Abstract
The objective of this research was to extend the Vitek MS fungal knowledge base version 2.0.0 to allow the robust identification of clinically relevant dermatophytes, using a variety of strains, incubation times, and growth conditions. First, we established a quick and reliable method for sample preparation to obtain a reliable and reproducible identification independently of the growth conditions. The Vitek MS V2.0.0 fungal knowledge base was then expanded using 134 well-characterized strains belonging to 17 species in the genera Epidermophyton, Microsporum, and Trichophyton. Cluster analysis based on mass spectrum similarity indicated good species discrimination independently of the culture conditions. We achieved a good separation of the subpopulations of the Trichophyton anamorph of Arthroderma benhamiae and of anthropophilic and zoophilic strains of Trichophyton interdigitale. Overall, the 1,130 mass spectra obtained for dermatophytes gave an estimated identification performance of 98.4%. The expanded fungal knowledge base was then validated using 131 clinical isolates of dermatophytes belonging to 13 taxa. For 8 taxa all strains were correctly identified, and for 3 the rate of successful identification was >90%; 75% (6/8) of the M. gypseum strains were correctly identified, whereas only 47% (18/38) of the African T. rubrum population (also called T. soudanense) were recognized accurately, with a large quantity of strains misidentified as T. violaceum, demonstrating the close relationship of these two taxa. The method of sample preparation was fast and efficient and the expanded Vitek MS fungal knowledge base reliable and robust, allowing reproducible dermatophyte identifications in the routine laboratory.
INTRODUCTION
Dermatophytes in the genera Epidermophyton, Microsporum, and Trichophyton are usually characterized and identified by cultural and morphological characters and physiological tests or, more recently, by sequencing (1). Morphological identification is time-consuming and complex, usually requiring expert mycological knowledge, while sequencing is comparatively expensive and at least 2 to 3 days elapse before sequencing results are available.
Matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry (MS) is a reliable technique for the identification and typing of microbial pathogens such as bacteria (2–7), yeasts (8–10), and filamentous fungi, including dermatophytes (7, 11–17). Recent studies confirm that this technique may be very attractive for dermatophyte identification (18–22). In one study, the rate of correct identification of isolates belonging to the T. mentagrophytes complex was 89% (19), and in others, the overall rates of successful identification of dermatophyte species reached 95.8% (18), 97.8% (21), and 99.3% (20), demonstrating the potential of MALDI-TOF MS to replace classical identification methods. The technique has now also been used for the direct identification of dermatophytes in clinical samples (23).
Technical challenges to its routine use include the simultaneous presence in a sample of different growth forms (mycelium or conidia), as well as the potential influence of duration of incubation, culture medium, preparation methods (solid or liquid cultures), the amount of biomass, and the type of instrument used (24, 25).
The objective of this work was to establish a simple and rapid sample preparation method suitable for routine laboratory work and to extend the Vitek MS fungal knowledge base version 2.0.0 to allow the robust identification of clinically relevant dermatophytes, using a variety of strains, incubation times, and growth conditions. The sample preparation method, based on previously described protocols for yeasts (9) and filamentous fungi (19, 21), combines organism inactivation and protein extraction and allows working with material taken directly from agar plates, thus avoiding any environmental or instrument contamination. We first established a spectrum database using 134 reference strains belonging to 17 species in the three anamorphic genera Epidermophyton, Microsporum, and Trichophyton. In the second step, 131 clinical strains not used to build the knowledge base and belonging to 13 taxa were identified using the new knowledge base to validate its performance. Results were compared with sequencing and/or previous MALDI-TOF MS analyses carried out using the SARAMIS system (18).
MATERIALS AND METHODS
Isolates studied. (i) Isolates used to build up the fungal knowledge base.
A total of 134 reference strains belonging to 17 species of the genera Epidermophyton, Microsporum, and Trichophyton and characterized by genetic (internal transcribed spacer [ITS] sequencing) and morphological (18) analyses were used for the Vitek MS knowledge base creation (Table 1). They originated from culture collections (CBS, Utrecht, The Netherlands; Micoteca da Universidade do Minho, Braga, Portugal), external quality control programs, and individual collections (Department of Dermatology, University Hospital of Zurich, Zurich, Switzerland; Laboratory of Applied Microbiology, University of Applied Sciences and Arts of Southern Switzerland, Bellinzona, Switzerland). Strains were inoculated on Sabouraud dextrose (SDA; bioMérieux, La Balme Les Grottes, France) and potato dextrose (PDA; Becton Dickinson, Franklin Lakes, NJ) agar plates and incubated at 30°C for 5 and 10 days to capture two distinct growing stages. Because of their low growth rates, Microsporum audouinii and Trichophyton violaceum were incubated for 10, 15, and 20 days to obtain enough biomass.
TABLE 1.
Performance of the expanded Vitek MS database for the identification of reference dermatophyte spectra
| Taxon | No. of strains | No. of spectra | % (no.) of correctly identified spectra (no.) | Discrimination |
% (no.) of unidentified spectra | % (no.) of spectra with discordant resultsd | Erroneous taxond | ||
|---|---|---|---|---|---|---|---|---|---|
| % (no.) with single-choice resulta | % (no.) with low-discrimination resultb | Second matchc | |||||||
| E. floccosum | 11 | 107 | 100 (107) | 100 (107) | |||||
| M. audouinii | 8 | 74 | 98.6 (73) | 94.6 (70) | 4 (3) | M. canis | 1.4 (1) | M. canis | |
| M. canis | 10 | 98 | 100 (98) | 100 (98) | |||||
| M. fulvum | 2 | 16 | 100 (16) | 100 (16) | |||||
| M. gypseum | 9 | 66 | 98.5 (65) | 98.5 (65) | 1.5 (1) | ||||
| M. persicolor | 5 | 38 | 97.4 (37) | 97.4 (37) | 2.6 (1) | ||||
| M. praecox | 1 | 11 | 100 (11) | 100 (11) | |||||
| Trichophyton sp. (A. benhamiae) | 14 | 81 | 95.1 (77) | 93.8 (76) | 1.2 (1) | T. erinacei | 3.7 (3) | 1.2 (1) | T. rubrum |
| T. erinacei | 7 | 53 | 100 (53) | 92.4 (49) | 7.6 (4) | A. benhamiae | |||
| T. equinum | 1 | 16 | 100 (16) | 100 (16) | |||||
| T. interdigitale (anthropophilic and zoophilic) | 19 | 185 | 99.5 (184) | 98.4 (182) | 1.1 (2) | T. tonsurans | 0.6 (1) | ||
| T. mentagrophytes | 2 | 25 | 88 (22) | 64 (16) | 24 (6) | T. equinum (3), T. interdigitale (3) | 8 (2) | 4 (1) | T. interdigitale |
| T. rubrum (African and sensu stricto) | 16 | 152 | 98.7 (150) | 93.4 (142) | 5.3 (8) | T. violaceum | 0.7 (1) | 0.7 (1) | T. violaceum |
| T. terrestre (A. quadrifidum) | 3 | 24 | 100 (24) | 100 (24) | |||||
| T. tonsurans | 10 | 82 | 97.6 (80) | 91.5 (75) | 6.1 (5) | T. interdigitale | 2.4 (2) | ||
| T. verrucosum | 3 | 24 | 95.8 (23) | 87.5 (21) | 8.3 (2) | T. erinacei | 4.2 (1) | ||
| T. violaceum | 13 | 78 | 97.4 (76) | 80.8 (63) | 16.7 (13) | T. rubrum | 1.3 (1) | 1.3 (1) | T. rubrum |
| Total | 134 | 1,130 | 98.4 (1,112) | 94.5 (1,068) | 3.9 (44) | 1.1 (13) | 0.5 (5) | ||
The mass spectrum matched the right species (high discrimination).
The mass spectrum matched the right species.
The mass spectrum also matched another, closely related species.
The mass spectrum matched another, unexpected species.
(ii) Clinical isolates used for testing.
The test of the expanded Vitek MS knowledge base was carried out on 131 isolates not used to build the knowledge base and including the Trichophyton anamorph of Arthroderma benhamiae (n = 3), Epidermophyton floccosum (n = 3), M. audouinii (n = 3), M. canis (n = 6), M. gypseum (n = 4), M. persicolor (n = 2), T. erinacei (n = 1), T. interdigitale (n = 38), T. rubrum sensu stricto (n = 34), African T. rubrum (T. soudanense) (n = 19), T. terrestre (n = 1), T. tonsurans (n = 5), and T. violaceum (n = 12) (Table 2). All of these originated from clinical material received from the Microbiology and Virology Unit, Papa Giovanni XXIII Hospital (Bergamo, Italy); the Department of Dermatology, University Hospital of Zurich (Zurich, Switzerland); and the Laboratory of Applied Microbiology, University of Applied Sciences and Arts of Southern Switzerland (Bellinzona, Switzerland). They had previously been characterized by morphology, ITS sequencing, and/or MALDI-TOF MS (SARAMIS system).
TABLE 2.
Identification of clinical strains using the expanded Vitek MS databasea
| Taxon | Total no. | Discrimination of strains grown on SDA |
Discrimination of strains grown on SGC2 |
||||||
|---|---|---|---|---|---|---|---|---|---|
| No. (%) positive | No. with single-choice result | No. with low-discrimination result | No. (%) with discordant result | No. (%) positive | No. with single-choice result | No. with low-discrimination result | No. (%) with discordant result | ||
| Trichophyton sp. (A. benhamiae) | 3 | 3 (100) | 3 | 3 (100) | 3 | ||||
| E. floccosum | 3 | 3 (100) | 3 | 3 (100) | 3 | ||||
| M. audouinii | 3 | 3 (100) | 3 | 3 (100) | 3 | ||||
| M. canis | 6 | 6 (100) | 6 | 6 (100) | 6 | ||||
| M. gypseum | 4 | 3 (75) | 3 | 1 (25) | 3 (75) | 3 | 1 (25) | ||
| M. persicolor | 2 | 2 (100) | 2 | 2 (100) | 2 | ||||
| T. erinacei | 1 | 1 (100) | 1 | 1 (100) | 1 | ||||
| T. interdigitale | 38 | 38 (100) | 37 | 1 | 37 (97.4) | 35 | 2 | 1 (2.6) | |
| T. rubrum sensu stricto | 34 | 34 (100) | 31 | 3 | 29 (85.3) | 25 | 4 | 5 (14.7) | |
| African T. rubrum | 19 | 15 (78.9) | 7 | 8 | 4 (21.1) | 3 (15.8) | 1 | 2 | 16 (84.2) |
| T. terrestre | 1 | 1 (100) | 1 | 1 (100) | 1 | ||||
| T. tonsurans | 5 | 5 (100) | 5 | 5 (100) | 5 | ||||
| T. violaceum | 12 | 11 (91.7) | 11 | 1 (8.3) | 11 (91.7) | 8 | 3 | 1 (8.3) | |
| Total | 131 | 125 (95.4) | 113 | 12 | 6 (4.6) | 107 (81.7) | 96 | 11 | 24 (18.3) |
For abbreviations and definitions, see the text.
Strains were grown on SDA plates and on Sabouraud gentamicin chloramphenicol 2 agar plates (SGC2; bioMérieux) to test the system's performance with cultures grown on a medium routinely used in clinical diagnostics laboratories. All strains were incubated at 30°C for 5 days, except for M. audouinii and T. violaceum samples, which were grown at 30°C for 9 to 10 days.
Sample preparation.
Sample preparation consisted of a combination of organism inactivation and protein extraction. It was based on a previously published method (9, 19, 21), with minor modifications. Fungal material containing all growth phases of the colony was scraped from a surface of approximately 1 cm2 using a wet swab and suspended in 300 μl of sterile deionized water. Nine hundred microliters of >99.8% (vol/vol) ethanol was added. After vortexing and centrifugation at 10,600 × g for 2 min, the supernatant was discarded, 40 μl of 70% formic acid was added to the resulting pellet, and 40 μl of acetonitrile was added after vortexing again. After a final vortexing and centrifugation step at 10,600 × g for 2 min, 1 μl of the supernatant was spotted in duplicate directly on a MALDI-TOF MS target plate (bioMérieux) and air dried. One microliter of α-cyano-4-hydroxy-cinnamic acid matrix (CHCA; bioMérieux) was added to each spot and air dried before MALDI-TOF MS analysis.
MALDI-TOF MS.
The bioMérieux Vitek MS Plus system used in this study is composed of an AXIMA (Shimadzu) mass spectrometer combined with both a closed database (Vitek MS V2.0.0) containing 755 species and an open database (SARAMIS version 4.09) comprising 1,202 species. Spectrum acquisition for knowledge base expansion was performed using the software LaunchPad version 2.9.3. Acquisition parameters such as laser power, detector voltage, etc., were set according to bioMérieux requirements for the Vitek MS system. Peak lists (m/z 2,000 to 20,000) were imported into the SARAMIS software (bioMérieux) for further taxonomic analyses. To be considered, the quality of the strains and spectra had to have a relative intraspecific similarity of >65%, a relative similarity between duplicates of >70%, and an absolute intraspecific similarity of >50 peaks. Spectra fulfilling these quality criteria were included in the Vitek MS dermatophyte spectral base.
Expanded Vitek MS knowledge base performance estimation by cross validation.
A 5-fold cross validation method (26) was used to estimate the performance of the expanded knowledge base. The whole Vitek MS spectrum base (including the new dermatophyte spectra) was divided into five subsets, four of which were used as references and the remaining one as a test subset (to predict identification). Dermatophyte spectra from the test subset were submitted to identification by comparing them with spectra from the four reference subsets. This process was repeated five times with permutations of the five subsets. The identification results of the five rounds were used to estimate the identification performance. A correct identification included (i) a single-choice result (high discrimination), meaning that the spectrum matched only the right species, and (ii) a low-discrimination result, when the spectrum matched the right species and another, closely related species.
RESULTS
Vitek MS fungal knowledge base extension.
A total of 1,130 dermatophyte mass spectra were retained and used for the Vitek MS fungal knowledge base extension (Table 1). The average number of strains and mass spectra per species were 7.9 and 66.5, respectively. Separation of all strains was achieved at least at the species level. In general, the mass spectra were very homogeneous when generated from material from SDA plates and less homogeneous for samples originating from PDA plates.
Microsporum sp. and Trichophyton anamorph of Arthroderma benhamiae.
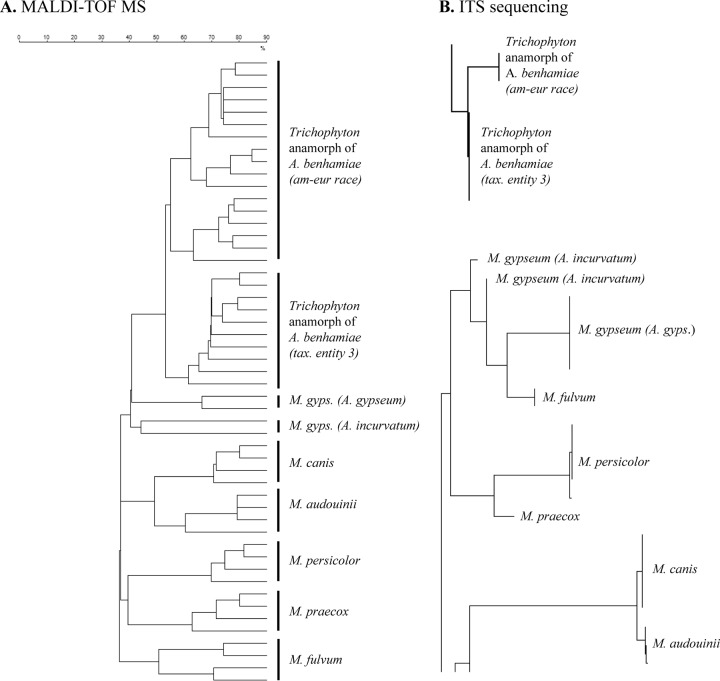
All Microsporum strains were well separated at the species level by MALDI-TOF MS (Fig. 1A). For M. audouinii, M. canis, M. persicolor, and M. praecox, MALDI-TOF MS results were similar to results obtained by ITS sequencing (Fig. 1B), with M. audouinii clustering together with M. canis and M. persicolor with M. praecox. Microsporum audouinii was separated clearly from M. canis by MALDI-TOF MS, although the ITS sequence similarity of the two species is >99.6% (18). The anamorphic M. gypseum strains used in this study belong to two teleomorphic species, Arthroderma gypseum and A. incurvatum, both well distinguished by MALDI-TOF MS (Fig. 1A).
FIG 1.
Microsporum and Trichophyton anamorph of A. benhamiae. (A) Dendrogram (Dice coefficient, single-linkage agglomerative clustering) based on the MALDI-TOF MS analysis of some of the reference strains used for the Vitek MS knowledge base creation (SDA and PDA plates; molecular mass range, 3 to 20 kDa). (B) Phylogenetic tree from ITS sequencing (reproduced from reference 18 with permission of Informa Healthcare).
Both ITS sequencing and MALDI-TOF MS were able to separate reliably the American-European race and taxonomic entity 3 of the Trichophyton anamorph of A. benhamiae (Fig. 1).
Trichophyton sp.
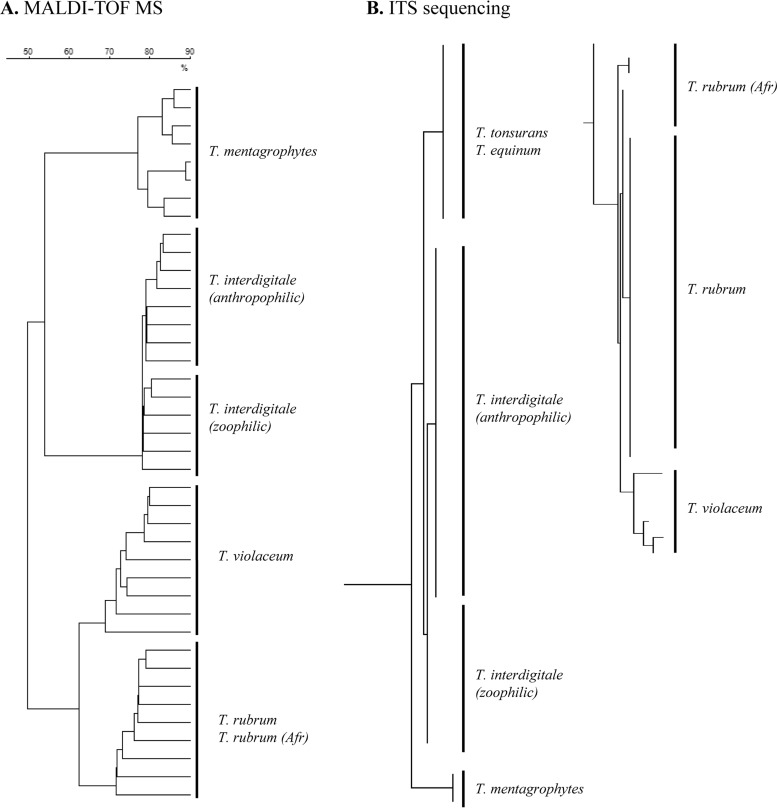
Previous ITS sequencing showed that T. violaceum differed from T. rubrum by only 7/538 nucleotides (Fig. 2B) (18); MALDI-TOF MS was able to distinguish the two species (Fig. 2A). T. rubrum sensu stricto and its African population are genetically very similar (sequence similarity of >99.5%) (18), and MALDI-TOF MS could not separate them.
FIG 2.
Trichophyton. (A) Dendrogram (Dice coefficient, single-linkage agglomerative clustering) based on the MALDI-TOF MS analysis of some of the reference strains used for the Vitek MS knowledge base creation (SDA plates; molecular mass range, 3 to 20 kDa). (B) Phylogenetic tree from ITS sequencing (reproduced from reference 18 with permission of Informa Healthcare).
Both T. interdigitale and T. mentagrophytes species could be identified unambiguously by MALDI-TOF MS. In addition, T. interdigitale spectra obtained from samples grown on SDA allowed separation of anthropophilic from zoophilic populations (Fig. 2A).
Performance of the expanded Vitek MS knowledge base.
Overall, the 1,130 dermatophyte mass spectra generated with the 134 reference strains gave a cross-validated performance of 98.4% (1,112 correctly identified spectra): 94.5% (1,068) of them provided a single-choice result (high discrimination) and 3.9% (44) a correct identification with low discrimination (Table 1). Only 1.1% (13) of the spectra could not be identified, and 0.5% (5) provided discordant identifications. One hundred percent of the spectra were correctly identified for 7, >95% for 9, and 88% for 1 (T. mentagrophytes) of the 17 species. For species with low-discrimination values or a discordant result, the second match was in general a taxon closely related to the correct one.
Evaluation of the expanded knowledge base with clinical samples.
The overall rate of correct identification of the 131 clinical isolates tested to estimate the knowledge base performance was 88.5% (95.4% for samples on SDA and 81.7% on SGC2) (Table 2). For 8 taxa, all strains were correctly identified, and for 3 the rate of successful identification was >90%; 75% of the M. gypseum strains were correctly identified, whereas only 47% (18/38) of the African T. rubrum population (T. soudanense) were recognized accurately, with a large rate of strains misidentified as T. violaceum, demonstrating the close relationship of these two taxa. Increasing the incubation time of cultures to 10 days and/or repeating the analyses did not improve results (data not shown).
DISCUSSION
Sample preparation for dermatophyte identification by Vitek MS.
The Vitek MS fungal knowledge base constructed using sample spectra obtained with the method described here proved to be reliable and robust, allowing reproducible dermatophyte identifications in the routine laboratory.
Obtaining high-quality MALDI-TOF MS spectra of fungi is more challenging than spectrum acquisition from most bacteria. Direct spotting does not provide reproducible results, and a careful standardization of the extraction and preparation method is necessary. This can be done only by taking into account culture media, incubation time, and amount and type of material (7, 25). Published extraction protocols include bead beating and the use of trifluoroacetic acid, acetonitrile, or formic acid treatments to disrupt the fungal cell walls, to increase protein extraction, and to provide high-quality mass spectra (27). In this study, we have used a newly developed protocol, combining microorganism inactivation and protein extraction, to obtain reliable, high-quality, and reproducible spectra from material grown on solid media. This protocol is rapid and safe, as it uses only nontoxic solvents and ensures inactivation of the molds, thus avoiding health hazards and the contamination of the instrument during measurements. It is also most convenient in the routing laboratory, where solid culture media are mainly used; for instance, nails and hairs suspected to contain dermatophytes are generally incubated on agar plates. Identification by MALDI-TOF MS can, therefore, be carried out immediately from material derived from the original culture, with no additional time-consuming steps described for other protocols (28). The use of this protocol allows also the user to visually identify distinct types of molds growing on the same agar plate, thus avoiding the potential pitfall of liquid cultures that could mask the presence of several organisms.
Extension of the Vitek MS knowledge base.
In this study, we used several strains per species (7.9 on average) and several culture conditions (different media and times of incubation) to capture the variability of each species and to create a robust knowledge base taking into account possible peak pattern variations in response to different cultivation conditions.
Spectral profiles obtained after incubation on SDA were in general very homogeneous. The presence of different growth stages on PDA with stage-specific spectral profiles resulted in more heterogeneous MALDI-TOF mass spectra than for organisms grown on SDA, on which sporulation generally is delayed. This made a separation of species grown on PDA possible, but often it did not allow distinguishing intraspecific populations or cryptic species.
To our knowledge, this is the first study to achieve a good separation of intraspecific taxa in some dermatophyte species. We obtained an excellent separation of the subpopulations of the Trichophyton anamorph of A. benhamiae grown on SDA or PDA (Fig. 1A) and of the anthropophilic and zoophilic strains of T. interdigitale grown on SDA (Fig. 2A). This confirms the high discriminatory power of the method used.
Identification of clinical strains.
Clinical strains were grown on SDA and on SGC2 agar plates, the latter being a medium frequently used in clinical diagnostic laboratories. This was done intentionally to ensure that the extraction and identification method would also work on material taken from a medium not used to construct the database. The identification of strains grown on SDA plates was very reliable, with the exception of some strains of the African population of T. rubrum (T. soudanense). Identification using spectra of material grown on SGC2, however, was less reliable for T. rubrum sensu stricto and unsatisfactory for the African population of T. rubrum. Identification of T. violaceum was less problematic, with only 1 out of 12 clinical strains wrongly identified as T. rubrum.
Gräser et al. (1) describe the T. rubrum complex to include only the two anthropophilic species T. rubrum sensu lato and T. violaceum, other taxa previously recognized being reclassified or synonymized as T. rubrum or T. violaceum (29, 30). Both T. violaceum and the African T. rubrum (T. soudanense) are endemic in Africa, whereas T. rubrum sensu stricto strains are cosmopolitan. T. rubrum sensu lato and T. violaceum are phylogenetically very close, even if morphologically quite distinct, and their taxonomic position has been under discussion for a long time (1, 29, 31–34). ITS sequence data would support T. soudanense as a synonym of T. violaceum (29, 35, 36); the two taxa share identical ITS2 sequences different from those of T. rubrum sensu stricto (36). In contrast, studies using microsatellite markers, known to evolve at a high mutation rate, are in disagreement with ITS data and suggest T. soudanense to be closely related to T. rubrum sensu stricto (33, 34). In our study, we followed the proposal of Gräser et al. to consider T. soudanense as belonging to T. rubrum sensu lato (1, 29, 33).
Six reference strains belonging to the African population of T. rubrum (T. soudanense) were used for the knowledge base creation and, as expected, were all closely related to T. rubrum sensu stricto. Identification of 19 clinical African T. rubrum strains was clearly less reliable than that of T. rubrum sensu stricto (78.9% for material grown on SDA and 15.8% on SGC2) (Table 2). Several strains were also misidentified as T. violaceum (4/19 strains grown on SDA and 16/19 on SGC2). De Respinis et al. (18) already showed a high degree of homology between T. violaceum and T. rubrum sensu lato by MALDI TOF MS. They could not achieve a direct identification and had to use the direct comparison of eight specific masses to reliably distinguish these two taxa. Other authors described similar problems in differentiating taxa from the T. rubrum complex. Nenoff et al. (20) reported a very high percentage (99.3%) of correct dermatophyte identifications, the only two strains that could not be identified belonging to T. violaceum. L'Ollivier et al. (21) reported that half of the T. violaceum strains they studied were misidentified as T. soudanense. The close relationship of taxa within the T. rubrum complex is confirmed by our results, and the high similarity between T. violaceum and African T. rubrum observed with MALDI-TOF MS is in accordance with the ITS sequence data (29, 35, 36).
Conclusions.
Our study has demonstrated that the inactivation-extraction method is rapid and reliable and that the newly built Vitek MS knowledge base provides a robust tool for the reproducible identification to the species (and in selected cases to the intraspecific) level of dermatophytes grown on common routine media such as SDA, SGC2, and PDA.
The quality of the spectra obtained is high and the method suitable for research and diagnostic work with dermatophytes and with filamentous fungi in general.
The overall identification performance of clinical strains was high, but strains identified as T. violaceum by MALDI-TOF MS should undergo additional morphological and/or molecular analysis to exclude any confusion with the African population of T. rubrum.
ACKNOWLEDGMENTS
This work was supported by bioMérieux SA, Unit Microbiology, R&D Microbiology, La Balme Les Grottes, France.
We thank Liliane E. Petrini (Breganzona, Switzerland) for checking the morphological identification of several dermatophyte strains.
Footnotes
Published ahead of print 8 October 2014
REFERENCES
- 1. Gräser Y, Scott J, Summerbell R. 2008. The new species concept in dermatophytes—a polyphasic approach. Mycopathologia 166:239–256. 10.1007/s11046-008-9099-y. [DOI] [PubMed] [Google Scholar]
- 2. Gaia V, Casati S, Tonolla M. 2011. Rapid identification of Legionella spp. by MALDI-TOF MS based protein mass fingerprinting. Syst. Appl. Microbiol. 34:40–44. 10.1016/j.syapm.2010.11.007. [DOI] [PubMed] [Google Scholar]
- 3. Hathout Y, Demirev PA, Ho YP, Bundy JL, Ryzhov V, Sapp L, Stutler J, Jackman J, Fenselau C. 1999. Identification of Bacillus spores by matrix-assisted laser desorption ionization-mass spectrometry. Appl. Environ. Microbiol. 65:4313–4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fenselau C, Demirev PA. 2001. Characterization of intact microorganisms by MALDI mass spectrometry. Mass Spectrom. Rev. 20:157–171. 10.1002/mas.10004. [DOI] [PubMed] [Google Scholar]
- 5. Bright JJ, Claydon MA, Soufian M, Gordon DB. 2002. Rapid typing of bacteria using matrix-assisted laser desorption ionisation time-of-flight mass spectrometry and pattern recognition software. J. Microbiol. Methods 48:127–138. 10.1016/S0167-7012(01)00317-7. [DOI] [PubMed] [Google Scholar]
- 6. Benagli C, Rossi V, Dolina M, Tonolla M, Petrini O. 2011. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for the identification of clinically relevant bacteria. PLoS One 6:e16424. 10.1371/journal.pone.0016424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Croxatto A, Prod'hom G, Greub G. 2012. Applications of MALDI-TOF mass spectrometry in clinical diagnostic microbiology. FEMS Microbiol. Rev. 36:380–407. 10.1111/j.1574-6976.2011.00298.x. [DOI] [PubMed] [Google Scholar]
- 8. Stevenson LG, Drake SK, Shea YR, Zelazny AM, Murray PR. 2010. Evaluation of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of clinically important yeast species. J. Clin. Microbiol. 48:3482–3486. 10.1128/JCM.00687-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bader O, Weig M, Taverne-Ghadwal L, Lugert R, Gross U, Kunhs M. 2011. Improved clinical laboratory identification of human pathogenic yeasts by MALDI-TOF MS. Clin. Microbiol. Infect. 17:1359–1365. 10.1111/j.1469-0691.2010.03398.x. [DOI] [PubMed] [Google Scholar]
- 10. Spanu T, Posteraro B, Fiori B, D'Inzeo T, Campoli S, Ruggeri A, Tumbarello M, Canu G, Trecarichi EM, Parisi G, Tronci M, Sanguinetti M, Fadda G. 2012. Direct MALDI-TOF mass spectrometry assay of blood culture broths for rapid identification of Candida species causing bloodstream infections: an observational study in two large microbiology laboratories. J. Clin. Microbiol. 50:176–179. 10.1128/JCM.05742-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li TY, Liu BH, Chen YC. 2000. Characterization of Aspergillus spores by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 14:2393–2400. . [DOI] [PubMed] [Google Scholar]
- 12. Hettick JM, Green BJ, Buskirk AD, Kashon ML, Slaven JE, Janotka E, Blachere FM, Schmechel D, Beezhold DH. 2008. Discrimination of Aspergillus isolates at the species and strain level by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry fingerprinting. Anal. Biochem. 380:276–281. 10.1016/j.ab.2008.05.051. [DOI] [PubMed] [Google Scholar]
- 13. De Respinis S, Vogel G, Benagli C, Tonolla M, Petrini O, Samuels GJ. 2010. MALDI-TOF MS of Trichoderma: a model system for the identification of microfungi. Mycol. Prog. 9:79–100. 10.1007/s11557-009-0621-5. [DOI] [Google Scholar]
- 14. Erhard M, Hipler UC, Burmester A, Brakhage AA, Wöstemeyer J. 2008. Identification of dermatophyte species causing onychomycosis and tinea pedis by MALDI-TOF mass spectrometry. Exp. Dermatol. 17:356–361. 10.1111/j.1600-0625.2007.00649.x. [DOI] [PubMed] [Google Scholar]
- 15. Theel ES, Hall L, Mandrekar J, Wengenack NL. 2011. Dermatophyte identification using matrix-assisted laser desorption/ionization-time of flight mass spectrometry. J. Clin. Microbiol. 49:4067–4071. 10.1128/JCM.01280-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Alshawa K, Beretti JL, Lacroix C, Feuilhade M, Dauphin B, Quesne G, Hassouni N, Nassif X, Bougnoux ME. 2012. Successful identification of clinical dermatophyte and Neoscytalidium species by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 50:2277–2281. 10.1128/JCM.06634-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schulthess B, Ledermann R, Mouttet F, Zbinden A, Bloemberg GV, Böttger EC, Hombach M. 2014. Use of the Bruker MALDI biotyper for identification of molds in the clinical mycology laboratory. J. Clin. Microbiol. 52:2797–2803. 10.1128/JCM.00049-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. De Respinis S, Tonolla M, Pranghofer S, Petrini L, Petrini O, Bosshard PP. 2013. Identification of dermatophytes by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Med. Mycol. 51:514–521. 10.3109/13693786.2012.746476. [DOI] [PubMed] [Google Scholar]
- 19. Packeu A, Hendrickx M, Beguin H, Martiny D, Vandenberg O, Detandt M. 2013. Identification of the Trichophyton mentagrophytes complex species using MALDI-TOF mass spectrometry. Med. Mycol. 51:580–585. 10.3109/13693786.2013.770605. [DOI] [PubMed] [Google Scholar]
- 20. Nenoff P, Erhard M, Simon JC, Muylowa GK, Herrmann J, Rata W, Gräser Y. 2013. MALDI-TOF mass spectrometry—a rapid method for the identification of dermatophyte species. Med. Mycol. 51:17–24. 10.3109/13693786.2012.685186. [DOI] [PubMed] [Google Scholar]
- 21. L'Ollivier C, Cassagne C, Normand AC, Bouchara JP, Contet-Audonneau N, Hendrickx M, Fourquet P, Coulibaly O, Piarroux R, Ranque S. 2013. A MALDI-TOF MS procedure for clinical dermatophyte species identification in the routine laboratory. Med. Mycol. 51:713–720. 10.3109/13693786.2013.781691. [DOI] [PubMed] [Google Scholar]
- 22. Packeu A, De Bel A, l'Ollivier C, Ranque S, Detandt M, Hendrickx M. 2014. Fast and acurate identification of dermatophytes by MALDI-TOF mass spectrometry: validation in the clinical lab. J. Clin. Microbiol. 52:3440–3443. 10.1128/JCM.01428-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hollemeyer K, Jager S, Altmeyer W, Heinzle E. 2005. Proteolytic peptide patterns as indicators for fungal infections and nonfungal affections of human nails measured by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal. Biochem. 338:326–331. 10.1016/j.ab.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 24. Bader O. 2013. MALDI-TOF-MS-based species identification and typing approaches in medical biology. Proteomics 13:788–799. 10.1002/pmic.201200468. [DOI] [PubMed] [Google Scholar]
- 25. Santos C, Paterson RRM, Venâncio A, Lima N. 2010. Filamentous fungal characterizations by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J. Appl. Microbiol. 108:375–385. 10.1111/j.1365-2672.2009.04448.x. [DOI] [PubMed] [Google Scholar]
- 26. Hastie T, Tibshirani R, Friedman J. 2009. Model assessment and selection, p 219–260 In Hastie T, Tibshirani R, Friedman J. (ed), The elements of statistical learning: data mining, interference, and prediction, 2nd ed. Springer, New York, NY. [Google Scholar]
- 27. Hettick JM, Green BJ, Buskirk AD, Slaven JE, Kashon ML, Beezhold DH. 2011. Discrimination of fungi by MALDI-TOF mass spectrometry, p 35–50 In Fenselau C, Demirev P. (ed), Rapid characterization of microorganisms by mass spectrometry. American Chemical Society, Washington, DC. [Google Scholar]
- 28. Schrödl W, Heydel T, Schwartze VU, Hoffmann K, Grosse-Herrenthey A, Walther G, Alastruey-Izquierdo A, Rodriguez-Tudela JL, Olias P, Jacobsen ID, de Hoog GS, Voigt K. 2012. Direct analysis and identification of pathogenic Lichtheimia species by matrix-assisted laser desorption ionization-time of flight analyzer-mediated mass spectrometry. J. Clin. Microbiol. 50:419–427. 10.1128/JCM.01070-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gräser Y, Kuijpers AFA, Presber W, De Hoog GS. 2000. Molecular taxonomy of the Trichophyton rubrum complex. J. Clin. Microbiol. 38:3329–3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cafarchia C, Iatta R, Latrofa MS, Gräser Y, Otranto D. 2013. Molecular epidemiology, phylogeny and evolution of dermatophytes. Infect. Genet. Evol. 20:336–351. 10.1016/j.meegid.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 31. Summerbell RC, Haugland RA, Li A, Gupta AK. 1999. rRNA gene internal transcribed spacer 1 and 2 sequences of asexual, anthropophilic dermatophytes related to Trichophyton rubrum. J. Clin. Microbiol. 37:4005–4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gräser Y, El Fari M, Vilgalys R, Kuijpers AFA, De Hoog GS, Presber W, Tietz H-J. 1999. Phylogeny and taxonomy of the family Arthrodermataceae (dermatophytes) using sequence analysis of the ribosomal ITS region. Med. Mycol. 37:105–114. 10.1080/02681219980000171. [DOI] [PubMed] [Google Scholar]
- 33. Gräser Y, Fröhlich J, Presber W, De Hoog S. 2007. Microsatellite markers reveal geographic population differentiation in Trichophyton rubrum. J. Med. Microbiol. 56:1058–1065. 10.1099/jmm.0.47138-0. [DOI] [PubMed] [Google Scholar]
- 34. Ohst T, De Hoog S, Presber W, Stavrakieva V, Gräser Y. 2004. Origins of microsatellite diversity in the Trichophyton rubrum-T. violaceum clade (dermatophytes). J. Clin. Microbiol. 42:4444–4448. 10.1128/JCM.42.10.4444-4448.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. de Hoog GS, Cuarro GJ, Figueras MJ. 2000. Atlas of clinical fungi, 2nd ed. Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands. [Google Scholar]
- 36. Li HC, Bouchara JP, Hsu MM-L, Barton R, Su S, Chang TC. 2008. Identification of dermatophytes by sequence analysis of the rRNA gene internal transcribed spacer regions. J. Med. Microbiol. 57:592–600. 10.1099/jmm.0.47607-0. [DOI] [PubMed] [Google Scholar]