Abstract
Despite the availability of standard methods for pneumococcal serotyping, there is room for improvement in the available methods, in terms of throughput, multiplexing capacity, and the number of serotypes identified. We describe a target enrichment-based next-generation sequencing method applied to nasopharyngeal samples for direct detection and serogroup prediction of all known serotypes of Streptococcus pneumoniae, 32 to the serotype level and the rest to the closely related serogroup level. The method was applied to detect and to predict the serogroups of pneumococci directly in clinical samples and from sweeps of primary culture DNA, with increased detection rates versus culture-based identification and agreement with the serotypes/serogroups determined by conventional serotyping methods. We propose this method, in conjunction with traditional serotyping methods, as an alternative to rapid detection and serotyping of pneumococci.
INTRODUCTION
The introduction of conjugate vaccination has dramatically altered the prevalence and community structure of pneumococcal serotypes, in both disease and carriage (1). As the serotype valency of conjugate vaccines increased from 7 to 13, the serotypes and their prevalence were subjected to dynamic changes to less common serotypes. Thus, there is a need for laboratory methods that are capable of identifying the maximum possible number of serotypes with a limited number of assays, in order to monitor serotype replacement and any emerging serotypes (2).
The Quellung reaction remains the standard method for identification of pneumococcal serotypes. This method is expensive and time-consuming and requires expertise (3). With the sequencing of the capsular biosynthesis loci of all 90 pneumococcal serotypes, new molecular methods for pneumococcal serotyping have been developed (4). The most widely used of these methods remain sequential multiplex PCRs. The Centers for Disease Control and Prevention (CDC) (Atlanta, GA) recommends a set of 8 multiplex PCRs that are capable of differentiating 40 seroidentities, 22 to the serotype level (5) (http://www.cdc.gov/streplab/pcr.html). More recently, a set of seven real-time PCR assays capable of differentiating 21 serotypes was also recommended by the CDC (6). However, the number of tests to be performed still remains relatively high, due to the limited multiplexing capabilities associated with gel-based differentiation and quencher dye combinations. Thus, there is a need for alternative serotyping methods capable of differentiating the greatest number of serotypes at least to closely related serogroups, with a minimal number of assays, in a rapid and cost-effective manner.
Next-generation sequencing (NGS) is an attractive alternative platform for the development of diagnostic methods. The high throughput, the increasingly simple and fast methods for sample preparation, and the ability to pool samples together make these platforms versatile for adaptation. Target enrichment-based sequencing through selective enrichment of the regions of interest enables the use of sequencing reads in a more cost-effective manner. While target enrichment and sequencing are commonly used for the diagnosis of cancers and hereditary diseases, their use in diagnostic microbiology is still emerging (7). We previously used a set of published primers to enrich the common pneumococcal serotypes included in the 23-valent pneumococcal polysaccharide vaccine (PPSV23) (8) and established cutoff values for the interpretation of serogroup/serotype data based on the NGS target reads. However, with the rapid changes in the serotype composition of Streptococcus pneumoniae globally, the methodology needs to be expanded to include other serotypes. We thus extended and validated the NGS protocol to enrich serotype-specific sequences from additional S. pneumoniae serotypes. This enabled the identification and detection of all current serogroups of S. pneumoniae, including 32 at the serotype level. We then applied this methodology to identify and to serotype S. pneumoniae directly from clinical samples from hospitalized children with pneumonia and from sweeps of primary cultures and compared the results with those of the conventional method of culture and serotyping of S. pneumoniae.
MATERIALS AND METHODS
Pneumococcal isolates and capsular typing.
Thirty-eight isolates of pneumococcal serotypes/serogroups included in the second enrichment PCR were used for the validation section. All isolates were serotyped as described previously, with multiplex PCR (5) (http://www.cdc.gov/streplab/pcr.html). DNA from bacterial isolates was prepared by boiling lysis of overnight cultures.
Processing of clinical samples.
Nasopharyngeal aspirate (NPA) samples collected from children hospitalized with pneumonia at a tertiary care pediatric department during a consecutive 9-month period were evaluated. NPA samples were stored in skim milk-glycerol-glucose-tryptone soy broth (STGG) at −80°C, thawed to room temperature, and vortex-mixed for 10 to 20 s. DNA was extracted from 200 μl using the Qiagen DNeasy blood and tissue kit, with modifications suggested by the CDC (http://www.cdc.gov/streplab/downloads/pcr-body-fluid-DNA-extract-strep.pdf). Ten microliters of each NPA sample was cultured on blood agar (BA) plates with gentamicin (5 μg/ml), the samples were incubated in 5% CO2 at 37°C for 24 h, and suspected S. pneumoniae colonies were identified by routine methods described previously (9). Plates with no growth were reincubated for an additional 24 h.
DNA extraction was also performed from sweeps of the primary cultures (sweep culture) as described by Turner et al. (10), with modifications. After a single colony was picked from the primary culture plate, a sweep of the remaining bacterial colonies was suspended in 500 μl to 1 ml of ultrapure water, and the turbidity was adjusted to a McFarland standard of 1. DNA was extracted from this suspension by simple boiling lysis.
Target enrichment-based next-generation sequencing.
The multiplex PCR for target enrichment of extended pneumococcal serotypes contained 33 previously described pairs of primers (11), of which 20 were serotype specific (13, 11F, 16F, 16A, 17A, 21, 23A, 23B, 27, 29, 31, 33C, 34, 35B, 36, 39, 43, 45, 47A, and 48) and 13 were serogroup specific (7B/7C/40, 10F/10A, 11B/11C, 15A/15F, 19B/19C, 24F/24A/24B, 25F/25A/38, 28F/28A, 32F/32A, 33B/33D/33C, 35F/47F, 35A/35C/42, and 41F/41A) (see Table S1 in the supplemental material). An 18-bp nucleotide adaptor was added to the 5′ end of the primers to enable sample pooling (8), and multiplex PCRs were optimized with or without the addition of a pair of primers targeting the streptococcal autolysin gene, with an intervening segment of specific sequence signatures (8), for the identification of S. pneumoniae. The remaining 24 common serotypes (those present in PPSV23 and serotype 6A) were enriched using the previously described PCR (8). The extended enrichment PCR used a 4-μl volume from a primer mixture containing serotype/serogroup-specific primers at 1 μM concentrations, without or with lytA-specific primers at 0.25 μM concentrations, with 2 μl of identified isolate DNA, 4 μl of primary culture DNA, or 8 μl of direct sample DNA as the template in a total reaction mixture of 25 μl, using a Platinum multiplex PCR kit (Life Technologies).
A modified step-out (MSO)-PCR used the sequence of the 18-nucleotide adaptor for the primer, with 10 unique 5-nucleotide indexes selected from an online list (http://cloud.github.com/downloads/faircloth-lab/edittag/edit_metric_tags.txt) at the 5′ end, to enable sample pooling (12, 13). The MSO-PCR used 4 μl of the purified products of multiplex PCR as the template in a 50-μl reaction mixture with other constituents as recommended for ActiTaq polymerase (Life Technologies), for 20 cycles with an annealing temperature of 53°C.
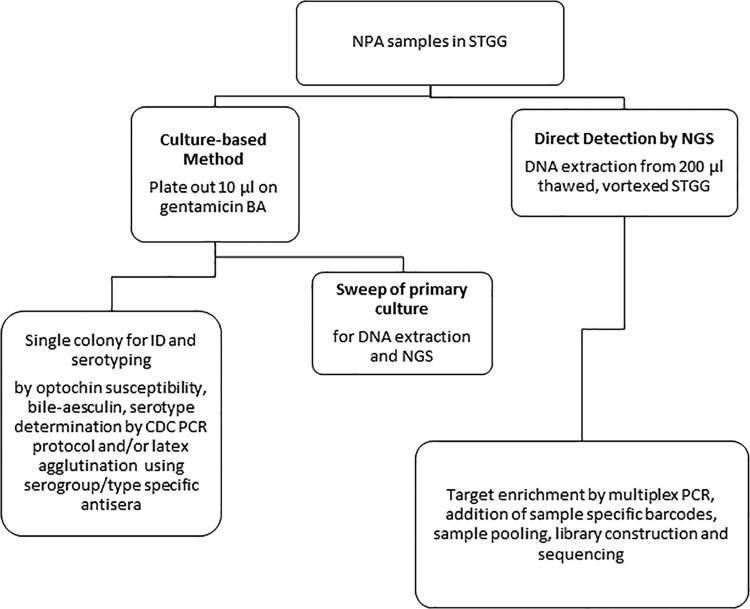
Figure 1 presents the workflow for sample preparation and the methods evaluated. The multiplex PCRs described above and described previously (8) were used in conjunction to determine the identification and pneumococcal serogroups/serotypes from DNA extracted directly from samples and from sweeps of primary cultures.
FIG 1.
Workflow for three methods for S. pneumoniae identification (ID) and prediction of serogroups/serotypes.
Library preparation and sequencing.
MSO-PCR products were analyzed visually for the presence of bands regardless of size, and DNA was purified using the QIAquick PCR product purification kit (Qiagen) and quantified using a Qubit fluorometer (Life Technologies). Purified PCR products from samples with 10 unique barcodes were pooled together in equal quantities to generate a single “index sample,” and library preparation was performed using the TruSeq DNA library preparation kit (version 2; Illumina), according to the manufacturer's instructions. Sequencing was performed with a MiSeq sequencer (Illumina), using 2 by 150-bp sequencing. The paired-end reads obtained from the sequencing run were demultiplexed for Illumina indexes with MiSeq reporter software, followed by quality filtering and demultiplexing for in-house indexes using the FASTX toolkit (http://hannonlab.cshl.edu/fastx_toolkit). Amplicons were aligned with reference sequences mentioned in the articles describing the original primers (11, 14) and with atypical and typical pneumococcal lytA gene sequences (GenBank accession numbers AJ419979.1 and AJ243407.1, respectively).
Confirmation of discrepant results between culture-based and target-based NGS methods.
For samples for which there were discrepancies in S. pneumoniae isolation or multiple serotypes were detected with the NGS method and not culture, the specimens were recultured using a 50-μl inoculum, and up to 20 to 50 colonies were picked for confirmation of the pneumococcal identities and serotyping by conventional methods. Additionally, the original DNA samples were tested with the CDC-recommended PCR, with a second round in which the product of the first round was used as a template.
Interpretation of results.
In the previous study for detection of 23-valent serotypes, we evaluated different criteria with variable stringency to correctly assign serotypes. We found that identification of a serotype for which >500 reads were mapped against the given serotype, accounting for >15% of reads mapped against serotype sequences, was a stringent criterion with 100% correct prediction of serotypes (8). Thus, the same criterion was used in this study. For the identification of S. pneumoniae in samples containing S. pneumoniae with a mixture of viridians streptococci, the percentage of reads mapped against the pneumococcus-specific lytA gene was identified as >10% of the total mapped reads for the given sample. Thus, in the validation using clinical samples versus pneumococcal isolates, only samples for which >10% of total reads were mapped against the typical lytA gene and >500 reads were mapped against serotype sequences were considered to contain pneumococci.
RESULTS
Target enrichment-based NGS detection and prediction of pneumococcal serotypes.
Table 1 shows the detailed results of the pneumococcal serotype/serogroup prediction by target-based NGS. The total numbers of reads mapped against each serotype/serogroup-specific sequence ranged from 1,667 to 18,106, while totals ranged from 1,902 to 18,701 reads in the results with inclusion of the lytA primers for detection of pneumococci. For the results with lytA primers, 316 to 14,760 reads were mapped against the typical lytA gene, which accounted for >10% of total mapped reads for a given sample. Considering all samples, the mean percentages of reads mapped against the correct serotype/serogroup-specific sequence were well above the defined cutoff value of >15% of reads at 80.6% (95% confidence interval [CI], 77.7 to 84.1%) and 80.9% (95% CI, 77.8 to 84.1%) with inclusion of the lytA gene. Applying the criteria derived from the previous study (>15% of all sequence reads and >500 total reads for a particular serotype sequence), all 38 serogroups/serotypes (100%) were correctly identified to the corresponding type; in the reaction including the lytA gene for pneumococcal identification, 37 of the 38 serogroups/serotypes (97.4%) were correctly identified. The remaining sample was also correctly identified to the original serotype but gave an additional serotype match based on the cutoff criterion.
TABLE 1.
Detection of S. pneumoniae and prediction of pneumococcal serotypes by target-based NGS
| Serotype | Serogroup/serotype prediction |
Pneumococcal identification and serogroup/serotype predictiona |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Total no. of reads mapped against serotype sequences | % of reads mapped against correct serotype | % of reads mapped against second match | Serotype(s) determined by target-based NGS | Total no. of reads mapped against serotype sequences | % of reads mapped against correct serotypeb | % of reads mapped against second matchb | No. of reads mapped for typical lytA | No. of reads mapped for atypical lytA | Serotype(s) determined by target-based NGS | |
| 7B | 1,667 | 74.8 | 7.7 | 7B/7C/40 | 3,700 | 72.4 | 11.1 | 4,830 | 0 | 7B/7C/40 |
| 7C | 7,036 | 64.8 | 9.6 | 7B/7C/40 | 6,601 | 65.1 | 14.8 | 10,432 | 0 | 7B/7C/40 |
| 10C | 14,929 | 78.1 | 6.8 | 10C/10F | 15,278 | 80.5 | 7.8 | 11,564 | 0 | 10C/10F |
| 10F | 7,382 | 60.3 | 12.7 | 10C/10F | 7,989 | 66.8 | 15.9 | 11,992 | 0 | 10C/10F (15A/15F) |
| 15F | 4,370 | 93 | 2.6 | 15A/15F | 9,554 | 94.9 | 9.1 | 3,385 | 0 | 15A/15F |
| 13 | 5,426 | 75.1 | 8 | 13 | 6,050 | 83.3 | 9.1 | 9,858 | 0 | 13 |
| 15A | 7,897 | 81.9 | 12.1 | 15A/15F | 18,701 | 95.7 | 1.8 | 7,575 | 0 | 15A/15F |
| 16F | 6,286 | 82.6 | 6.5 | 16F | 7,819 | 87.3 | 5 | 4,173 | 0 | 16F |
| 39 | 9,738 | 96.1 | 1.4 | 39 | 3,974 | 96.7 | 1.1 | 1,515 | 0 | 39 |
| 17A | 2,721 | 75.5 | 4.2 | 17A | 4,556 | 80.5 | 4 | 4,710 | 0 | 17A |
| 19B/19C | 8,756 | 85.2 | 3 | 19B/19C | 6,179 | 80.9 | 4.2 | 5,249 | 0 | 19B/19C |
| 21 | 10,255 | 69.5 | 8.6 | 21 | 9,786 | 80.2 | 5.4 | 14,760 | 0 | 21 |
| 23A | 6,086 | 69.8 | 8.7 | 23A | 7,970 | 82.1 | 5.4 | 18,645 | 0 | 23A |
| 24F | 4,247 | 90 | 2.2 | 24A/24B/24F | 1,902 | 64.7 | 8.8 | 6,025 | 0 | 24A/24B/24F |
| 23B | 6,992 | 70.8 | 12.6 | 23B | 11,687 | 88.9 | 2.9 | 6,502 | 0 | 23B |
| 25A | 6,677 | 81 | 6.2 | 25A/24F/38 | 5,269 | 62.3 | 9.2 | 10,103 | 0 | 25A/24F/38 |
| 27 | 2,353 | 88.4 | 2.9 | 27 | 1,914 | 82.3 | 6.3 | 1,805 | 0 | 27 |
| 35A | 2,549 | 73.4 | 8.5 | 35A/35C/42 | 1,938 | 76.7 | 5.1 | 2,254 | 0 | 35A/35C/42 |
| 28F | 10,356 | 76.1 | 8.4 | 28A/28F | 13,502 | 80.1 | 4.7 | 4,696 | 0 | 28A/28F |
| 29 | 10,893 | 76.2 | 3.8 | 29 | 11,204 | 76.4 | 5.7 | 10,271 | 0 | 29 |
| 31 | 13,968 | 89.5 | 3.8 | 31 | 6,383 | 80.2 | 5.6 | 4,251 | 0 | 31 |
| 33B | 4,163 | 97.1 | 0.1 | 33B/33C/33D | 1,362 | 88.9 | 3.2 | 1,163 | 0 | 33B/33C/33D |
| 32A | 4,796 | 94.8 | 1.7 | 32A/32F | 6,976 | 93.0 | 2.2 | 847 | 0 | 32A/32F |
| 33C | 4,678 | 52.7 | 4.2 | 33B/33C/33D | 3,470 | 72.3 | 12.9 | 4,833 | 0 | 33B/33C/33D |
| 34 | 14,260 | 99 | 0.3 | 34 | 4,117 | 94.1 | 2 | 6,032 | 0 | 34 |
| 41F | 6,475 | 85.3 | 3.2 | 41A/41F | 2,114 | 67.6 | 8.8 | 316 | 0 | 41A/41F |
| 35B | 18,106 | 78 | 6.1 | 35B | 12,078 | 81.0 | 6 | 9,415 | 0 | 35B |
| 35F | 15,598 | 81.2 | 4.2 | 35F/47F | 13,292 | 85.0 | 3.4 | 3,507 | 0 | 35F/47F |
| 36 | 8,061 | 73.9 | 8.4 | 36 | 5,014 | 68.0 | 11.2 | 12,799 | 0 | 36 |
| 41F | 3,117 | 88.2 | 4.2 | 41A/41F | 3,227 | 83.6 | 5.4 | 1,960 | 0 | 41A/41F |
| 28A | 8,378 | 85.6 | 4.7 | 28A/28F | 5,028 | 88.6 | 3.1 | 6,341 | 0 | 28A/28F |
| 40 | 4,260 | 75.6 | 10.5 | 7B/7C/40 | 3,723 | 74.1 | 11.3 | 4,045 | 0 | 7B/7C/40 |
| 48 | 10,193 | 96.3 | 0.1 | 48 | 8,084 | 96.6 | 0.7 | 3,247 | 0 | 48 |
With inclusion of lytA primers for pneumococcal identification.
Percentage of reads against serotype sequences for the given sample.
Comparison of serotypes determined by target-based NGS versus conventional culture-based serotyping.
Of 155 respiratory samples, 22.6% of the samples (35 samples) yielded S. pneumoniae in culture. The serotypes of these isolates are listed in Table 2. DNA was prepared from sweeps of bacterial colonies from 81 of 155 samples that revealed bacterial growth in the primary cultures. Of these, 40 samples were positive after PCR enrichment and were subjected to sequencing by NGS, and the results are presented in Table 2. Of the 40 samples, 38 fulfilled the criteria for pneumococcal identification, giving a pneumococcal positivity rate of 24.5%. Thirty-seven of these samples gave predicted serotypes; 36 samples contained a single serotype with >500 reads mapped against a serotype accounting for >15% of reads, while one sample (sample 27) had two serotypes predicted based on the same criteria for serotype allocation. One sample (sample 95) was identified as containing S. pneumoniae based on the lytA criteria but did not fulfill the criteria for serotype allocation.
TABLE 2.
Detailed results for pneumococcal identification and serotype prediction in respiratory samples
| Sample no. | Serotype(s) determined by culture | Sweep of primary culture |
Direct detection by NGS |
Pneumococcal identification and serotype prediction |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total no. of reads mapped against serotype sequences | Serotype(s)/serogroup(s) with greatest proportion of reads mapped | % of reads mapped against correct serotypea | % of reads mapped against second matcha | % of reads mapped against pneumococcal lytA sequenceb | Total no. of reads mapped against serotype sequences | Serotype(s)/serogroup(s) with greatest proportion of reads mapped | % of reads mapped against correct serotypea | % of reads mapped against second matcha | % of reads mapped against pneumococcal lytA sequenceb | Sweep of primary culture | Direct detection by NGS | ||
| 1 | Oral flora | 5,167 | 39 | 38.5 | 29.8 (9N/9L) and 27.3 (19B/19C) | 3.0 | 2,221 | 9N/9L | 34.8 | 30.1 (39) and 19 (19B/19C) | 8.8 | No pneumococci | No pneumococci |
| 6 | 15A/15F | 6,175 | 15A/15F | 91.5 | 5.7 | 43.2 | 9,005 | 15A/15F | 82.9 | 7.9 | 50.5 | 15A/15F | 15A/15F |
| 9 | 19F | 6,012 | 19F | 97.4 | 0.9 | 31.4 | 4,578 | 19F | 80.9 | 8.8 | 73.9 | 19F | 19F |
| 22 | 3 | 3,691 | 3 | 85.2 | 6.1 | 76.2 | 2,789 | 3 | 69.7 | 16.6 (23A) | 80.9 | 3 | 3 |
| 23 | 23A | 7,049 | 23A | 97.6 | 1.2 | 35.9 | 6,904 | 23A | 87.1 | 2.2 | 26.2 | 23A | 23A |
| 25 | 23A | 1,166 | 23A | 87.3 | 5.2 | 71.4 | 3,535 | 23A | 91.5 | 5.6 | 62.6 | 23A | 23A |
| 26 | 6D | 3,461 | Sgc 6 | 95.7 | 2.2 | 43.9 | 8,691 | Sg 6 | 94.8 | 3.3 | 40.5 | Sg 6 | Sg 6 |
| 27 | 6A | 3,370 | Sg 6 | 74.2 | 19.3 (34) | 80.1 | 3,595 | Sg 6 | 92.0 | 5 | 54.9 | Sg 6 with minor cocolonizer | Sg 6 |
| 28 | 15B/15C | 2,000 | 15B/15C | 93.4 | 2.2 | 64.5 | 2,562 | 15B/15C | 87.6 | 9 | 63.8 | 15B/15C | 15B/15C |
| 29 | 23A | 8,213 | 23A | 97.7 | 1.1 | 34.6 | 7,663 | 23A | 97.0 | 0.7 | 39.9 | 23A | 23A |
| 30 | 3 | 1,511 | 3 | 86.1 | 3.4 | 68.7 | 2,278 | 3 | 88.3 | 4.8 | 71.7 | 3 | 3 |
| 32 | 6A | 2,292 | Sg 6 | 95.1 | 2.2 | 59.4 | 2,916 | Sg 6 | 66.9 | 14.7 | 30.9 | Sg 6 | Sg 6 |
| 33 | 23A | 1,459 | 23A | 85.4 | 7.1 | 77.6 | 7,900 | 23A | 86.2 | 8.4 | 69.3 | 23A | 23A |
| 34 | 6C | 6,328 | Sg 6 | 96.5 | 2 | 59.6 | 12,069 | Sg 6 | 98.1 | 0.7 | 37.4 | Sg 6 | Sg 6 |
| 35 | 6C | 527 | Sg 6 | 79.3 | 12.5 | 42.0 | 4,140 | Sg 6 | 98.0 | 1 | 25.4 | Sg 6 | Sg 6 |
| 36 | 10A | 758 | 10A/10B | 76 | 16.5 | 74.5 | 2,654 | 10A/10B | 84.9 | 8.3 | 72.5 | 10A/10B | 10A/10B |
| 37 | 15B/15C | 1,658 | 15B/15C | 90.9 | 3.7 | 46.3 | 2,801 | 15B/15C | 87.6 | 7.5 | 73.6 | 15B/15C | 15B/15C |
| 44 | Oral flora | 2,949 | 3 | 93.6 | 5.2 | 58.9 | 2,949 | 3 | 93.6 | 5.1 | 58.9 | 3 | 3 |
| 71 | No growth | 6,062 | Sg 6 | 98.4 | 0.5 | 36.0 | No pneumococci | Sg 6 | |||||
| 73 | Oral flora | 148 | 28F | 19.6 | 15.5 (41F) | 90.65 | 169 | Sg 6 | 34.3 | 24.3 (3) | 0.0 | No pneumococci | No pneumococci |
| 94 | 3 | 1,833 | 3 | 81.1 | 8.2 | 80.7 | 1,810 | 3 | 82.7 | 5.7 | 81.8 | 3 | 3 |
| 95 | 10A | 1,002 | 10A/10B | 39.9 | 28.9 (19A) | 62.1 | 1,490 | 23A | 24.4 | 16.6 (19A), 13.6 (10A/10B) | 87.0 | Pneumococcus identified; serotype not predicted | Pneumococcus identified; serotype not predicted |
| 99 | 17F | 2,056 | 17F | 77.7 | 9.4 | 85.3 | 4,030 | 17F | 78.3 | 7.8 | 81.1 | 17F | 17F |
| 100 | Oral flora | 893 | 19B/19C | 75.6 | 9.6 | 10.3 | 2,194 | 19B/19C | 65.0 | 15.4 | 18.7 | 19B/19C | 19B/19C |
| 102 | 3 | 1,903 | 3 | 78.6 | 15.5 (34) | 79.2 | 1,587 | 3 | 86.4 | 4.4 | 76.7 | 3 | 3 |
| 103 | 15B/15C | 1,968 | 15B/15C | 94.6 | 1.2 | 68.8 | 4,798 | 15B/15C | 90.4 | 4.4 | 66.7 | 15B/15C | 15B/15C |
| 104 | No growth | 4,569 | 23A | 81.4 | 6.8 | 72.5 | No pneumococci | 23A | |||||
| 106 | 6C | 2,933 | Sg 6 | 92.9 | 2.8 | 60.7 | 2,868 | Sg 6 | 92.3 | 4 | 59.4 | Sg 6 | Sg 6 |
| 111 | 15A/15F | 2,029 | 15A/15F | 78.8 | 10 | 74.8 | 3,330 | 15A/15F | 92.4 | 5.2 | 55.1 | 15A/15F | 15A/15F |
| 112 | 19A | 13,724 | 19A | 96.2 | 2.2 | 56.5 | 9,256 | 19A | 98.7 | 0.4 | 25.5 | 19A | 19A |
| 113 | 19A | 3,958 | 19A | 95.4 | 3.7 | 54.0 | 2,986 | 19A | 79.6 | 11.8 | 66.7 | 19A | 19A |
| 114 | 19F | 6,736 | 19F | 79.8 | 10.6 | 68.9 | 7,337 | 19F | 79.7 | 14.4 | 59.7 | 19F | 19F |
| 116 | Oral flora | 9,932 | 11B/11C | 76.8 | 12.1 | 7.5 | No pneumococci | No pneumococci | |||||
| 117 | 3 | 1,528 | 3 | 80.6 | 6.2 | 82.6 | 3,089 | 3 | 63.7 | 31.3 (15A/15F) | 80.0 | 3 | 3 with minor cocolonizer |
| 125 | 15A/15F | 8,546 | 15A/15F | 97.6 | 0.5 | 28.1 | 18,404 | 15A/15F | 99.1 | 0.2 | 14.5 | 15A/15F | 15A/15F |
| 128 | 15A/15F | 7,360 | 15A/15F | 84.3 | 6.9 | 68.9 | 25,102 | 15A/15F | 98.9 | 0.4 | 18.3 | 15A/15F | 15A/15F |
| 132 | 14 | 1,772 | 14 | 84 | 6.9 | 67.5 | 2,854 | 14 | 72.0 | 23.4 (15A/15F) | 66.1 | 14 | 14 with minor cocolonizer |
| 137 | No growth | 12 | Oral | NA | NA | 29.4 | No pneumococci | No pneumococci | |||||
| 142 | 15A/15F | 10,385 | 15A/15F | 82.9 | 10.7 | 57.2 | 9,362 | 15A/15F | 97.3 | 1 | 19.6 | 15A/15F | 15A/15F |
| 143 | Oral flora | 3,052 | 15B/15C | 82.7 | 14.2 | 69.2 | 2,400 | 15B/15C | 93.7 | 3.4 | 52.0 | 15B/15C | 15B/15C |
| 145 | 23A | 5,942 | 23A | 96 | 1.2 | 39.5 | 5,865 | 23A | 93.0 | 4.2 | 50.8 | 23A | 23A |
| 146 | 23A | 2,686 | 23A | 85.5 | 6.5 | 66.7 | 11,800 | 23A | 91.4 | 4.9 | 47.4 | 23A | 23A |
| 150 | 6A | 8,332 | Sg 6 | 93.3 | 3.5 | 61.6 | 14,680 | Sg 6 | 96.5 | 1.9 | 27.1 | Sg 6 | Sg 6 |
| 152 | 6C | 2,811 | Sg 6 | 90.9 | 7.1 | 38.8 | 11,209 | Sg 6 | 95.1 | 2.6 | 8.3 | Sg 6 | No pneumococci |
Predicted serotype based on previously established criteria for assignment to a serotype (10), i.e., >500 reads mapped to a particular serotype sequence and representing >15% of all reads for the sample.
For pneumococcal identification, each sample should have >10% of its mapped reads aligned against the typical pneumococcal lytA gene for further consideration for serotyping.
Sg, serogroup.
Of all 155 samples tested with the direct NGS method, 44 had amplified products after multiplex PCR for target enrichment and were subjected to NGS sequencing; 39 samples fulfilled the criteria for pneumococcal identification, giving a positivity rate of 25.2%. Thirty-eight of 39 samples fulfilled the criteria for prediction of serotypes and were mapped to a predominant serotype. In addition, two of these samples (samples 117 and 132) had sufficient numbers and percentages of reads to map against a second serotype. One sample (sample 95) had 24.4% of reads mapped against serotype 23A, 16.6% against serotype 19A, and 13.6% against serotype 10A/10B; however, as none of the serotypes had >500 reads mapped against the given type, this sample did not fulfill the criteria for serotype allocation.
All 35 samples yielding S. pneumoniae from cultures were positive for S. pneumoniae by sweeps of primary culture DNA for NGS, and the results also corroborated the serotype predictions. However, only 34 of 35 samples were considered positive by the direct sample DNA NGS method (yielding a sensitivity of 97.4% versus sweep culture identification), as one sample (sample 152) did not meet the cutoff criterion and had only 8.3% of total mapped reads aligned to the typical lytA gene. Sample 95 did not meet the criteria for serotype allocation by either primary culture or the direct sample DNA NGS method.
Sweep culture DNA NGS identified 3 additional samples as containing S. pneumoniae, and these were also identified and confirmed by the direct sample NGS method (Table 3). Two of these three samples (samples 44 and 143) yielded pneumococcal isolation of the corresponding serotypes on repeat culture. Another two samples (samples 71 and 104) were identified as containing pneumococci by the direct sample NGS method. All five samples were confirmed to contain S. pneumoniae by repeat testing of the original DNA samples, with a second-round PCR using the CDC PCR method for prediction of serotypes. Of the three samples identified as having a second serotype (Table 3), two from the direct NGS method were confirmed to have the second serotype with reanalysis of the original DNA with additional PCR cycles using the CDC PCR method.
TABLE 3.
Discrepancies between conventional culture and NGS methods for pneumococcal identification and serotype prediction
| Sample no. | Serotype identified |
Repeat culture with 50 μl of NPA sample and identification of up to 200 colonies |
Sweep culture DNA for serotype prediction based on CDC PCRs |
Direct sample DNA for serotype prediction based on CDC PCRs |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Culture | Sweep culture DNA for NGS | Direct sample DNA for NGS | Pneumococcal identification | Serotype identified | Pneumococcal identification | Serotype identified | Pneumococcal identification | Serotype identified | |
| Additional samples identified as having S. pneumoniae | |||||||||
| 44 | Oral flora | 3 | 3 | + | 3 | + | 3 | + | 3 |
| 71 | No growth | No growth | Sga 6 | − | NAb | − | NA | + | Sg 6 |
| 100 | Oral flora | 19B/19C | 19B/19C | − | NA | + | 19B/C | + | 19B/19C |
| 104 | No growth | No growth | 23A | − | NA | − | NA | + | 23A |
| 143 | Oral flora | 15B/15C | 15B/15C | + | 15B/15C | + | 15B/15C | + | 15B/15C |
| Samples identified as having second serotype | |||||||||
| 27 | 6A | Sg 6 plus 34 | Sg 6 | + | 6A | + | 6A | + | 6A |
| 117 | 3 | 3 | 3 plus 15A/15F | + | 3 | + | 3 | + | 3 plus 15A/15F |
| 132 | 14 | 14 | 14 plus 15A/15F | + | 14 | + | 14 | + | 14 plus 15A/15F |
| Samples with other discrepancies | |||||||||
| 95 | 10A | 10A/10B (400 reads) plus 19A (290 reads) | 23A (364 reads) plus 19A (248 reads) | + | 10A, 19A, 23A | + | 10A, 19A | + | 10A, 23A |
| 73 | Oral flora | 1,412 (90%) reads against pneumococcal lytA with <500 reads for serotype sequences | Oral flora | − | NA | − | NA | − | NA |
| 116 | Oral flora | No pneumococci | <10% of reads for pneumococcal lytA but >500 reads for serotype 11B | − | NA | − | NA | + | 11B |
| 1 | Oral flora | <10% of reads for pneumococcal lytA but >500 reads for serotypes, with 39, 9N/9L, and 19B/19C | <10% of reads for pneumococcal lytA but >500 reads for serotypes, with 39, 9N/9L, and 19B/19C | + | 9N/9L | + | 9N/9L, 39 | + | 9N/9L, 39 |
Sg, serogroup.
NA, not applicable.
For sample 95, which was identified as having pneumococci but did not fulfill the criteria for serotype allocation, the original respiratory sample was recultured on 3 plates in 50-μl aliquots, and the progeny of 50 colonies were serotyped. Thirty-six of the colonies belonged to serotype 10A, 9 to serotype 19A, and 5 to serotype 23A (Table 3). The results of reanalysis of the samples with <10% of total reads mapped against the pneumococcal lytA gene with >500 reads mapped for sequence-specific reads are presented in Table 3.
DISCUSSION
The target-based NGS method described herein was capable of identifying all serogroups, including 32 serotypes at the serotype level. NGS offers a more versatile, high-throughput alternative to detection with previously described detection methods (11, 14).
Recently, a number of novel methods to increase the capacity to detect a broader range of pneumococcal serotypes have been described. These methods include a multiplexed PCR coupled to an automated microarray assay differentiating 22 serotypes and 24 other serotypes to the subgroup level, the sequetyping method, which relies on sequencing products of a single consensus pair of primers capable of amplifying products of 84 serotypes and differentiating 46, a 5-plex multiplex PCR followed by ionization mass spectrometry, which is capable of differentiating 45 serotypes, and a set of three multiplex PCRs with 40 pairs of previously described primers followed by fragment analysis using automated fluorescence-based capillary electrophoresis, which is capable of differentiating 39 serotype/serogroups (15–17). These methods are similar to our current method in terms of serotype resolution; however, to the best of our knowledge this is the first instance in which NGS coupled with target enrichment has been used to determine S. pneumoniae with serogroups directly from clinical specimens, although recently a similar method was used to identify other bacterial isolates (18). The potential advantages of whole-genome sequencing (WGS) over the current method include the potential ability to detect novel serotypes and the ability to distinguish more serogroups to the serotype level. In this study, however, the identification of lytA positivity in the absence of an identified serotype may suggest novel serotypes that deserve reinvestigation of the original samples. Also, the current method is much more affordable, in terms of per-isolate or per-sample costs, and enables greater throughput than WGS.
The rates of detection by direct NGS from clinical samples (25.2%) and sweep culture DNA identification (24.5%) were both higher than the rate of culture-based identification (22.6%). Culture-independent methods for pneumococcal detection have been shown to increase the detection rates in both colonization studies and assessments of sterile samples (19). It might be presumed that S. pneumoniae identified from only direct sample DNA (samples 71 and 104) and not from cultures or sweep cultures reflected remnant DNA of S. pneumoniae with serotypes predicted from the sequences. Thus, this method could potentially detect nonviable organisms or organisms from specimens from patients already receiving antibiotic treatment.
A drawback of molecular methods for detecting S. pneumoniae directly from clinical samples is the potential misidentification of nonpneumococcal isolates with similar genetic make-ups. Viridans group streptococci have been found to harbor a large number of genes originally identified as pneumococcal genes (20, 21). Our assay incorporated a specific pair of primers that amplified a signature of the streptococcal autolysin gene that differentiated S. pneumoniae from nonpneumococcal isolates. The presence of specific lytA gene sequences but not serotype-specific sequences at the given cutoff criteria could potentially be used to indicate novel serotypes. This could also be due to the presence of multiple nondominant serotypes in low abundance, as exemplified by the results of reanalyzing sample 95.
One key factor in the successful application of the current method is the determination of cutoff read numbers and proportions to consider a sample for serotype allocation. Pooling multiple samples into one index sample, while helping to reduce costs, could potentially introduce false positivity of minor serotypes due to chimera formation-related issues. Further evaluations of the cutoff values in relation to the number of samples pooled to distinguish a true minor serotype versus a sample pooling artifact are needed. The number of samples pooled together is best kept uniform for a given diagnostic assay after full validation (22).
Detection of multiple serotypes in colonization is one key area in pneumococcal research that is of increasing importance, due to changes in the capsular types with the use of vaccines (2). Multiplex PCRs, microarrays, and latex agglutination assays have been used to detect multiple serotypes in colonization, with different success rates (23–25). The use of culture for detection of multiple serotypes is limited by the number of colonies that need to be identified in order to have a realistic probability of identifying a minor serotype (26, 27). The method described offers an attractive alternative to detection of multiple serotypes in colonization from primary cultures or direct sample DNA.
This method is sufficiently versatile to be applied to pneumococcal isolates, sweep cultures, and direct clinical samples. As the enrichment requires only two multiplex PCRs optimized to the same thermocycling conditions, the method is amenable to automation, and NGS could be adapted to different sequencing platforms with modified library preparation protocols. With the emergence of competitively priced kits for rapid library preparation, this method can be carried out in a relatively short time. The bioinformatics pipeline used for this method is simple. Alternatively, on-instrument data-processing pipelines may be used for analysis.
Siira et al. (28) indicated that the best way to utilize different methodologies for serotyping is to use them in a complementary manner, whereby molecular methods are used to rapidly screen large numbers of samples in a high-throughput manner and then the more precise but costly standard methods are used for further identification where needed (28). We propose our target enrichment-based sequencing method as a versatile adaptable method that can be used in conjunction with standard methods.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Jerris Chang and Wei Chi Wang for technical bioinformatics assistance.
Footnotes
Published ahead of print 1 October 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.02397-14.
REFERENCES
- 1. Weinberger DM, Malley R, Lipsitch M. 2011. Serotype replacement in disease after pneumococcal vaccination. Lancet 378:1962–1973. 10.1016/S0140-6736(10)62225-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Satzke C, Turner P, Virolainen-Julkunen A, Adrian PV, Antonio M, Hare KM, Henao-Restrepo AM, Leach AJ, Klugman KP, Porter BD, Sa-Leao R, Scott JA, Nohynek H, O'Brien KL, WHO Pneumococcal Carriage Working Group 2013. Standard method for detecting upper respiratory carriage of Streptococcus pneumoniae: updated recommendations from the World Health Organization Pneumococcal Carriage Working Group. Vaccine 32:165–179. 10.1016/j.vaccine.2013.08.062. [DOI] [PubMed] [Google Scholar]
- 3. John J, Gopalkrishnan S, Marie MAM, Gowda KL. 2013. Historical development of typing methods for Streptococcus pneumoniae. Rev. Med. Microbiol. 25:27–33. 10.1097/MRM.0000000000000001. [DOI] [Google Scholar]
- 4. Bentley SD, Aanensen DM, Mavroidi A, Saunders D, Rabbinowitsch E, Collins M, Donohoe K, Harris D, Murphy L, Quail MA, Samuel G, Skovsted IC, Kaltoft MS, Barrell B, Reeves PR, Parkhill J, Spratt BG. 2006. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet. 2:e31. 10.1371/journal.pgen.0020031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pai R, Gertz RE, Beall B. 2006. Sequential multiplex PCR approach for determining capsular serotypes of Streptococcus pneumoniae isolates. J. Clin. Microbiol. 44:124–131. 10.1128/JCM.44.1.124-131.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pimenta FC, Roundtree A, Soysal A, Bakir M, du Plessis M, Wolter N, von Gottberg A, McGee L, Carvalho MG, Beall B. 2013. Sequential triplex real-time PCR assay for detecting 21 pneumococcal capsular serotypes that account for a high global disease burden. J. Clin. Microbiol. 51:647–652. 10.1128/JCM.02927-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Arena F, Rolfe PA, Doran G, Conte V, Gruszka S, Clarke T, Adesokan Y, Giani T, Rossolini GM. 2014. Rapid resistome fingerprinting and clonal lineage profiling of carbapenem-resistant Klebsiella pneumoniae by targeted next-generation sequencing. J. Clin. Microbiol. 52:987–990. 10.1128/JCM.03247-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liyanapathirana V, Ang I, Tsang D, Fung K, Ng TK, Zhou H, Ip M. 2014. Application of a target enrichment-based next-generation sequencing protocol for identification and sequence-based prediction of pneumococcal serotypes. BMC Microbiol. 14:60. 10.1186/1471-2180-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ip M, Chau SSL, Lai LS, Ma H, Chan PKS, Nelson EAS. 2013. Increased nasopharyngeal carriage of serotypes 6A, 6C, and 6D Streptococcus pneumoniae after introduction of childhood pneumococcal vaccination in Hong Kong. Diagn. Microbiol. Infect. Dis. 76:153–157. 10.1016/j.diagmicrobio.2013.02.036. [DOI] [PubMed] [Google Scholar]
- 10. Turner P, Hinds J, Turner C, Jankhot A, Gould K, Bentley SD, Nosten F, Goldblatt D. 2011. Improved detection of nasopharyngeal co-colonization by multiple pneumococcal serotypes by use of latex agglutination or molecular serotyping by microarray. J. Clin. Microbiol. 49:1784–1789. 10.1128/JCM.00157-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhou F, Kong F, Tong Z, Gilbert GL. 2007. Identification of less-common Streptococcus pneumoniae serotypes by a multiplex PCR-based reverse line blot hybridization assay. J. Clin. Microbiol. 45:3411–3415. 10.1128/JCM.01076-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Faircloth BC, Glenn TC. 2012. Not all sequence tags are created equal: designing and validating sequence identification tags robust to indels. PLoS One 7:e42543. 10.1371/journal.pone.0042543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Matz M, Shagin D, Bogdanova E, Britanova O, Lukyanov S, Diatchenko L, Chenchik A. 1999. Amplification of cDNA ends based on template-switching effect and step-out PCR. Nucleic Acids Res. 27:1558–1560. 10.1093/nar/27.6.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kong F, Brown M, Sabananthan A, Zeng X, Gilbert GL. 2006. Multiplex PCR-based reverse line blot hybridization assay to identify 23 Streptococcus pneumoniae polysaccharide vaccine serotypes. J. Clin. Microbiol. 44:1887–1891. 10.1128/JCM.44.5.1887-1891.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Leung MH, Bryson K, Freystatter K, Pichon B, Edwards G, Charalambous BM, Gillespie SH. 2012. Sequetyping: serotyping Streptococcus pneumoniae by a single PCR sequencing strategy. J. Clin. Microbiol. 50:2419–2427. 10.1128/JCM.06384-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Massire C, Gertz RE, Svoboda P, Levert K, Reed MS, Pohl J, Kreft R, Li F, White N, Ranken R, Blyn LB, Ecker DJ, Sampath R, Beall B. 2012. Concurrent serotyping and genotyping of pneumococci by use of PCR and electrospray ionization mass spectrometry. J. Clin. Microbiol. 50:2018–2025. 10.1128/JCM.06735-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Raymond F, Boucher N, Allary R, Robitaille L, Lefebvre B, Tremblay C, Corbeil J, Gervaix A. 2013. Serotyping of Streptococcus pneumoniae based on capsular genes polymorphisms. PLoS One 8:e76197. 10.1371/journal.pone.0076197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Arena F, Rolfe PA, Doran G, Conte V, Gruszka S, Clarke T, Adesokan Y, Giani T, Rossolini GM. 2014. Rapid resistome fingerprinting and clonal lineage profiling of carbapenem-resistant Klebsiella pneumoniae isolates by targeted next-generation sequencing. J. Clin. Microbiol. 52:987–990. 10.1128/JCM.03247-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Azzari C, Moriondo M, Indolfi G, Cortimiglia M, Canessa C, Becciolini L, Lippi F, de Martino M, Resti M. 2010. Realtime PCR is more sensitive than multiplex PCR for diagnosis and serotyping in children with culture negative pneumococcal invasive disease. PLoS One 5:e9282. 10.1371/journal.pone.0009282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Johnston C, Hinds J, Smith A, van der Linden M, Van Eldere J, Mitchell TJ. 2010. Detection of large numbers of pneumococcal virulence genes in streptococci of the mitis group. J. Clin. Microbiol. 48:2762–2769. 10.1128/JCM.01746-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Carvalho MG, Jagero G, Bigogo GM, Junghae M, Pimenta FC, Moura I, Roundtree A, Li Z, Conklin L, Feikin DR, Breiman RF, Whitney CG, Beall B. 2012. Potential nonpneumococcal confounding of PCR-based determination of serotype in carriage. J. Clin. Microbiol. 50:3146–3147. 10.1128/JCM.01505-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gargis AS, Kalman L, Berry MW, Bick DP, Dimmock DP, Hambuch T, Lu F, Lyon E, Voelkerding KV, Zehnbauer BA, Agarwala R, Bennett SF, Chen B, Chin EL, Compton JG, Das S, Farkas DH, Ferber MJ, Funke BH, Furtado MR, Ganova-Raeva LM, Geigenmuller U, Gunselman SJ, Hegde MR, Johnson PL, Kasarskis A, Kulkarni S, Lenk T, Liu CS, Manion M, Manolio TA, Mardis ER, Merker JD, Rajeevan MS, Reese MG, Rehm HL, Simen BB, Yeakley JM, Zook JM, Lubin IM. 2012. Assuring the quality of next-generation sequencing in clinical laboratory practice. Nat. Biotechnol. 30:1033–1036. 10.1038/nbt.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bronsdon MA, O'Brien KL, Facklam RR, Whitney CG, Schwartz B, Carlone GM. 2004. Immunoblot method to detect Streptococcus pneumoniae and identify multiple serotypes from nasopharyngeal secretions. J. Clin. Microbiol. 42:1596–1600. 10.1128/JCM.42.4.1596-1600.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Carvalho MG, Pimenta FC, Jackson D, Roundtree A, Ahmad Y, Millar EV, O'Brien KL, Whitney CG, Cohen AL, Beall BW. 2010. Revisiting pneumococcal carriage by use of broth enrichment and PCR techniques for enhanced detection of carriage and serotypes. J. Clin. Microbiol. 48:1611–1618. 10.1128/JCM.02243-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rivera-Olivero IA, Blommaart M, Bogaert D, Hermans PWM, de Waard JH. 2009. Multiplex PCR reveals a high rate of nasopharyngeal pneumococcal 7-valent conjugate vaccine serotypes co-colonizing indigenous Warao children in Venezuela. J. Med. Microbiol. 58:584–587. 10.1099/jmm.0.006726-0. [DOI] [PubMed] [Google Scholar]
- 26. Hare KM, Morris P, Smith-Vaughan H, Leach AJ. 2008. Random colony selection versus colony morphology for detection of multiple pneumococcal serotypes in nasopharyngeal swabs. Pediatr. Infect. Dis. J. 27:178–180. 10.1097/INF.0b013e31815bb6c5. [DOI] [PubMed] [Google Scholar]
- 27. Huebner RE, Dagan R, Porath N, Wasas AD, Klugman KP. 2000. Lack of utility of serotyping multiple colonies for detection of simultaneous nasopharyngeal carriage of different pneumococcal serotypes. Pediatr. Infect. Dis. J. 19:1017–1020. 10.1097/00006454-200010000-00019. [DOI] [PubMed] [Google Scholar]
- 28. Siira L, Kaijalainen T, Lambertsen L, Nahm MH, Toropainen M, Virolainen A. 2012. From Quellung to multiplex PCR, and back when needed, in pneumococcal serotyping. J. Clin. Microbiol. 50:2727–2731. 10.1128/JCM.00689-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.