Abstract
Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS)-based species identification has become a reliable and fast tool for use in clinical diagnostics, including in mycology. To identify yeasts in the MALDI Biotyper system, a multistep extraction protocol, which is also used to generate the reference spectra, is recommended. Sample preparation by on-target lysis (OTL) requires significantly less hands-on time and is therefore highly desirable, but it results in too-low MALDI Biotyper log score values to allow automated species identification. To overcome this problem, we developed a procedure for generating and validating an OTL spectrum data set for the most relevant and frequently occurring yeast species in clinical specimens. The performance was evaluated against a set of OTL spectra derived during clinical routine procedures and from a set of closely related yeasts. In the diagnostic setting, the OTL procedure significantly decreased the workload but allowed species identification with high specificity and sensitivity. False identifications were not observed. The use of in-house-generated OTL reference spectra can highly accelerate MALDI-TOF MS-based yeast species identification using the MALDI Biotyper.
INTRODUCTION
For species identification from culture material, matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) is a reliable and fast tool for use in clinical diagnostics, including in mycology (1, 2). To prepare bacterial samples for species identification, it is generally sufficient to transfer a small amount of intact cells from the agar plate onto the MALDI target plate and overlay the cells with a small amount of matrix solution. This smear preparation protocol is appropriate for most bacterial species, but due to their stronger cell walls, fungal cells do not necessarily lyse to the same extent and may not sufficiently release their intracellular contents under these conditions, resulting in an insufficient number of peaks above background level.
To identify yeast species in the MALDI Biotyper system, the manufacturer recommends the use of an extraction protocol, which also has been used to generate the reference spectrum database. Briefly, cells are harvested and washed in 70% ethanol, dried, and then lysed in 70% formic acid, followed by the addition of acetonitrile. The debris is removed by centrifugation and the extract spotted onto a steel target. This method generally leads to an increased number of distinct peaks, which are sufficient for species identification. However, this multistep procedure delays the workflow during routine diagnostic procedures.
Between the simple smear preparation protocol and the full extraction, several variants of the extraction steps have been described in the literature. In the simplest version, the smear is overlaid with 70% formic acid and left to dry before overlaying with the matrix. Here, this is referred to as on-target lysis (OTL); in the literature, it is also called short extraction or formic acid overlay (3–7). Since this particular sample preparation method is significantly faster and requires less hands-on time than extraction, several studies have evaluated its usefulness in routine diagnostic procedures with the MALDI Biotyper (3, 4, 6–11). The technical problem to be overcome here is that compared to full extraction, this method generates (i) partially different and (ii) fewer peaks with a signal-to-noise ratio above the specified threshold ratio. As outlined above, the MALDI Biotyper reference spectra have been generated using the full extraction protocol and, therefore, the OTL spectra are matched only at decreased log scores (2). A log score is the measure of quality of the database hit, where log scores of ≥2.000 are classified as species-level identifications, and log scores between 1.700 and 1.999 are classified as genus-level identifications only. Nevertheless, it has been found that lower log scores starting from 1.800 (10, 11), 1.700 (3, 4, 7, 9), or even 1.500 (6, 12) can be accepted as species specific for yeasts if certain criteria are met (4, 8, 12).
To achieve an automated species-level identification in the current V3 MALDI Biotyper client series, all spectrum scores of ≥2.000 must be of the same species and all spectrum scores of ≥1.700 of the same genus. In the absence of automated implementation of the modified algorithms, laborious manual inspection of the hit lists for each sample and a manual adjustment of the identification level to the species level must be performed if the criteria are met.
Obviously, the best preparation method to use for the system at hand is the same method with which the spectra for the database have been generated, since this will lead to the highest concordance. To streamline the diagnostic procedures for the identification of yeast species with the MALDI Biotyper, it was recently shown that reference spectra added to the database from yeast strains prepared with the quick OTL method can lead to significant score values (13).
To overcome the need to generate and curate large amounts of OTL reference spectra into the MALDI Biotyper database, we have tested what can be achieved with a minimal set of spectra for the clinically most frequently occurring species only.
MATERIALS AND METHODS
Yeast strains and culture.
For long-term storage, yeasts were kept as Cryobank stocks (Mast Diagnostica, Reinfeld, Germany) at −70°C. Once thawed, the strains were kept on Sabouraud agar slants supplemented with 0.5% peptone (casein), 0.5% peptone (meat), and 2% glucose. The isolates from diagnostic procedures were grown on either Columbia blood, chocolate, or Sabouraud agar (all from Oxoid, Wesel, Germany). The isolates selected for this study were from Candida albicans (n = 122), Candida dubliniensis (n = 10), Candida tropicalis (n = 25), Candida lusitaniae (n = 14), Candida parapsilosis (n = 20), Candida metapsilosis (n = 2), Candida orthopsilosis (n = 7), Candida glabrata (n = 27), Candida nivariensis (n = 1), Candida palmioleophila (n = 7), Candida guilliermondii (n = 16), and Candida krusei (n = 14).
The isolates eventually chosen for making the six final reference entries (see below) were deposited at the German Collection of Microorganisms and Cell Cultures (DSMZ) under DSM 28718 (C. glabrata), DSM 28719 (C. albicans), DSM 28720 (C. tropicalis), DSM 28721 (C. krusei), DSM 28722 (C. parapsilosis), and DSM 28723 (C. dubliniensis).
Sample preparation and spectrum acquisition.
The samples were prepared by three different methods. First, smear preparation was done by careful streaking of a small amount of cells obtained from a fresh colony onto the steel target plate and overlaying it with matrix solution (saturated aqueous solution of α-cyano-4-hydroxycinnamic acid in 50% acetonitrile, 0.125% trifluoroacetic acid). Second, OTL was performed similarly to smear preparation, except that the cells were lysed by adding 1 μl of 70% formic acid and left to dry prior to adding matrix solution (2, 14). Third, full extraction was performed according to the protocol provided by the manufacturer, extracting proteins from ethanolized cells by sequential treatment with 70% formic acid and acetonitrile.
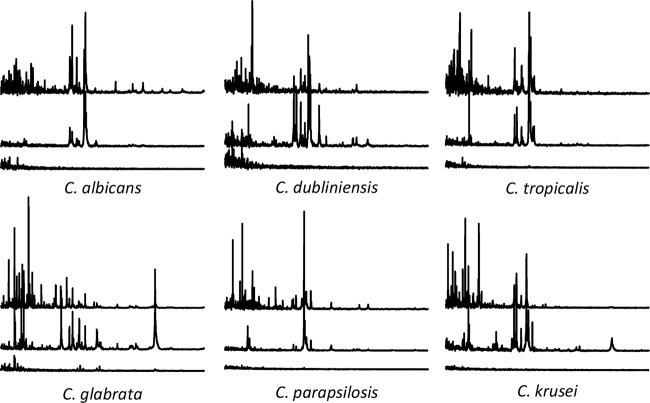
All OTL sample preparations were done in triplicate. The spectra from the samples prepared with either full extraction or OTL were recorded using the automated acquisition mode of the MALDI Biotyper 3.1 software on an Autoflex III mass spectrometer (both from Bruker Daltonics, Bremen, Germany). The spectra from the smear preparations were acquired by manually adding 600 spectra from spot regions where at least a minimal set of distinct peaks was recordable. The smear preparation spectra served only in a qualitative demonstration of the differences by the three methods (Fig. 1).
FIG 1.
Comparison of example mass spectra obtained by the different preparation methods. Top spectra, full extraction; middle spectra, OTL method; bottom spectra, smear preparation. The mass range of each spectrum is 2 to 14 kDa (x axis), and the peak intensities (y axis) are set to 15,000 arbitrary units. The acquisition of smear spectra was performed manually, whereas that of OTL and extraction spectra was by automatic mode.
Generation of MALDI Biotyper database spectra.
For the generation of MALDI Biotyper database entries (mass spectra [MSP]), strains were selected from those occurring during routine diagnostics. For less frequently occurring species, such as C. dubliniensis, C. orthopsilosis, C. metapsilosis, C. nivariensis, and C. palmioleophila, isolates were additionally taken from the 2001–2013 collection of the German National Reference Center for Systemic Mycoses Göttingen. The species of all isolates selected for MSP generation were confirmed by internal transcribed spacer (ITS) sequencing (15) and the MALDI Biotyper (current V3 database with 4,613 spectra) using the full extraction method. Prior to the preparation of samples, strains were cultivated overnight on Sabouraud agar at 35°C.
Before spectrum acquisition for the generation of MSP, the instrument was calibrated with Bruker Test Standard (BTS). For each isolate, 30 individual spots were prepared using the OTL method. The spectra were acquired using the automated acquisition mode and an MSP generated to include masses occurring in ≥20% of the spectrum set. For all procedures, the standard settings as implemented in the MALDI Biotyper were used.
RESULTS
Over a period of 1 month, we collected all yeasts differentiated in our clinical diagnostic laboratory. This amounted to 122 C. albicans, 25 C. tropicalis, 20 C. parapsilosis, 27 C. glabrata, 10 C. dubliniensis, and 14 C. krusei isolates, as well as several single isolates from rare species. These six species also represented the most frequently occurring yeasts in our clinical specimen over longer periods of time. The mass spectra of samples prepared with the extraction method were visually different from those prepared with the OTL method (Fig. 1). The OTL spectra generally contained fewer (∼70 to 90) but more intense mass signals above the signal-to-noise ratio than those of the extraction spectra, for which we regularly obtained >150 mass signals (data not shown). These OTL spectra resulted in log scores in the genus-level range of 1.800 to 1.999, as previously reported by others (10, 11). Only for C. tropicalis, ∼50% of the scores were >2.000 (Fig. 2A, Biotyper column for C. tropicalis). The best-effort mass spectra obtained manually from the samples prepared by smear preparation gave no useful mass signals and were insufficient for either genus or species identification (Fig. 1).
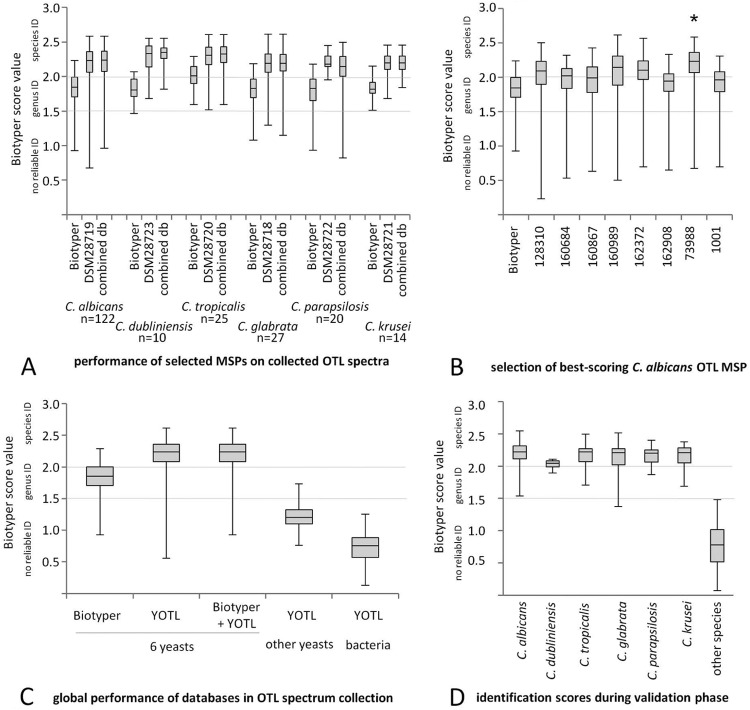
FIG 2.
Performance of the YOTL database. Quantile plots of MALDI Biotyper score values depicting database performances. (A) MALDI Biotyper database versus final selected best-performing MSP of the six most frequent yeast species. The use of YOTL or the combined YOTL and MALDI Biotyper database results in higher log scores from samples with OTL preparation. (B) Selection process for OTL mass spectra, using C. albicans as an example. *, Best-scoring MSP (minimum >75% species identifications in retrospective OTL spectrum collection). (C) Different spectrum set-MSP database combinations. Columns 1 to 3, log score values from samples prepared by OTL are much lower if the MALDI Biotyper database is used compared to YOTL or the combined database. Column 4, yeast species not present in the YOTL database give no reliable identification in YOTL. Column 5, bacterial smear spectra do not give rise to false hits in the YOTL database. (D) In-house validation. Quantile plots of log score values obtained during the validation phase, stratified per species represented in YOTL. ID, identification; db, database.
Generation of MSP entries for epidemiologically frequent yeasts.
Different isolates of C. albicans (n = 8), C. tropicalis (n = 3), C. parapsilosis (n = 4), C. glabrata (n = 2), C. krusei (n = 2), and C. dubliniensis (n = 2) were selected from among our routine isolates and reference MSP generated from samples prepared by OTL (see below). The identification levels obtained by these novel MSP were calculated retrospectively on all OTL spectra derived from routine diagnostics during the initial study period (acquired with the automated “smart” mode). The MSP of one species showed different matching levels to the respective set of routine OTL spectra, as outlined, for example, for C. albicans (Fig. 2B). For each species, we selected the one MSP that resulted in the highest rates of correct OTL spectrum identifications (≥75% of scores at the species level) and entered them into a novel database, the Yeast On-Target Lysis (YOTL) database. The YOTL database, as well as the combination of YOTL with the MALDI Biotyper database, allowed efficient and correct matching of the OTL spectra. When the log score values initially remained <2.000, this was generally amended by repeating the preparation or by preparing the sample in duplicate (data not shown).
Quality control.
Although the MSP generated were efficiently able to match OTL spectra correctly, it was a major concern that due to the reduced peak numbers, their discriminatory power might be reduced and consequently also their ability to discriminate correctly between closely related species. We therefore introduced a control group consisting of several fungal species that were closely related to the species in the YOTL database. For C. albicans, this was C. dubliniensis (already included in the database); for C. glabrata, this was C. nivariensis; and for C. parapsilosis, we included C. orthopsilosis and C. metapsilosis. Also, C. lusitaniae, C. guilliermondii, and C. palmioleophila were included, as these species are notoriously difficult to tell apart, and only MALDI-TOF has recently facilitated easy differentiation between them (16, 17).
While the C. dubliniensis MSP efficiently matched all respective isolates (Fig. 2A), there was no cross-matching with the C. albicans MSP or those of any other species (data not shown). The OTL spectra of all other species were summarized into a single spectrum group and evaluated against the set of six MSP. There were no matches to YOTL with scores of >1.800 (Fig. 2C). Only the spectrum of C. nivariensis matched with the C. glabrata MSP, with a score of 1.732 (at the genus level). Similarly, the testing of a set of 300 consecutive routine bacterial smear spectra did not result in any matches within the YOTL database above the genus-level significance threshold of 1.500 (Fig. 2C).
In-house validation.
For validation of the YOTL database, all yeasts occurring in our routine diagnostics over the following 2 months were prepared by both OTL and extraction, side-by-side and in duplicate. Excluding isolates processed during an initial familiarization phase of 1 week, during which technicians were trained in the OTL protocol, this amounted to 208 C. albicans, 22 C. tropicalis, 15 C. parapsilosis, 54 C. glabrata, 7 C. dubliniensis, and 9 C. krusei isolates, as well as 41 isolates of species not included in YOTL, as determined by the MALDI Biotyper extraction method as the gold standard.
Using the recommended cutoff at a log score value of 2.000, there were no false species identifications using YOTL only or the combined YOTL and MALDI Biotyper databases. The yeast isolates whose species were represented in YOTL scored mostly >2.000 (Fig. 2D). The isolates of those species not represented in YOTL did not produce log score values of >1.5 and thus were not within the relevant range (Fig. 2D). Of the isolates whose species were contained in YOTL, 84% and 88% were correctly identified on the first attempt at the species level using YOTL only or the combined YOTL and MALDI Biotyper databases, respectively (Table 1). In 25 cases, we analyzed whether repeating the analysis after replating or extending the incubation time for plates with small colonies had been successful. Indeed, this was the case in 23 of 25 (92%) cases, achieving an estimated overall level of 96% correct species identifications (Table 1).
TABLE 1.
Overall performance of different databases alone and in combination
| ID levela | 1st attempt with single database |
Combined databases |
||||||
|---|---|---|---|---|---|---|---|---|
| Biotyper |
YOTL |
1st attempt |
2nd attemptb |
|||||
| No. | % | No. | % | No. | % | No. | % | |
| Species | 124 | 39 | 266 | 84 | 278 | 88 | 302 | 96 |
| Genus | 150 | 48 | 46 | 15 | 35 | 11 | 13 | 4 |
| None | 41 | 13 | 3 | 1 | 2 | 1 | 0 | 0 |
ID, identification.
Estimated cumulative positive rate calculated from a test set of 25 isolates.
DISCUSSION
Mass spectra generated from yeasts prepared with the OTL method generally give rise to classifications with score values of <2.000 using the regular MALDI Biotyper database. The addition of our yeast OTL spectra (YOTL) to the regular MALDI Biotyper database significantly improved the recognition of such samples and circumvented the need for specific algorithms to classify low-scoring matches (2). Using the combined YOTL and MALDI Biotyper databases, we achieved 88% correct identifications for the six species on the first attempt and up to 96% (cumulative) when we repeated the analysis after replating the isolates or extending the growth time. Incidentally, on the first attempt, several correct matches were observed at the genus level that were incorrect at the species level. This indicates that once YOTL spectra are incorporated, any other algorithms accepting genus-level identifications as species specific (3, 4, 6–11) must not be used.
In contrast to others (13), our auxiliary YOTL database contained only a single OTL MSP for each of the six most frequently occurring yeasts in clinical specimens. The YOTL database significantly improved the diagnostic workflow of MALDI-TOF MS-based yeast species identifications in our routine laboratory, as shown by our in-house validation. Our procedure of making such OTL reference spectra is technically simple and can be accomplished by most laboratories that are generally familiar with the technology. In the absence of manufacturer-provided OTL spectra, such in-house-generated spectra can be integrated into the database, as long as proper quality control measures are met. Due to the absence of cross-matching at the species level with other closely related yeast species, it can be assumed that such databases can easily be enlarged to encompass other species, if needed. The YOTL spectrum set is freely available upon request.
ACKNOWLEDGMENTS
We thank Agnieszka Goretzki and Iris Iben for ITS sequencing, Madlen Targatz, Jana Lamloumi, Doreen Meinhard, Dunja Koch, and Uta Vasiljevic for conducting the routine MALDI identification process, and Katrin Gunka for critically reading the manuscript.
M.B. did the experiments, M.B., M.W., and O.B. wrote the manuscript, A.E.Z. helped to evaluate results, and U.G., M.W., and O.B. conceived the study.
O.B. has received speaker's fees and travel reimbursements from Bruker Daltonics. The other authors declare no conflicts of interest.
Footnotes
Published ahead of print 17 September 2014
REFERENCES
- 1.Bader O, Weig M, Taverne-Ghadwal L, Lugert R, Gross U, Kuhns M. 2011. Improved clinical laboratory identification of human pathogenic yeasts by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Microbiol. Infect. 17:1359–1365. 10.1111/j.1469-0691.2010.03398.x. [DOI] [PubMed] [Google Scholar]
- 2.Bader O. 2013. MALDI-TOF-MS-based species identification and typing approaches in medical mycology. Proteomics 13:788–799. 10.1002/pmic.201200468. [DOI] [PubMed] [Google Scholar]
- 3.Van Herendael BH, Bruynseels P, Bensaid M, Boekhout T, De Baere T, Surmont I, Mertens AH. 2012. Validation of a modified algorithm for the identification of yeast isolates using matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry (MALDI-TOF MS). Eur. J. Clin. Microbiol. Infect. Dis. 31:841–848. 10.1007/s10096-011-1383-y. [DOI] [PubMed] [Google Scholar]
- 4.Goyer M, Lucchi G, Ducoroy P, Vagner O, Bonnin A, Dalle F. 2012. Optimization of the pre-analytical steps of matrix-assisted laser desorption ionization–time of flight mass spectrometry identification provides a flexible and efficient tool for identification of clinical yeast isolates in medical laboratories. J. Clin. Microbiol. 50:3066–3068. 10.1128/JCM.06381-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goffinet J, Hendrickx M, Gras J, Mullier F, Boreas A, Delmee M, Rodriguez-Villalobos H. 2010. Treatment method for yeast identification with MALDI-TOF MS; presentation M-407/429. 50th Intersci. Conf. Antimicrob. Agents Chemother. (ICAAC), 12 to 15 September 2010, Boston, MA. [Google Scholar]
- 6.Theel ES, Schmitt BH, Hall L, Cunningham SA, Walchak RC, Patel R, Wengenack NL. 2012. Formic acid-based direct, on-plate testing of yeast and Corynebacterium species by Bruker Biotyper matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 50:3093–3095. 10.1128/JCM.01045-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cassagne C, Cella AL, Suchon P, Normand AC, Ranque S, Piarroux R. 2013. Evaluation of four pretreatment procedures for MALDI-TOF MS yeast identification in the routine clinical laboratory. Med. Mycol. 51:371–377. 10.3109/13693786.2012.720720. [DOI] [PubMed] [Google Scholar]
- 8.Steensels D, Verhaegen J, Lagrou K. 2011. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for the identification of bacteria and yeasts in a clinical microbiological laboratory: a review. Acta Clin. Belg. 66:267–273. [DOI] [PubMed] [Google Scholar]
- 9.Pinto A, Halliday C, Zahra M, van Hal S, Olma T, Maszewska K, Iredell JR, Meyer W, Chen SC. 2011. Matrix-assisted laser desorption ionization-time of flight mass spectrometry identification of yeasts is contingent on robust reference spectra. PLoS One 6:e25712. 10.1371/journal.pone.0025712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stevenson LG, Drake SK, Shea YR, Zelazny AM, Murray PR. 2010. Evaluation of matrix-assisted laser desorption ionization–time of flight mass spectrometry for identification of clinically important yeast species. J. Clin. Microbiol. 48:3482–3486. 10.1128/JCM.00687-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dhiman N, Hall L, Wohlfiel SL, Buckwalter SP, Wengenack NL. 2011. Performance and cost analysis of matrix-assisted laser desorption ionization–time of flight mass spectrometry for routine identification of yeast. J. Clin. Microbiol. 49:1614–1616. 10.1128/JCM.02381-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schubert S, Weinert K, Wagner C, Gunzl B, Wieser A, Maier T, Kostrzewa M. 2011. Novel, improved sample preparation for rapid, direct identification from positive blood cultures using matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry. J. Mol. Diagn. 13:701–706. 10.1016/j.jmoldx.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Carolis E, Vella A, Vaccaro L, Torelli R, Posteraro P, Ricciardi W, Sanguinetti M, Posteraro B. 2014. Development and validation of an in-house database for matrix-assisted laser desorption ionization–time of flight mass spectrometry-based yeast identification using a fast protein extraction procedure. J. Clin. Microbiol. 52:1453–1458. 10.1128/JCM.03355-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Posteraro B, De Carolis E, Vella A, Sanguinetti M. 2013. MALDI-TOF mass spectrometry in the clinical mycology laboratory: identification of fungi and beyond. Expert Rev. Proteomics 10:151–164. 10.1586/epr.13.8. [DOI] [PubMed] [Google Scholar]
- 15.Chen YC, Eisner JD, Kattar MM, Rassoulian-Barrett SL, LaFe K, Yarfitz SL, Limaye AP, Cookson BT. 2000. Identification of medically important yeasts using PCR-based detection of DNA sequence polymorphisms in the internal transcribed spacer 2 region of the rRNA genes. J. Clin. Microbiol. 38:2302–2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jensen RH, Arendrup MC. 2011. Candida palmioleophila: characterization of a previously overlooked pathogen and its unique susceptibility profile in comparison with five related species. J. Clin. Microbiol. 49:549–556. 10.1128/JCM.02071-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Desnos-Ollivier M, Ragon M, Robert V, Raoux D, Gantier JC, Dromer F. 2008. Debaryomyces hansenii (Candida famata), a rare human fungal pathogen often misidentified as Pichia guilliermondii (Candida guilliermondii). J. Clin. Microbiol. 46:3237–3242. 10.1128/JCM.01451-08. [DOI] [PMC free article] [PubMed] [Google Scholar]