Abstract
Quantitative lytA real-time PCR (rtPCR) results from nasopharyngeal (NP) swabs distinguish community-acquired pneumococcal pneumonia (CAP) from asymptomatic colonization. The use of an optimized cutoff value improved pneumococcal etiology determination compared to that of traditional diagnostic methods. Here, we compare the utility of lytA rtPCR from induced sputum and from NP swabs. Pneumococcus was considered the cause of CAP in HIV-infected South African adults if blood culture, induced-sputum culture or Gram stain, urine antigen test, or whole-blood lytA rtPCR revealed pneumococcus or if lytA rtPCR from NP swabs gave a result of >8,000 copies/ml. lytA rtPCR was also performed on induced sputum. Pneumococcus was detected by lytA rtPCR from sputum in 149 (67.1%) of 222 patients with available induced sputum, whereas the results of either Gram stain or culture of sputum were positive in 105 of 229 patients (45.9%; P < 0.001). The mean copy numbers from sputum were higher when the sputum cultures were positive than when the sputum cultures were negative (7.9 versus 5.6 log10 copies/ml; P < 0.001). Against the composite diagnostic standard, a cutoff value of 10,000 copies/ml for good-quality sputum lytA rtPCR had a sensitivity of 78.1% and a specificity of 80.0%. This cutoff value performed similarly to the previously identified cutoff value of 8,000 copies/ml for NP swab lytA rtPCR (area under the curve receiver operating characteristic [AUC-ROC], 80.4% for sputum of any quality versus 79.6% for NP swabs). The AUC-ROC for good-quality sputum was 83.2%. Overall, lytA rtPCR performs similarly well on induced sputum as on NP swabs for most patients but performs slightly better if good-quality sputum can be obtained. Due to the ease of specimen collection, NP swabs may be preferable for the diagnosis of pneumococcal pneumonia.
INTRODUCTION
Streptococcus pneumoniae is the leading etiology of community-acquired pneumonia (CAP) in adults (1), including those in Africa (2), but it is believed to be underdiagnosed (3). There is increasing interest in the application of molecular techniques for the diagnosis of respiratory tract infections. Advantages of nonculture techniques include increased sensitivity and shorter turnaround time. We and others previously reported quantitative nasopharyngeal (NP) colonization density as a novel diagnostic tool for determining pneumococcal etiology in HIV-infected adults with pneumonia (4, 5). Testing of sputum samples with quantitative PCR was initially reported with the less specific pneumococcal target ply (6–8). Subsequently, the pneumococcus-specific gene target lytA in sputum samples was very sensitive in comparison to culture results in adult patients with pneumonia from New Zealand (9). Most recently, Stralin et al. performed a direct comparison of sputum and nasopharyngeal aspirates using lytA and another pneumococcus-specific gene target (Spn9802) to identify pneumococcal pneumonia in Swedish adults with quantitative PCR (qPCR). In their study, sputum lytA qPCR had the best sensitivity and specificity with a cutoff value of 105 DNA copies/ml (10). The prevalence and density of pneumococcal nasopharyngeal colonization vary with age, with HIV serostatus, and in different geographic areas (11–13). It is unknown whether pneumococcal loads in sputum vary in different populations, as they have only been reported for adults in high-income countries. However, this information is essential if specific cutoff values are to be generalized for use as diagnostic tools.
In this study, we compared the diagnostic utility of lytA real-time PCR (rtPCR) from different qualities of induced sputum and nasopharyngeal swabs in adults hospitalized with pneumonia in South Africa. Since only 11% of those for whom HIV status was available were HIV uninfected (4), we focused primarily on HIV-infected patients in our analysis.
(This work was presented in part at the 52nd Interscience Conference on Antimicrobial Agents and Chemotherapy, San Francisco, CA, 2012.)
MATERIALS AND METHODS
Details of the studied patient cohort and applied standard microbiological methods were previously described (4). In brief, adult patients (age, ≥18 years) were enrolled on admission to Chris Hani Baragwanath Academic Hospital in Soweto, South Africa, for acute radiologically confirmed community-acquired pneumonia (CAP) between December 2005 and September 2007. Exclusion criteria included active tuberculosis and current tuberculostatic treatment. S. pneumoniae was identified from specimens (i.e., blood, sputum, nasopharyngeal swab) by traditional methods, including colony morphology, optochin susceptibility, and bile solubility. Sputum samples were cultured semiquantitatively by serial plating and scored 1+, 2+, or 3+. The detection limit for pneumococcal culture was 100 CFU/ml. The immunochromatographic BinaxNOW S. pneumoniae test for pneumococcal C-polysaccharide was applied to unconcentrated urine according to the manufacturer's instructions.
NP swabs were obtained from a single nostril with Dacron swabs (Medical Wire and Equipment Co.), placed in 1 ml of skim milk, tryptone, glucose, and glycerin (STGG) medium, and stored at 4°C for ≤12 h before processing. Sputum was induced within 12 h of hospitalization. For this, patients were nebulized using 5 ml of sterile hypertonic (5%) saline via a jet nebulizer attached to wall oxygen at a flow rate of 3 to 5 liters/min. Nebulization was continued for 15 min or until all the solution was nebulized, whichever occurred first. Physiotherapy was performed on the patient's chest by percussion to optimize the mobilization of secretions. Sputum was assessed according to Bartlett's criteria and considered to be of good quality if there were >25 neutrophils and <10 epithelial cells per high-power field; otherwise, it was considered to be of suboptimal quality (14).
Quantitative lytA rtPCR was performed on NP swabs and induced sputum by blinded study personnel as described previously (4). For whole-blood testing, nucleic acids were extracted from 200 μl of whole blood with the QIAamp DNA blood minikit (Qiagen, Netherlands) and eluted in 100 μl of elution buffer. Five microliters of extracted DNA was then tested with a triplex rtPCR for pneumococcus (lytA), Haemophilus influenzae type b, and Staphylococcus aureus with the iQ Multiplex Powermix (Bio-Rad, USA).
The etiology was defined as pneumococcal if S. pneumoniae was detected by blood culture, whole-blood lytA rtPCR, the urinary BinaxNOW test, or sputum culture, if Gram-positive cocci were identified in pairs on Gram stain (composite diagnostic standard), or if any of the above tests or NP swab lytA rtPCR gave a result of >8,000 copies/ml (expanded composite diagnostic standard) (4).
Continuous variables were compared with 2-sided pooled t tests or the Mann-Whitney-Wilcoxon test, as appropriate. Pearson's correlation coefficient was used for correlations between log10-transformed 1-step lytA counts from induced sputum and from NP swabs and for comparisons between different qualities of sputum samples. P values of ≤0.05 were considered significant. Paired t tests were used to compare lytA rtPCR values from sputum and from NP swabs. The optimal cutoff value for the sputum lytA rtPCR was identified based on the best combination of sensitivity and specificity (Youden index) against the composite diagnostic standard. Diagnostic accuracy for detecting pneumococcal pneumonia was calculated with area under the curve receiver operating characteristic (AUC-ROC) curves. To assess the effect of immunological status on the quantitative lytA rtPCR from sputum and NP swabs, Spearman's correlation coefficient (for CD4 counts) and the Mann-Whitney-Wilcoxon test (for the presence or absence of antiretroviral therapy and co-trimoxazole) were used. Analyses were performed with SAS software version 9.2 (SAS Institute, Cary, NC, USA) and OpenEpi.
This study was approved by the ethics committees of the University of the Witwatersrand and Emory University. Each patient provided written informed consent.
RESULTS
Pneumonia was radiologically confirmed in 370 of 514 patients, 320 of whom had a known HIV status. Analyses were restricted to those 280 patients with CAP who were diagnosed with HIV infection, of whom 229 (81.8%) had an induced-sputum sample available for culture and Gram staining. A good-quality sputum sample was obtained from only 48 (17.1%) patients, and 201 (82.9%) patients had suboptimal sputum quality. Induced sputum from 222 (79.3%) patients was available for lytA rtPCR. Demographic data are presented in Table S1 in the supplemental material. The performances of the individual tests in those 222 patients are shown in Table 1. The number of positive results from sputum Gram stain and culture for pneumococcus was significantly higher in patients with good-quality sputum than in patients with sputum of any quality or of suboptimal quality (Table 2). While there was still a trend for a higher number of positive results from good- versus suboptimal-quality sputum with lytA rtPCR, this trend was not significant (Table 2). Qualitative lytA rtPCR was significantly more frequently positive than either positive sputum Gram staining or culture (66.7% versus 41.4%, respectively; P < 0.001) for suboptimal-quality sputum but not significant for good-quality sputum (69.1% versus 62.5%, respectively; P = 0.26; Table 2).
TABLE 1.
Performance of diagnostic pneumococcal assays
| Testa | No. positive/no. tested (%)a |
|---|---|
| Composite diagnostic standard | 81/222 (36.5) |
| Expanded composite diagnostic standard | 126/222 (56.8) |
| Blood culture | 19/214 (8.9) |
| Urine BinaxNOW | 43/204 (21.1) |
| Sputum culture | |
| Good quality | 19/42 (45.2) |
| Any quality | 77/222 (36.5) |
| Gram stain | |
| Good quality | 19/42 (45.2) |
| Any quality | 45/222 (20.3) |
| lytA rtPCR | |
| Sputum | 149/222 (67.1) |
| NP swab (>8,000 copies/ml) | 109/222 (49.1) |
| Whole blood | 44/222 (19.8) |
Positive test denotes growth of pneumococcus from culture, visualization of Gram-positive diplococci from Gram stain, and detection of lytA by rtPCR from whole blood and detection of lytA in rtPCR at fewer than 35 cycles (i.e., cycle threshold [CT] < 35) from sputum. Results are shown for the 222 patients with available sputum and nasopharyngeal (NP) swab for lytA rtPCR. The composite diagnostic standard was considered positive if blood culture, induced-sputum culture or Gram stain, urine antigen, or whole-blood lytA rtPCR revealed pneumococci. The result for the expanded composite diagnostic standard was considered positive if blood culture, induced sputum culture or Gram stain, urine antigen, or whole-blood lytA rtPCR revealed pneumococcus or if NP swab lytA rtPCR was >8,000 copies/ml.
TABLE 2.
Performance of diagnostic assays on sputum
| Assay type | No. of positive results/total no. (%)a from sputum of: |
Pb | Pc | ||
|---|---|---|---|---|---|
| Any quality | Good quality | Suboptimal quality | |||
| Culture | 77/229 (33.6) | 21/48 (43.8) | 56/181 (30.9) | 0.19 | 0.10 |
| Gram stain | 51/229 (22.3) | 22/48 (45.8) | 29/181 (16.0) | <0.001 | <0.001 |
| Culture or Gram stain | 105/229 (45.9)d | 30/48 (62.5)e | 75/181 (41.4)f | 0.04 | 0.01 |
| lytA rtPCR | 149/222 (67.1)d | 29/42 (69.1)e | 120/180 (66.7)f | 0.82 | 0.78 |
| lytA rtPCR (highly positive [i.e., CT < 35]) | 116/222 (52.3) | 27/42 (64.3) | 89/180 (49.4) | 0.16 | 0.09 |
Positive test denotes growth of pneumococci from culture, visualization of Gram-positive diplococci from Gram stain and detection of lytA by rtPCR, and detection of lytA rtPCR at fewer than 35 cycles (i.e., cycle threshold [CT] < 35), respectively.
Comparing good-quality with any-quality sputum.
Comparing good-quality with suboptimal-quality sputum.
Comparing qualitative lytA rtPCR with either culture or Gram stain (P < 0.001).
Comparing qualitative lytA rtPCR with either culture or Gram stain (P = 0.26).
Comparing qualitative lytA rtPCR with either culture or Gram stain (P < 0.001).
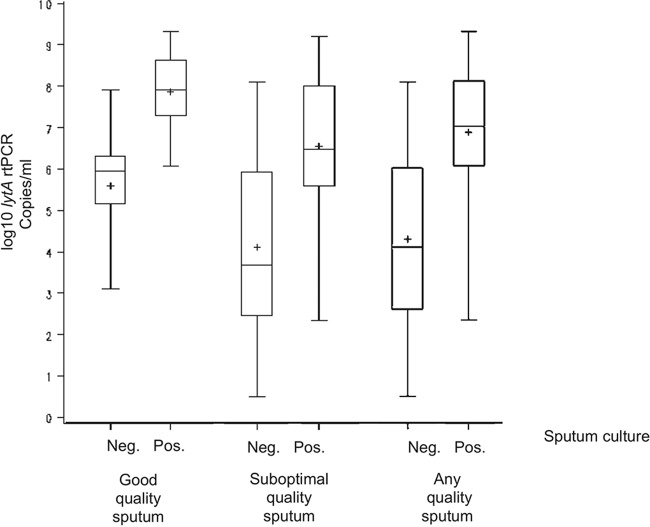
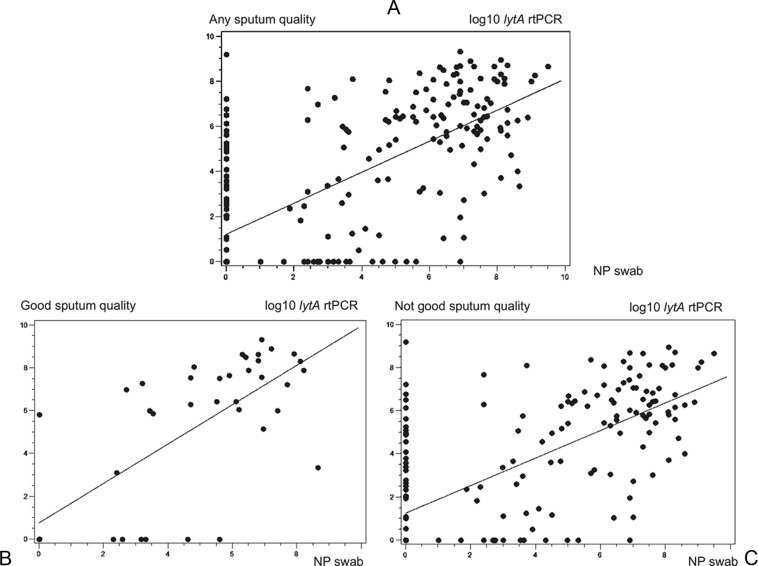
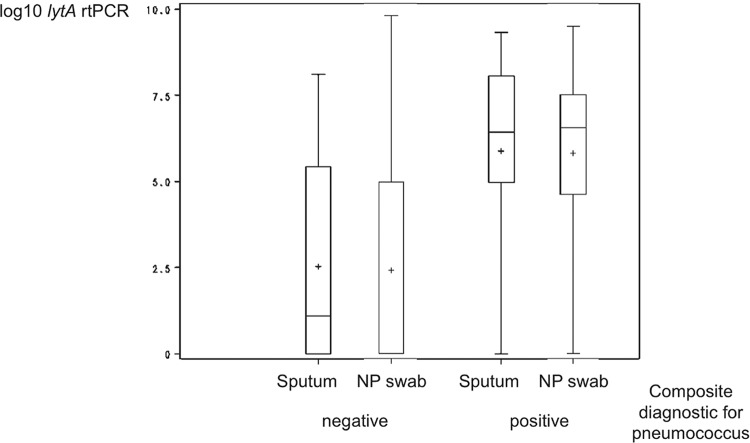
Patients with a positive sputum culture had significantly higher genomic bacterial loads (lytA rtPCR) from sputum (for good-quality or suboptimal-quality sputum) than those with a negative sputum culture (P < 0.001, P = 0.008, and P = 0.002, respectively; Fig. 1). The mean pneumococcal density as measured by lytA rtPCR was more than 1 log higher in patients with good-quality sputum than in those with suboptimal-quality or any quality sputum, both for patients with a positive pneumococcal sputum culture (7.9 versus 6.6 versus 6.9 log10/ml, respectively) and for patients without pneumococcus in the sputum culture (5.6 versus 4.1 versus 4.3 log10/ml, respectively). There was a good correlation between quantitative genomic bacterial loads (lytA rtPCR) from sputum and from NP swabs (r = 0.68 and P < 0.001 for sputum of any quality; r = 0.72 and P < 0.001 for good-quality sputum; r = 0.68 and P < 0.001 for suboptimal-quality sputum) (Fig. 2A to C). Overall and among patients with or without a pneumococcal etiology based on the composite diagnostic standard, there were no significant differences between quantitative lytA rtPCR values for sputum and NP swabs (P = 0.76, P = 0.85, P = 0.81, respectively; Fig. 3).
FIG 1.
Quantitative lytA rtPCR results in relation to those of culture in patients with different qualities of sputum. Plus signs represent means; lengths of boxes, interquartile ranges between the 25th and 75th percentiles; horizontal lines in boxes, medians; and whiskers, minimum and maximum values.
FIG 2.
Correlation between quantitative lytA rtPCR results from sputum and from nasopharyngeal swabs.
FIG 3.
Quantitative lytA rtPCR from sputum and nasopharyngeal swabs in relation to pneumococcal and nonpneumococcal pneumonia. Plus signs represent means; lengths of boxes, interquartile ranges between the 25th and 75th percentiles; horizontal lines in boxes, medians; and whiskers, minimum and maximum values. NP, nasopharyngeal; composite diagnostic standard, positive blood culture, induced-sputum culture or Gram stain, urine antigen test, or whole-blood lytA rtPCR.
The optimal cutoff value for sputum lytA rtPCR was identified as 10,000 copies/ml. Using this cutoff value, good-quality sputum had a sensitivity of 78.1% and a specificity of 80.0% for detecting pneumococcal etiology (expanded composite diagnostic standard), while suboptimal-quality sputum had a slightly lower sensitivity (73.4%; see Table S2 in the supplemental material). The diagnostic performance of this sputum cutoff value in determining pneumococcal etiology was compared to that of the previously proposed cutoff value for NP swabs (>8,000 copies/ml for lytA rtPCR) against the composite diagnostic standard (i.e., without the lytA rtPCR > 8,000 copies/ml from NP swabs criterion) in the 222 patients who had both tests performed (4). The sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and additional yield over the composite diagnostic were similar for the sputum and the NP swab cutoff values regardless of whether any-quality, suboptimal-quality, or only good-quality sputum was assessed. Sputum of suboptimal quality resulted in a lower positive predictive value compared to that of good-quality sputum (Table 3). The lytA rtPCR from good-quality sputum had a significantly higher PPV (P = 0.004) but also a significantly lower NPV (P = 0.015) than the sputum of suboptimal quality (Table 3).
TABLE 3.
Comparison between genomic loads measured by lytA rtPCR from sputum and from nasopharyngeal swabs
| Performance | No. of lytA rtPCR results of >10,000/ml/total no. (%) in sputum ofa: |
No. of NP swab lytA rtPCR results of >8,000/ml/total no. (%) (n = 222) | ||
|---|---|---|---|---|
| Any quality (n = 222) | Good quality (n = 42) | Suboptimal quality (n = 180) | ||
| Sensitivity | 63/81 (77.8) | 22/28 (78.6) | 41/53 (77.4) | 64/81 (79.0) |
| Specificity | 94/141 (66.7) | 9/14 (64.3) | 85/127 (66.9) | 96/141 (68.1) |
| PPVb | 63/110 (57.3) | 22/27 (81.5) | 41/83 (49.4) | 64/109 (58.7) |
| NPV | 94/112 (83.9) | 9/15 (60.0) | 85/97 (87.6) | 96/113 (85.0) |
| Additional yield | 47/141 (33.3) | 5/14 (35.7) | 42/127 (32.3) | 45/141 (31.9) |
Composite diagnostic standard: positive blood culture, induced-sputum culture or Gram stain, urine antigen, or whole-blood lytA rtPCR; for this comparison, the cutoff value of >8,000 copies/ml for NP swab lytA rtPCR criterion was not included in the composite diagnostic.
PPV, positive predictive value; NPV, negative predictive value; additional yield, 1−specificity (i.e., patients with sputum lytA rtPCR cutoff value of >10,000/ml or NPS lytA rtPCR cutoff value of >8,000/ml and a negative composite diagnostic standard represent false positives).
The diagnostic accuracies (AUC-ROC) for lytA rtPCR were 83.2% for good-quality sputum, 79.9% for suboptimal-quality sputum, 80.4% for any-quality sputum, and 79.6% for NP swabs (see Fig. S1 in the supplemental material).
Of the 35 patients who were HIV uninfected and had induced-sputum and NP swab samples available for lytA rtPCR, the lytA rtPCR results from sputum were positive for 65.7% of them. Comparisons of lytA rtPCR results from sputum and NP swabs for HIV-uninfected patients are shown in Table S3 in the supplemental material.
None of the variables examined (CD4 count, current antiretroviral therapy, and current co-trimoxazole prophylaxis) were significantly predictive of lytA copy numbers from sputum (r = 0.05 and P = 0.48, z = 0.59 and P = 0.56, and z = 1.59 and P = 0.11, respectively) or from NP swabs (r = 0.05 and P = 0.46, z = −0.24 and P = 0.81, and z = −1.06 and P = 0.29, respectively).
DISCUSSION
This study shows that pneumococcal genomic loads from induced sputum and from NP swabs had similar diagnostic accuracies of approximately 80% in diagnosing pneumococcal pneumonia in HIV-infected South African adults. lytA rtPCR performs slightly better on good-quality sputum specimens, which were obtained from only a minority of patients. The optimal cutoff values for sputum (10,000 copies/ml) and NP swabs (8,000 copies/ml) were almost identical and resulted in comparable diagnostic performances in detecting pneumococcal etiology in patients with pneumonia.
While the diagnostic accuracy of our optimal sputum lytA rtPCR cutoff value was 83% in patients with good-quality sputum, it was still lower than that recently published by Stralin et al. in a Swedish pneumonia cohort (10). Of note, their optimal cutoff value was comparable to and only 1 log higher than ours and within the same range as that reported from a New Zealand study by Werno et al. (9). The most likely explanation for this slight difference is the different patient populations and the sputum being induced in our study; however, differences in extraction and rtPCR methods (as described in reference 4) cannot be ruled out. A limitation of all these studies is that relatively few patients had good-quality sputum available, which reflects the difficulties not only in clinical practice but also during research studies to obtain sufficiently good-quality sputum from pneumonia patients. However, in our study, there was only a relatively small difference in the sputum lytA rtPCR performance between patients with good-quality sputum and those with sputum of any quality. The higher PPV and lower NPV of good-quality sputum compared to those of suboptimal-quality sputum indicate that the benefit of obtaining a good-quality sputum sample lies in more true positives at the cost of more false negatives. This observation of minor differences is therefore of practical importance, as it would also allow for the testing of suboptimal-quality sputum samples with lytA rtPCR to reliably diagnose pneumococcal pneumonia. One might speculate that without sputum induction, there would be an even greater proportion of patients with suboptimal sputum quality. Our results indirectly suggest that lytA rtPCR can be applied to those specimens as well, as shown by Stralin, who used expectorated sputum and likely had a higher proportion of suboptimal-quality samples (10). We initially used NP swabs as a diagnostic tool to diagnose pneumococcal pneumonia, in part because of the ease of obtaining these noninvasive specimens and because many pneumonia patients have a dry cough and are unable to produce sputum. Musher et al. demonstrated that 31 of 105 patients with pneumococcal pneumonia were unable to provide a sputum sample, and the delivered specimens from another 15 patients were inadequate by their criteria (15), resulting in 56% with adequate sputum samples, which is still higher than the proportions in our and Stralin's studies.
Reasons for the worse performance of the sputum and NP assays in our study compared to that in Stralin's study may relate primarily to the study populations; our patients were young (mean age, 36.5 ± 9.7 years) HIV-infected South African adults with a relatively higher mortality rate (13.3%) (4), and there was a high proportion of coinfections with Mycobacterium tuberculosis (our unpublished data) in our study, whereas the Swedish population was much older (median age, 71 years) and had a lower mortality rate (2.6%) (16). Interestingly, the poorer performance in our population was despite using induced sputum. In HIV-uninfected patients, the lytA rtPCR results from sputum were positive in 65.7%, which was a proportion similar to that in HIV-infected patients. In HIV-uninfected patients, lytA rtPCR results from sputum had a higher sensitivity but a lower specificity compared to those from NP swabs, but this comparison was limited by the small number of patients. A limitation is that the majority of the patients in our cohort were HIV-infected adults from a single site in South Africa. However, it is reassuring that other studies found similar cutoff values or performance of rtPCR in other HIV-uninfected adults (5, 6, 9). As pneumococci are found in NP swabs from healthy young children with a higher frequency than in adults (11), and although density was higher in radiologically confirmed pneumonia cases than in controls, no useful cutoff value was found for NP colonization density in Vietnamese children to use for diagnosing pneumococcal pneumonia (17), so our results cannot be directly applied to children.
In addition, the optimal lytA rtPCR cutoff values for NP specimens differed in the two studies, with 8,000 copies/ml for NP swabs in our study (4) and 102 copies/ml for NP aspirates in Stralin's study (10). Even though there might be some dilution with aspirates compared to swabs, this alone does not explain an almost 3-log difference in the cutoff values. However, a previous study by Stralin's group reported an optimal cutoff value of 104 copies/ml for the Spn9802 rtPCR from NP aspirates (5). In addition, in our study, not only were the performances of lytA rtPCR from sputum and NP swabs comparable for identifying a pneumococcal etiology, but the optimal cutoff values were also very similar, and in individual patients there was a good correlation between sputum and NP swab lytA rtPCR results. This suggests that, at least in our population, what specimen to choose might depend on the convenience of specimen collection.
Due to the absence of accepted gold standards for pneumococcal diagnosis, the specificity of an assay is difficult to interpret. Instead, latent class analysis using multiple diagnostic tests (without molecular assays) has shown that the true pneumococcal etiology in adult pneumonia in Kenya was approximately 46% (18), which is roughly the yield of our composite diagnostic (36.5%) plus lytA from sputum (total pneumococcal etiology, 58.7%) or NP swabs (total pneumococcal etiology, 56.8%). Therefore, we believe that these tests, rather than lacking specificity, indeed have an additional yield beyond the standard diagnostic tests.
In conclusion, our study generally confirms the very promising diagnostic value of lytA rtPCR from sputum but with slightly poorer diagnostic performance as reported by Stralin et al. Among HIV-infected South African adults, its performance varied little in relation to quality of the induced-sputum sample, which may make this test particularly appealing, not only for research studies but also for clinical routine, probably without the need for induced sputum. However, we also did not find it superior to lytA rtPCR from NP swabs, which might be an even easier specimen to collect and might provide similar performance as a diagnostic tool. Confirmation of our results should be sought in additional international pneumonia studies.
Supplementary Material
ACKNOWLEDGMENTS
The C-polysaccharide antigen BinaxNOW S. pneumoniae test was provided free of charge by Binax.
We are indebted to Ping Zhao, Mini Sidhu, and Michael W. Pride of Pfizer Vaccine Research for expert technical assistance in performing the lytA rtPCR from sputum and NP swabs.
W.C.A. received an honorarium from GlaxoSmithKline (GSK) and support from BRAHMS Thermo Fisher and bioMérieux to attend meetings and fulfilled speaking engagements. S.A.M. received research funding and honoraria from Pfizer Vaccines and GSK and institutional grant support from Wyeth. K.P.K. received consulting and research funding from Pfizer Vaccines and consulting funding from GSK. The other authors report no conflicts of interest.
K.P.K. received funding from the Center for AIDS Research/NIH (P30 A1050409).
Footnotes
Published ahead of print 24 September 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.01553-14.
REFERENCES
- 1. Woodhead M, Blasi F, Ewig S, Garau J, Huchon G, Ieven M, Ortqvist A, Schaberg T, Torres A, van der Heijden G, Read R, Verheij TJ. 2011. Guidelines for the management of adult lower respiratory tract infections—full version. Clin. Microbiol. Infect. 17(Suppl):E1–E59. 10.1111/j.1469-0691.2011.03672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Scott JA, Hall AJ, Muyodi C, Lowe B, Ross M, Chohan B, Mandaliya K, Getambu E, Gleeson F, Drobniewski F, Marsh K. 2000. Aetiology, outcome, and risk factors for mortality among adults with acute pneumonia in Kenya. Lancet 355:1225–1230. 10.1016/S0140-6736(00)02089-4. [DOI] [PubMed] [Google Scholar]
- 3. Bartlett JG. 2011. Diagnostic tests for agents of community-acquired pneumonia. Clin. Infect. Dis. 52(Suppl):S296–S304. 10.1093/cid/cir045. [DOI] [PubMed] [Google Scholar]
- 4. Albrich WC, Madhi SA, Adrian PV, van Niekerk N, Mareletsi T, Cutland C, Wong M, Khoosal M, Karstaedt A, Zhao P, Deatly A, Sidhu M, Jansen KU, Klugman KP. 2012. Use of a rapid test of pneumococcal colonization density to diagnose pneumococcal pneumonia. Clin. Infect. Dis. 54:601–609. 10.1093/cid/cir859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Abdeldaim GM, Stralin K, Olcen P, Blomberg J, Herrmann B. 2008. Toward a quantitative DNA-based definition of pneumococcal pneumonia: a comparison of Streptococcus pneumoniae target genes, with special reference to the Spn9802 fragment. Diagn. Microbiol. Infect. Dis. 60:143–150. 10.1016/j.diagmicrobio.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 6. Yang S, Lin S, Khalil A, Gaydos C, Nuemberger E, Juan G, Hardick J, Bartlett JG, Auwaerter PG, Rothman RE. 2005. Quantitative PCR assay using sputum samples for rapid diagnosis of pneumococcal pneumonia in adult emergency department patients. J. Clin. Microbiol. 43:3221–3226. 10.1128/JCM.43.7.3221-3226.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Murdoch DR, Anderson TP, Beynon KA, Chua A, Fleming AM, Laing RT, Town GI, Mills GD, Chambers ST, Jennings LC. 2003. Evaluation of a PCR assay for detection of Streptococcus pneumoniae in respiratory and nonrespiratory samples from adults with community-acquired pneumonia. J. Clin. Microbiol. 41:63–66. 10.1128/JCM.41.1.63-66.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Abdeldaim G, Herrmann B, Korsgaard J, Olcen P, Blomberg J, Stralin K. 2009. Is quantitative PCR for the pneumolysin (ply) gene useful for detection of pneumococcal lower respiratory tract infection? Clin. Microbiol. Infect. 15(6):565–570. 10.1111/j.1469-0691.2009.02714.x. [DOI] [PubMed] [Google Scholar]
- 9. Werno AM, Anderson TP, Murdoch DR. 2012. Association between pneumococcal load and disease severity in adults with pneumonia. J. Med. Microbiol. 61:1129–1135. 10.1099/jmm.0.044107-0. [DOI] [PubMed] [Google Scholar]
- 10. Stralin K, Herrmann B, Abdeldaim G, Olcen P, Holmberg H, Molling P. 2014. Comparison of sputum and nasopharyngeal aspirate samples and of the PCR gene targets lytA and Spn9802 for quantitative PCR for rapid detection of pneumococcal pneumonia. J. Clin. Microbiol. 52:83–89. 10.1128/JCM.01742-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bogaert D, De Groot R, Hermans PW. 2004. Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect. Dis. 4:144–154. 10.1016/S1473-3099(04)00938-7. [DOI] [PubMed] [Google Scholar]
- 12. Gill CJ, Mwanakasale V, Fox MP, Chilengi R, Tembo M, Nsofwa M, Chalwe V, Mwananyanda L, Mukwamataba D, Malilwe B, Champo D, Macleod WB, Thea DM, Hamer DH. 2008. Impact of human immunodeficiency virus infection on Streptococcus pneumoniae colonization and seroepidemiology among Zambian women. J. Infect. Dis. 197:1000–1005. 10.1086/528806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Roca A, Bottomley C, Hill PC, Bojang A, Egere U, Antonio M, Darboe O, Greenwood BM, Adegbola RA. 2012. Effect of age and vaccination with a pneumococcal conjugate vaccine on the density of pneumococcal nasopharyngeal carriage. Clin. Infect. Dis. 55:816–824. 10.1093/cid/cis554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bartlett RC. 1974. Medical microbiology: quality, cost, and clinical relevance. John Wiley & Sons, New York, NY. [Google Scholar]
- 15. Musher DM, Montoya R, Wanahita A. 2004. Diagnostic value of microscopic examination of Gram-stained sputum and sputum cultures in patients with bacteremic pneumococcal pneumonia. Clin. Infect. Dis. 39:165–169. 10.1086/421497. [DOI] [PubMed] [Google Scholar]
- 16. Stralin K, Olcen P, Tornqvist E, Holmberg H. 2010. Definite, probable, and possible bacterial aetiologies of community-acquired pneumonia at different CRB-65 scores. Scand. J. Infect. Dis. 42:426–434. 10.3109/00365540903552353. [DOI] [PubMed] [Google Scholar]
- 17. Vu HTT, Yoshida LM, Suzuki M, Nguyen HAT, Nguyen CDL, Nguyen ATT, Oishi K, Yamamoto T, Watanabe K, Vu TD, SChmidt W-P, Phan HTL, Morimoto K, Le TH, Yanai H, Kilgore PE, Dang AD, Ariyoshi K. 2011. Association between nasopharyngeal load of Streptococcus pneumoniae, viral coinfection, and radiologically confirmed pneumonia in Vietnamese children. Pediatr. Infect. Dis. J. 30:11–18. 10.1097/INF.0b013e3181f111a2. [DOI] [PubMed] [Google Scholar]
- 18. Jokinen J, Scott JA. 2010. Estimating the proportion of pneumonia attributable to pneumococcus in Kenyan adults: latent class analysis. Epidemiology 21:719–725. 10.1097/EDE.0b013e3181e4c4d5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.