 27 August 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.01957-14.
REFERENCES
- 1.Koh WJ, Kwon OJ, Jeon K, Kim TS, Lee KS, Park YK, Bai GH. 2006. Clinical significance of nontuberculous mycobacteria isolated from respiratory specimens in Korea. Chest 129:341–348. 10.1378/chest.129.2.341. [DOI] [PubMed] [Google Scholar]
- 2.Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F, Holland SM, Horsburgh R, Huitt G, Iademarco MF, Iseman M, Olivier K, Ruoss S, von Reyn CF, Wallace RJ, Jr, Winthrop K. 2007. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am. J. Respir. Crit. Care Med. 175:367–416. 10.1164/rccm.200604-571ST. [DOI] [PubMed] [Google Scholar]
- 3.Griffith DE. 2010. Nontuberculous mycobacterial lung disease. Curr. Opin. Infect. Dis. 23:185–190. 10.1097/QCO.0b013e328336ead6. [DOI] [PubMed] [Google Scholar]
- 4.Patel JB, Leonard DG, Pan X, Musser JM, Berman RE, Nachamkin I. 2000. Sequence-based identification of Mycobacterium species using the MicroSeq 500 16S rDNA bacterial identification system. J. Clin. Microbiol. 38:246–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yam WC, Yuen KY, Kam SY, Yiu LS, Chan KS, Leung CC, Tam CM, Ho PO, Yew WW, Seto WH, Ho PL. 2006. Diagnostic application of genotypic identification of mycobacteria. J. Med. Microbiol. 55:529–536. 10.1099/jmm.0.46298-0. [DOI] [PubMed] [Google Scholar]
- 6.Makinen J, Sarkola A, Marjamaki M, Viljanen MK, Soini H. 2002. Evaluation of genotype and LiPA MYCOBACTERIA assays for identification of Finnish mycobacterial isolates. J. Clin. Microbiol. 40:3478–3481. 10.1128/JCM.40.9.3478-3481.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quezel-Guerraz NM, Arriaza MM, Avila JA, Sanchez-Yebra Romera WE, Martinez-Lirola MJ, Indal TBG. 2010. Evaluation of the Speed-oligo(R) Mycobacteria assay for identification of Mycobacterium spp. from fresh liquid and solid cultures of human clinical samples. Diagn. Microbiol. Infect. Dis. 68:123–131. 10.1016/j.diagmicrobio.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 8.Kim BJ, Lee KH, Park BN, Kim SJ, Bai GH, Kim SJ, Kook YH. 2001. Differentiation of mycobacterial species by PCR-restriction analysis of DNA (342 base pairs) of the RNA polymerase gene (rpoB). J. Clin. Microbiol. 39:2102–2109. 10.1128/JCM.39.6.2102-2109.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim JU, Cha CH, An HK. 2012. Multiplex real-time PCR assay and melting curve analysis for identifying Mycobacterium tuberculosis complex and nontuberculous mycobacteria. J. Clin. Microbiol. 50:483–487. 10.1128/JCM.06155-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chakravorty S, Kothari H, Aladegbami B, Cho EJ, Lee JS, Roh SS, Kim H, Kwak H, Lee EG, Hwang SH, Banada PP, Safi H, Via LE, Cho SN, Barry CE, III, Alland D. 2012. Rapid, high-throughput detection of rifampin resistance and heteroresistance in Mycobacterium tuberculosis by use of sloppy molecular beacon melting temperature coding. J. Clin. Microbiol. 50:2194–2202. 10.1128/JCM.00143-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim K, Lee H, Lee MK, Lee SA, Shim TS, Lim SY, Koh WJ, Yim JJ, Munkhtsetseg B, Kim W, Chung SI, Kook YH, Kim BJ. 2010. Development and application of multiprobe real-time PCR method targeting the hsp65 gene for differentiation of Mycobacterium species from isolates and sputum specimens. J. Clin. Microbiol. 48:3073–3080. 10.1128/JCM.00939-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clinical and Laboratory Standards Institute. 2008. MM18-AE. Interpretive criteria for identification of bacteria and fungi by DNA target sequencing. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 13.Telenti A, Marchesi F, Balz M, Bally F, Bottger EC, Bodmer T. 1993. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J. Clin. Microbiol. 31:175–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frothingham R, Wilson KH. 1993. Sequence-based differentiation of strains in the Mycobacterium avium complex. J. Bacteriol. 175:2818–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kendall BA, Winthrop KL. 2013. Update on the epidemiology of pulmonary nontuberculous mycobacterial infections. Semin. Respir. Crit. Care Med. 34:87–94. 10.1055/s-0033-1333567. [DOI] [PubMed] [Google Scholar]
- 16.Koh WJ, Jeon K, Lee NY, Kim BJ, Kook YH, Lee SH, Park YK, Kim CK, Shin SJ, Huitt GA, Daley CL, Kwon OJ. 2011. Clinical significance of differentiation of Mycobacterium massiliense from Mycobacterium abscessus. Am. J. Respir. Crit. Care Med. 183:405–410. 10.1164/rccm.201003-0395OC. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


