Abstract
Norovirus (NoV) is considered a major cause of nonbacterial gastroenteritis among people of all ages worldwide, but the natural course of infection is incompletely known. In this study, the pattern of circulation of NoVs was studied among 146 children and 137 adults in a small community in southwestern Cameroon. The participants provided monthly fecal samples during a year. NoV RNA was detected in at least one sample from 82 (29%) of the participants. The partial VP1 region could be sequenced in 36 NoV GII-positive samples. Three different genotypes were identified (GII.1, GII.4, and GII.17), with each genotype circulating within 2 to 3 months and reappearing after a relapse period of 2 to 3 months. Most infections occurred once, and 2 episodes at most within a year were detected. No difference in the frequency of NoV infection between children and adults was recorded. The same genotype was detected for a maximum of 2 consecutive months in 3 children only, suggesting that a less than 30-day duration of viral shedding in natural infection was common. Reinfection within a year with the same genotype was not observed, consistent with short-term homotypic immune protection. The study revealed that NoV strains are circulating with a limited duration of viral shedding both in the individuals and the population as part of their natural infection. The results also provide evidence of cross-protective immunity of limited duration between genotypes of the same genogroup.
INTRODUCTION
Norovirus (NoV) has emerged as a major cause of nonbacterial gastroenteritis in developed countries, with an overwhelming peak of outbreak of the disease occurring during the winter season (1, 2). In tropical regions, infections are relatively common in the rainy season (3). The virus is a small nonenveloped, single-stranded, positive-sense RNA virus with a genome of about 7.5 kb in size classified in the Caliciviridae family (4). There are at least 35 NoV genotypes classified within 5 genogroups (GI to GV). Genogroups GI, GII, and GIV primarily infect humans, while bovine and murine NoVs are classified into genogroups GIII and GV, respectively (5, 6). A sixth genogroup has been proposed after the discovery of a new canine norovirus (7).
NoV is highly infectious, with just 10 to 100 virions required to induce a disease (8), and transmission occurs mainly by ingestion of the virus in contaminated food and drink or by person-to-person spread (9). Despite the efficient mode of transmission, asymptomatic infections are common, and most symptomatic cases generally are self-limiting. That is, the duration of symptoms such as diarrhea and vomiting, when present, usually resolve within 2 to 3 days (1). On the other hand, severe forms of the disease may occur in children, the elderly, and in malnourished or immunosuppressed individuals (10).
There also have been reports of prolonged NoV shedding in immunosuppressed individuals (11), but only a limited number of studies have investigated excretion in healthy persons during the natural course of infection (12, 13). Most previous NoV epidemiologic and immunologic studies were based on outbreak investigation or human challenge studies with filtrate of Norwalk-like viruses (14–17), while longitudinal studies of natural NoV infection are lacking from developing country settings. Also, the finding that NoV is a common gastroenteritis-causing agent is often extrapolated globally from results of studies performed in western countries, and recent investigations in developing countries, with healthy controls included, show a much milder scenario (13, 18, 19). This study extends knowledge on the frequency of infection or reinfection, viral turnover in the population, and the duration of excretion of NoV strains in healthy individuals during the natural course of infection. It provides a report on the pattern of circulation of NoVs investigated in fecal samples from children and adults sampled monthly over a period of 1 year in a developing country setting.
MATERIALS AND METHODS
Study location and setting.
This prospective study was undertaken in Limbe, Cameroon, a semiurban city located at the foot of Mount Cameroon, close to the Atlantic Ocean, with a population of about 84,000 inhabitants. Participants lived in Mile-one, with a population of about 2,000 inhabitants who are mostly merchants, civil servants, and students. Sources of drinking water are mainly tap water and borehole wells. Participants were selected randomly within the community in their respective homes, were taught aseptic sample collection techniques, and were requested to provide their monthly samples at the collection centers.
Participants and clinical samples.
We conducted a 1-year longitudinal prospective study. Enrollment began in September 2011 and was completed in August 2012. Enrolled participants (consisting of a child and adult from the same household) were examined and questioned for the presence of >3 episodes of diarrhea/day within the last month. Inclusion criteria involved a lack of any signs or symptoms of gastroenteritis within the preceding month prior to enrollment. A total of 283 individuals, 146 children aged 1 to 17 years (median, 6 years) and 137 adults aged 18 to 69 years (median, 32 years), met the inclusion criteria and were prospectively followed up regularly in Limbe, Cameroon. The participants were sampled monthly over a period of 12 months. Although participants were asked to provide a fecal sample each month from September 2011 to August 2012, some participants were unavailable during some sampling periods. However, 76 of the 146 (52%) children and 80 of the 137 (58%) adults provided 10 or more fecal samples during the study period. Fecal samples were provided on the first day of every month. The surveillance protocol was reviewed and approved by the southwest regional delegation of public health in Cameroon, and participants (parents or guardians of children) provided written or oral informed consent.
Stool preparation and nucleic acid extraction.
Stool suspensions were prepared as described elsewhere (20). Briefly, 1 g of stool was added to 5 ml of phosphate-buffered saline containing 1 g of glass beads (Corning Inc., Corning, NY). The mixture was shaken for 2 min and centrifuged at 2,000 × g for 15 min at 4°C. Fecal suspensions (130 μl) were mixed with 220 μl lysis buffer, and total nucleic acids were extracted into an elution volume of 100 μl by using the MagNA pure liquid chromatography (LC) instrument (MagNA pure LC total nucleic acid isolation kit; Roche Diagnostic GmbH, Manheim, Germany) according to the manufacturer's instructions.
Real-time PCR.
NoV RNA was detected by real-time PCR as described previously (3). Briefly, the assay was performed in 25-μl reaction volumes containing 5 μl nucleic acid, 13.5 μl 2× reaction mix with ROX (Invitrogen Ltd., Paisley, United Kingdom), 0.5 μl Superscript III platinum one-step quantitative reverse transcription-PCR (RT-PCR) mix, 20 U RNaseOUT, and 0.5 μM primers and probes published elsewhere (21). After a reverse transcription step at 48°C for 25 min and an initial denaturation at 95°C for 10 min, the reaction was run for 45 cycles of two steps (95°C for 15 s and 60°C for 60 s) in an ABI 7300 real-time PCR platform (Applied Biosystems, Foster City, CA).
Reverse transcription.
Reverse transcription (cDNA synthesis) was carried out as described previously (3). Briefly, 5 μl of nucleic acid was added to 45 μl of a reaction mix consisting of 100 μM each deoxynucleoside triphosphate (dNTP; Amersham Biosciences, Piscataway, NJ), 2 μl 5× reaction buffer (Invitrogen, Carlsbad, CA), 0.01 M dithiothreitol, 20 U RNasin (Promega Corp., Madison, WI), 100 U SuperScript II reverse transcriptase (Invitrogen), and random hexamer primers (Promega, United Kingdom). The reaction was carried out at 65°C for 5 min, 25°C for 5 min, 37°C for 60 min, and finally 70°C for 10 min.
PCR and sequencing.
Primers targeting partial RNA-dependent RNA polymerase and partial VP1 were used for PCR. For each sample, 45 μl master mix was prepared containing 5 μl of 10× PCR buffer (Roche Applied Science), 200 μM each dNTP, 10 pmol of forward and reverse primers, and 2.5 U Taq DNA polymerase (Roche Applied Science). Primers JV12yF and G2SKR were used for the first-round PCR, and primers NiF, G2SKF, G2SKR, and G1SKR were used for the nested PCR (22, 23). The program for both PCRs included an initial denaturation at 95°C for 5 min, followed by 40 cycles of 95°C for 60 s, 54°C for 60 s, and 72°C for 90 s. The PCR-amplified products (∼600 bp) were purified using a QIAquick gel extraction kit (Qiagen) and sequenced in both directions using 1 pmol of each primer used in the nested PCR. Cycle sequencing was performed using BigDye Terminator v3.1 (Applied Biosystems, Foster City, CA, USA) according to the manufacturer's instructions. Considering the risk of cross contamination, precautions were taken in order to avoid PCR contamination. The preparation of reagents, processing of stool samples, thermal cycling, nested PCR, and analysis of PCR products all were performed in safety cabinets in separate laboratories and were subjected to strict rules for laboratory practice. Negative controls were included in each PCR analysis.
Sequence analysis and phylogenetic analysis.
Chromatogram sequencing files were visually inspected and contigs were prepared. The obtained partial RNA polymerase and partial VP1 sequences were compared to sequences available in GenBank using BLAST (http://www.ncbi.nlm.nih.gov/BLAST/). Multiple sequence alignments were prepared using CLUSTAL X (24), followed by visual correction. The tree was constructed by the maximum likelihood algorithm implemented in the MEGA program using the Tamura-Nei model. The NoV sequences were classified into genotypes by BLAST and their clustering in the phylogenetic trees.
Statistical analysis.
Monthly prevalence was calculated as the number of monthly cases of infection divided by the sample size multiplied by 100. The frequency of NoV infection was compared in children and adults by means of two-sided Fisher's exact test. Odd ratios (ORs) and 95% confidence intervals (CIs) of risks of NoV infection in children and adults were calculated using the SPSS software package, v. 17.0, for Mac (SPSS Inc., USA).
Nucleotide sequence accession numbers.
Sequences obtained in this work have been deposited in GenBank under accession numbers KJ946387 to KJ946411.
RESULTS
In all, 2,458 fecal samples were obtained for NoV RNA investigation from the 283 participants. This includes 1,244 samples from the children and 1,214 from the adults (Table 1). NoV RNA was identified in 100 (4%) of all samples, 57 (4.6%) from children and 43 (3.5%) from adults (Table 1). Out of the 100 NoV-positive samples, only 3 (3%) coincided with diarrhea. About one-third of children (45/146; 30.8%) and adults (37/137; 27.0%) had at least one episode of NoV infection during the 1-year study period (Tables 1 and 2). The monthly prevalence of NoV RNA in the fecal samples was between 1% and 15% in children and 0% and 12% in adults. Fifty-five percent of NoVs were identified as belonging to genogroup GII and 45% to genogroup GI by genogroup-specific real-time PCR. The viral loads of all NoV GI- and 19 NoV GII-positive samples were low; this rendered them difficult to sequence. There was no significant difference in the prevalence of NoV infection in children and adults (P > 0.05) (Table 1).
TABLE 1.
Number of samples obtained monthly and number and percent positive for norovirus by real-time PCR in Limbe, Cameroon, September 2011 to August 2012
| Date of collectionb (mo/yr) | No. of samples | No. (%) of RT-PCR positive children | No. of samples | No. (%) RT-PCR positive of adults | OR (95% CI)/P value |
|---|---|---|---|---|---|
| Sep 2011 | 89 | 2 (2.2) | 89 | 2 (2.2) | |
| Oct 2011 | 114 | 2 (1.7) | 99 | 1 (1.0) | |
| Nov 2011 | 109 | 2 (1.8) | 96 | 2 (2.0) | |
| Dec 2011 | 105 | 2 (1.9) | 99 | 0 (0) | |
| Jan 2012 | 108 | 1 (0.9) | 106 | 2 (1.9) | |
| Feb 2012 | 102 | 7 (6.8) | 108 | 7 (6.4) | |
| Mar 2012 | 111 | 2 (1.8) | 111 | 3 (2.7) | |
| Apr 2012 | 108 | 4 (3.7) | 110 | 1 (0.9) | |
| May 2012 | 109 | 2 (1.8) | 111 | 2 (1.8) | |
| Jun 2012 | 89 | 14 (15.7) | 88 | 11 (12.5) | |
| Jul 2012 | 98 | 9 (9.2) | 100 | 6 (6.0) | |
| Aug 2012 | 102 | 10 (9.8) | 99 | 6 (6.0) | |
| Total no. of samples | 1,244 | 57 (4.6) | 1,214 | 43 (3.5) | 1.3 (0.8–1.9)/0.2 |
| No. of persons providing samples | 146 | 45 (30.8)a | 137 | 37 (27.0)a | 1.2 (0.7–2.0)/0.5 |
Number/percentage of persons with at least one norovirus-positive sample.
Sep, September; Oct, October; Nov, November; Dec, December; Jan, January; Feb, February; Mar, March; Apr, April; Jun, June; Jul, July; Aug, August.
TABLE 2.
Number of episodes of norovirus infection among participants from September 2011 to August 2012 in Limbe, Cameroon
| No. of episodes of NoV infection | No. (%) of participants infected |
Total | |
|---|---|---|---|
| Children | Adults | ||
| 0 | 101 (69) | 100 (73) | 201 (71) |
| 1 | 38 (26) | 30 (22) | 68 (24) |
| 2 | 4 (3) | 4 (3) | 8 (3) |
| 1aa | 3 (2) | 3 (2) | 6 (2) |
| Total | 146 | 137 | 283 |
One or two norovirus infection episodes (the exact number of episodes is unknown due to one untyped sample).
Norovirus phylogenetic clustering, seasonality, and pattern of circulation.
The sequencing of partial RNA-dependent RNA polymerase and partial VP1 could be achieved in 36 NoV GII RNA-positive samples, 20 from children and 16 from adults (Tables 3 and 4). Three different genotypes were identified, consisting of 20 strains belonging to GII.1, 12 to GII.4, and 4 to GII.17 (Table 3). The major peak of NoV prevalence was seen during the rainy season between June and August, while the rest of the months were associated with infection at relatively low levels (Table 3). Most GII genotypes circulated for 2 to 3 months, with a relapse period of 2 to 3 months before reemergence. Genotype GII.1 was detected consecutively for 2 months (February-March) and then reemerged in July-August after a period of absence of 3 months. Genotype GII.4 circulated from November to January and reappeared in the population in April and May (Table 3 and Fig. 1).
TABLE 3.
Norovirus genotypes circulating in Limbe, Cameroon, in relation to month of sampling
| Type | No. of samples positive for infection in: |
Total | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sep 11 | Oct 11 | Nov 11 | Dec 11 | Jan 12 | Feb 12 | Mar 12 | Apr 12 | May 12 | Jun 12 | Jul 12 | Aug 12 | ||
| GII.1 | 0 | 0 | 0 | 0 | 0 | 2 | 3 | 0 | 0 | 0 | 6 | 9 | 20 |
| GII.4 | 0 | 0 | 1 | 1 | 2 | 0 | 0 | 4 | 4 | 0 | 0 | 0 | 12 |
| GII.17 | 3 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 |
| Untyped GII | 1 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 8 | 7 | 19 |
| Untyped GI | 0 | 3 | 2 | 1 | 1 | 9 | 2 | 1 | 0 | 25 | 1 | 0 | 45 |
| Total | 4 | 3 | 4 | 2 | 3 | 14 | 5 | 5 | 4 | 25 | 15 | 16 | 100 |
TABLE 4.
Individual timeline of norovirus infection in the longitudinal follow-up of adults from September 2011 to August 2012 in Limbe, Cameroon
| Adult | Norovirus genotype detected ina: |
Totalb | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sep | Oct | Nov | Dec | Jan | Feb | Mar | Apr | May | Jun | Jul | Aug | ||
| A2 | GII.17 | – | – | * | – | – | – | – | – | – | – | – | 1 |
| A5 | – | – | * | – | – | – | – | – | – | – | – | GII | 2 |
| A8 | – | – | * | – | – | – | – | – | – | – | – | GII.1 | 1 |
| A13 | – | – | * | – | – | GI | – | – | – | – | – | – | 1 |
| A14 | – | – | * | – | – | GI | – | – | – | – | GII.1 | – | 2 |
| A16 | – | – | – | – | – | – | – | – | – | – | – | GII.1 | 1 |
| A18 | – | – | * | – | – | – | – | – | – | – | – | GII.1 | 1 |
| A43 | – | – | – | – | GII.4 | – | – | – | – | * | – | – | 1 |
| A45 | GII | – | – | – | – | – | * | – | * | – | – | – | 1 |
| A47 | – | GI | – | – | – | – | – | – | – | – | * | * | 1 |
| A53 | – | – | – | * | – | – | GII.1 | – | – | – | * | * | 1 |
| A56 | – | – | – | * | * | – | – | GII.4 | – | – | – | – | 1 |
| A60 | – | * | – | – | – | – | – | – | GII.4 | – | – | – | 1 |
| A66 | – | – | – | – | – | – | – | – | – | GI | – | – | 1 |
| A69 | – | – | – | – | GI | GI | – | – | – | – | * | * | 1ac |
| A72 | – | – | – | – | – | – | – | – | – | GI | – | – | 1 |
| A77 | – | – | – | – | – | – | GII.1 | – | – | GI | – | – | 2 |
| A79 | – | – | – | – | – | – | GII.1 | – | – | GI | – | – | 2 |
| A80 | – | – | – | – | – | – | – | – | – | GI | – | – | 1 |
| A83 | – | – | GII.4 | – | – | – | – | – | – | – | – | – | 1 |
| A84 | – | – | – | – | – | – | – | – | GII.4 | – | – | GII | 1a |
| A92 | – | * | * | – | – | – | – | – | – | – | – | GII.1 | 1 |
| A97 | – | * | – | – | – | GI | – | – | – | – | – | – | 1 |
| A104 | – | * | * | – | – | – | – | – | – | – | GI | – | 1 |
| A105 | – | – | – | – | – | – | – | – | – | GI | – | – | 1 |
| A107 | – | – | – | – | – | – | – | – | – | GI | – | – | 1 |
| A114 | – | – | – | – | – | – | – | – | – | GI | – | – | 1 |
| A130 | – | – | – | – | – | GII | – | – | – | – | GII | – | 1a |
| A132 | – | – | – | – | – | GII | – | – | – | – | – | – | 1 |
| A135 | – | – | – | – | – | – | – | – | – | GI | – | – | 1 |
| A136 | – | – | – | – | – | GII | – | – | – | – | – | – | 1 |
| A138 | – | – | – | – | – | – | – | – | – | GI | – | – | 1 |
| A141 | – | – | – | – | – | – | – | – | – | – | GII | – | 1 |
| A142 | – | – | – | – | – | – | – | – | – | GI | – | – | 1 |
| A144 | – | – | – | – | – | – | – | – | – | – | GII.1 | – | 1 |
| A147 | – | – | GII.17 | – | – | – | – | – | – | – | – | – | 1 |
| A151 | – | – | * | – | – | – | – | – | – | – | GII | – | 1 |
Entries in boldface were from diarrhea episodes. *, no sample; –, samples negative for NoV by real-time PCR.
Total number of episodes of infection.
1a indicates the occurrence of one or two norovirus infection episodes (the exact number of episodes is unknown due to samples being untyped).
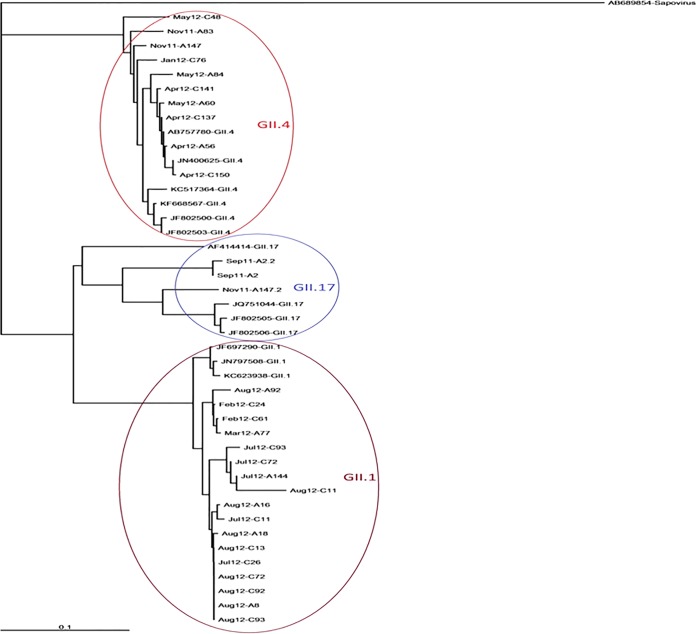
FIG 1.
Phylogenetic tree of noroviruses for a 600-nucleotide fragment of the partial RNA polymerase and VP1 region. Thirty novel sequences are included and are designated according to the month of sampling and laboratory identifier. C and A in the laboratory identifiers represent strains originating from children and adults, respectively. Reference sequences from GenBank also were included and are designated according to accession number and genotype. The tree was constructed by the maximum likelihood algorithm implemented in the MEGA program using the Tamura-Nei model.
Frequency of norovirus infection and duration of RNA shedding in feces.
Episodes of NoV infection were defined as NoV RNA-positive samples with different genotypes separated by NoV RNA-negative samples. However, the occurrence of multiple episodes of infection was similar in both children and adults (Table 4 and 5). About one-third (29%) of the participants had at least one episode of NoV infection in a year, with a maximum of 2 episodes in 7 children and 7 adults (5%). Most of the infections (24%) occurred only once. The same genotype was identified in a maximum of two consecutive samples in 3 children only, suggesting that the duration of excretion usually was less than 1 month (Table 5). The duration of NoV excretion in adults similarly was short, since infection with a particular genotype was detected just once in all subjects (Table 4).
TABLE 5.
Individual timeline of norovirus infection in the longitudinal follow-up of children from September 2011 to August 2012 in Limbe, Cameroon
| Child | Norovirus genotype detected ina: |
Totalb | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sep | Oct | Nov | Dec | Jan | Feb | Mar | Apr | May | Jun | Jul | Aug | ||
| C1 | – | – | – | GI | – | – | – | – | – | – | – | – | 1 |
| C8 | – | – | – | – | – | – | – | – | – | GI | – | – | 1 |
| C11 | – | * | * | – | – | GI | – | – | – | – | GII.1 | GII.1 | 2 |
| C13 | – | – | – | – | – | GI | – | – | – | – | GII | GII.1 | 2 |
| C14 | – | – | – | – | – | GI | – | – | – | – | – | – | 1 |
| C17 | – | * | * | – | – | – | – | – | – | – | GII | – | 1 |
| C18 | – | – | – | – | – | – | – | – | – | – | GII | – | 1 |
| C19 | – | – | GI | – | – | – | – | – | – | – | – | – | 1 |
| C24 | – | – | – | – | – | GII.1 | – | – | – | – | – | GII | 1ac |
| C25 | – | – | * | – | – | – | – | – | – | – | – | GII | 1 |
| C26 | * | * | – | – | – | – | – | – | – | – | GII.1 | GII | 1 |
| C28 | – | – | – | – | – | – | – | – | – | GI | – | – | 1 |
| C30 | GII.17 | – | – | – | – | – | – | * | * | – | – | – | 1 |
| C44 | – | – | – | – | – | – | – | – | – | GI | – | – | 1 |
| C45 | – | – | – | * | – | – | GI | – | – | – | – | – | 1 |
| C46 | – | – | – | – | – | – | – | – | – | GI | – | – | 1 |
| C47 | – | – | – | – | * | * | – | – | – | – | GII | – | 1 |
| C48 | – | – | – | – | – | – | – | – | GII.4 | GI | – | – | 2 |
| C53 | GII.17 | – | – | – | – | – | – | – | – | – | – | – | 1 |
| C55 | – | – | – | – | – | – | – | – | – | GI | – | – | 1 |
| C56 | * | – | – | – | – | – | – | – | – | – | GII | GII | 1a |
| C61 | – | * | – | – | – | GII.1 | – | – | – | – | * | * | 1 |
| C72 | – | – | – | * | * | – | – | – | – | – | GII.1 | GII.1 | 1 |
| C76 | – | – | – | – | GII.4 | – | – | – | – | * | * | * | 1 |
| C77 | – | * | – | GII.4 | – | – | – | – | – | – | – | – | 1 |
| C78 | – | – | – | – | – | – | – | – | – | GI | – | – | 1 |
| C79 | – | * | – | – | – | – | – | – | – | GI | – | – | 1 |
| C91 | – | – | * | * | – | – | – | – | – | – | – | GII | 1 |
| C92 | – | – | * | * | – | – | – | – | – | – | – | GII.1 | 1 |
| C93 | – | – | * | * | – | – | – | – | – | – | GII.1 | GII.1 | 1 |
| C97 | – | – | – | * | – | GI | – | – | – | – | – | – | 1 |
| C99 | – | – | * | * | – | GI | – | – | – | – | – | – | 1 |
| C101 | – | – | GI | – | – | – | – | – | – | – | – | – | 1 |
| C104 | – | GI | – | – | – | – | – | – | – | GI | – | – | 1a |
| C105 | – | – | – | – | – | – | – | – | – | GI | – | – | 1 |
| C117 | – | – | * | – | – | – | – | GI | – | – | – | – | 1 |
| C134 | – | – | * | – | – | – | – | – | – | GI | – | – | 1 |
| C135 | – | GI | – | – | – | – | – | – | – | – | – | – | 1 |
| C137 | – | – | – | – | – | – | – | GII.4 | – | – | – | – | 1 |
| C140 | – | – | – | – | – | – | – | – | – | GI | – | – | 1 |
| C141 | – | – | – | – | * | – | – | GII.4 | – | GI | – | – | 2 |
| C143 | – | – | – | – | – | – | – | – | – | GI | – | – | 1 |
| C145 | – | – | – | – | – | – | GI | – | – | – | – | * | 1 |
| C150 | – | – | – | – | – | – | – | GII.4 | – | – | – | – | 1 |
| C155 | – | * | – | – | – | – | – | – | GII.4 | – | – | – | 1 |
Entries in boldface were from diarrhea episodes. *, no sample; –, samples negative for NoV by real-time PCR.
Total number of episodes of infection.
1a indicates the occurrence of one or two norovirus infection episodes (the exact number of episodes is unknown due to samples being untyped).
DISCUSSION
This NoV longitudinal survey in children and adults provides an account of the natural course of NoV infection in a low-income country. The frequency of NoV infections was similar in children and adults, with monthly prevalences in children ranging between 1 and 15%. This detection rate is compatible with those of other previous studies where NoV was detected in 4 to 11% of asymptomatic participants (25–27). NoVs are considered an important cause of viral gastroenteritis in all age groups. Out of 100 identified NoV infections, only three coincided with symptoms of gastroenteritis. This finding suggests that some degree of immunity is acquired, and that most of the norovirus infections in these settings are asymptomatic. The possibility of a prolonged postsymptomatic shedding at very low viral load is unlikely, since participants all were healthy and reported the absence of gastroenteritis 1 month before enrollment. To investigate whether the NoV-positive cases detected in the limited population were due to circulating infections of different genotypes or prolonged infection of the same genotype, the sequencing of NoV RNA-positive samples was performed. It was observed that both children and adults had similar rates of NoV infections, with a maximum of 2 infection episodes per year. A prolonged shedding of NoV GII.1 for up to a month was observed in three children (mean age, 6 years) without any apparent clinical manifestation. However, in the absence of a high-resolution analysis of samples (28), the possibility of long-term carriage at undetectable levels cannot be ruled out. Prolonged shedding of NoVs has been observed in infants less than 6 months of age, in the elderly, and in hospitalized and immunosuppressed patients (12, 29, 30).
Another important observation was that reinfection with genotypes within the same NoV genogroup was rare. This might be due to immune cross protection between genotypes of the same genogroup. However, the exact duration of such immune protection is unclear, since the study was limited to 1 year. Overall, the frequency of NoV infection and duration of RNA shedding is in contrast to our earlier observation for enterovirus infection, where up to 5 episodes of different enterovirus infections were observed within a year and a prolonged viral shedding of up to 8 months was reported in asymptomatic children (20).
To the best of our knowledge, this is the first study using longitudinal analysis to describe the natural course of NoV infections over time in children and adults.
The relative increase in the number of NoV-positive subjects detected in the rainy season may reflect differential transmission between the tropical and temperate regions, considering that NoV infections typically are common in the winter in temperate regions (31, 32). This finding suggests an association of contaminated water with increased transmission of NoVs in the study area. Also, the high prevalence of NoV GII.1 strains with high sequence similarity in the month of August likely is the result of an outbreak.
The focus of the current study has been on NoV GII, since most NoV GI-positive samples were untyped due to very low viral load. However, the individual time line analysis clearly suggests that the risk of reinfection with the same genotype/genogroup was rare and that the majority of infected individuals had just one encounter with NoV infection within a year. The 1-month sampling interval might be a long enough period to allow that infection, resolution of symptoms, and viral excretion to go unnoticed; therefore, the episodes of infection presented here may be an underestimate of the true frequency. Despite these limitations, this prospective study based on PCR detection and sequencing of partial RNA-dependent RNA polymerase VP1 extends our knowledge on the natural course of NoV infection. The results reveal new insights on the frequency of infection, duration of viral shedding, and dynamics of circulation of NoV GII genotypes among inhabitants in a small community in Cameroon. The study suggests that the majority of natural NoV infections in this population is asymptomatic due to low viral load and/or herd immunity. A similar prospective study performed in temperate western countries hard hit by NoV would be of interest to search for differences in the natural infection of NoVs.
ACKNOWLEDGMENTS
We thank Maria Andersson for assisting in real-time PCR assays. The study was approved by the southwest regional delegation of public health in Cameroon. The study was sponsored by the Swedish International Development Agency, the FAS foundation, and the ALF Foundation at Sahlgrenska University Hospital.
We have no competing interests to declare.
Footnotes
Published ahead of print 1 October 2014
REFERENCES
- 1. Glass RI, Parashar UD, Estes MK. 2009. Norovirus gastroenteritis. N. Engl. J. Med. 361:1776–1785. 10.1056/NEJMra0804575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Patel MM, Widdowson MA, Glass RI, Akazawa K, Vinje J, Parashar UD. 2008. Systematic literature review of role of noroviruses in sporadic gastroenteritis. Emerg. Infect. Dis. 14:1224–1231. 10.3201/eid1408.071114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ayukekbong JA, Andersson ME, Vansarla G, Tah F, Nkuo-Akenji T, Lindh M, Bergstrom T. 2013. Monitoring of seasonality of norovirus and other enteric viruses in Cameroon by real-time PCR: an exploratory study. Epidemiol. Infect. 142:1393–1402. 10.1017/S095026881300232X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Xi JN, Graham DY, Wang KN, Estes MK. 1990. Norwalk virus genome cloning and characterization. Science 250:1580–1583. 10.1126/science.2177224. [DOI] [PubMed] [Google Scholar]
- 5. Zheng DP, Ando T, Fankhauser RL, Beard RS, Glass RI, Monroe SS. 2006. Norovirus classification and proposed strain nomenclature. Virology 346:312–323. 10.1016/j.virol.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 6. Hsu CC, Riley LK, Livingston RS. 2007. Molecular characterization of three novel murine noroviruses. Virus Genes 34:147–155. 10.1007/s11262-006-0060-1. [DOI] [PubMed] [Google Scholar]
- 7. Mesquita JR, Barclay L, Nascimento MS, Vinje J. 2010. Novel norovirus in dogs with diarrhea. Emerg. Infect. Dis. 16:980–982. 10.3201/eid1606.091861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Teunis PF, Moe CL, Liu P, Miller SE, Lindesmith L, Baric RS, Le Pendu J, Calderon RL. 2008. Norwalk virus: how infectious is it? J. Med. Virol. 80:1468–1476. 10.1002/jmv.21237. [DOI] [PubMed] [Google Scholar]
- 9. Chapman AS, Witkop CT, Escobar JD, Schlorman CA, DeMarcus LS, Marmer LM, Crum ME. 2011. Norovirus outbreak associated with person-to-person transmission, U.S. Air Force Academy, July 2011. MSMR 18:2–5. [PubMed] [Google Scholar]
- 10. Goller JL, Dimitriadis A, Tan A, Kelly H, Marshall JA. 2004. Long-term features of norovirus gastroenteritis in the elderly. J. Hosp. Infect. 58:286–291. 10.1016/j.jhin.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 11. Henke-Gendo C, Harste G, Juergens-Saathoff B, Mattner F, Deppe H, Heim A. 2009. New real-time PCR detects prolonged norovirus excretion in highly immunosuppressed patients and children. J. Clin. Microbiol. 47:2855–2862. 10.1128/JCM.00448-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tu ET, Bull RA, Kim MJ, McIver CJ, Heron L, Rawlinson WD, White PA. 2008. Norovirus excretion in an aged-care setting. J. Clin. Microbiol. 46:2119–2121. 10.1128/JCM.02198-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ayukekbong J, Lindh M, Nenonen N, Tah F, Nkuo-Akenji T, Bergstrom T. 2011. Enteric viruses in healthy children in Cameroon: viral load and genotyping of norovirus strains. J. Med. Virol. 83:2135–2142. 10.1002/jmv.22243. [DOI] [PubMed] [Google Scholar]
- 14. Nordgren J, Kindberg E, Lindgren PE, Matussek A, Svensson L. 2010. Norovirus gastroenteritis outbreak with a secretor-independent susceptibility pattern, Sweden. Emerg. Infect. Dis. 16:81–87. 10.3201/eid1601.090633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Parrino TA, Schreiber DS, Trier JS, Kapikian AZ, Blacklow NR. 1977. Clinical immunity in acute gastroenteritis caused by Norwalk agent. N. Engl. J. Med. 297:86–89. 10.1056/NEJM197707142970204. [DOI] [PubMed] [Google Scholar]
- 16. Kaplan JE, Schonberger LB, Varano G, Jackman N, Bied J, Gary GW. 1982. An outbreak of acute nonbacterial gastroenteritis in a nursing home. Demonstration of person-to-person transmission by temporal clustering of cases. Am. J. Epidemiol. 116:940–948. [DOI] [PubMed] [Google Scholar]
- 17. Atmar RL, Opekun AR, Gilger MA, Estes MK, Crawford SE, Neill FH, Graham DY. 2008. Norwalk virus shedding after experimental human infection. Emerg. Infect. Dis. 14:1553–1557. 10.3201/eid1410.080117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Huynen P, Mauroy A, Martin C, Savadogo LG, Boreux R, Thiry E, Melin P, De Mol P. 2013. Molecular epidemiology of norovirus infections in symptomatic and asymptomatic children from Bobo Dioulasso, Burkina Faso. J. Clin. Virol. 58:515–521. 10.1016/j.jcv.2013.08.013. [DOI] [PubMed] [Google Scholar]
- 19. Garcia C, DuPont HL, Long KZ, Santos JI, Ko G. 2006. Asymptomatic norovirus infection in Mexican children. J. Clin. Microbiol. 44:2997–3000. 10.1128/JCM.00065-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ayukekbong JA, Fobisong C, Lindh M, Nkuo-Akenji T, Bergstrom T, Norder H. 14 March 2014. Molecular analysis of enterovirus in Cameroon by partial 5′UTR-VP4 gene sequencing reveals a high genetic diversity and frequency of infections. J. Med. Virol. 10.1002/jmv.23926. [DOI] [PubMed] [Google Scholar]
- 21. Nenonen NP, Hannoun C, Olsson MB, Bergström T. 2009. Molecular analysis of an oyster-related norovirus outbreak. J. Clin. Virol. 45:105–108. 10.1016/j.jcv.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 22. Vennema H, de Bruin E, Koopmans M. 2002. Rational optimization of generic primers used for Norwalk-like virus detection by reverse transcriptase polymerase chain reaction. J. Clin. Virol. 25:233–235. 10.1016/S1386-6532(02)00126-9. [DOI] [PubMed] [Google Scholar]
- 23. Kojima S, Kageyama T, Fukushi S, Hoshino FB, Shinohara M, Uchida K, Natori K, Takeda N, Katayama K. 2002. Genogroup-specific PCR primers for detection of Norwalk-like viruses. J. Virol. Methods 100:107–114. 10.1016/S0166-0934(01)00404-9. [DOI] [PubMed] [Google Scholar]
- 24. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882. 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kabayiza JC, Andersson ME, Welinder-Olsson C, Bergstrom T, Muhirwa G, Lindh M. 2013. Comparison of rectal swabs and faeces for real-time PCR detection of enteric agents in Rwandan children with gastroenteritis. BMC Infect. Dis. 13:447. 10.1186/1471-2334-13-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Trainor E, Lopman B, Iturriza-Gomara M, Dove W, Ngwira B, Nakagomi O, Nakagomi T, Parashar U, Cunliffe N. 2013. Detection and molecular characterisation of noroviruses in hospitalised children in Malawi, 1997–2007. J. Med. Virol. 85:1299–1306. 10.1002/jmv.23589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bucardo F, Nordgren J, Carlsson B, Kindberg E, Paniagua M, Mollby R, Svensson L. 2010. Asymptomatic norovirus infections in Nicaraguan children and its association with viral properties and histo-blood group antigens. Pediatr. Infect. Dis. J. 29:934–939. 10.1097/INF.0b013e3181ed9f2f. [DOI] [PubMed] [Google Scholar]
- 28. Allen DJ, Gray JJ, Gallimore CI, Xerry J, Iturriza-Gomara M. 2008. Analysis of amino acid variation in the P2 domain of the GII-4 norovirus VP1 protein reveals putative variant-specific epitopes. PLoS One 3:e1485. 10.1371/journal.pone.0001485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Murata T, Katsushima N, Mizuta K, Muraki Y, Hongo S, Matsuzaki Y. 2007. Prolonged norovirus shedding in infants ≤6 months of age with gastroenteritis. Pediatr. Infect. Dis. J. 26:46–49. 10.1097/01.inf.0000247102.04997.e0. [DOI] [PubMed] [Google Scholar]
- 30. Siebenga JJ, Beersma MF, Vennema H, van Biezen P, Hartwig NJ, Koopmans M. 2008. High prevalence of prolonged norovirus shedding and illness among hospitalized patients: a model for in vivo molecular evolution. J. Infect. Dis. 198:994–1001. 10.1086/591627. [DOI] [PubMed] [Google Scholar]
- 31. McSwiggan DA, Cubitt D, Moore W. 1978. Calicivirus associated with winter vomiting disease. Lancet i:1215. [DOI] [PubMed] [Google Scholar]
- 32. Mounts AW, Ando T, Koopmans M, Bresee JS, Noel J, Glass RI. 2000. Cold weather seasonality of gastroenteritis associated with Norwalk-like viruses. J. Infect. Dis. 181(Suppl 2):S284–S287. 10.1086/315586. [DOI] [PubMed] [Google Scholar]