Abstract
A single-tube method, ligation-mediated real-time PCR high-resolution melt analysis (LMqPCR HRMA), was modified for the rapid typing of Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp. (ESKAPE) pathogens. A 97% agreement (60/62 isolates) was achieved in comparison to pulsed-field gel electrophoresis (PFGE) results, which indicates that LMqPCR HRMA is a rapid and accurate screening tool for monitoring nosocomial outbreaks.
TEXT
Increasing antibiotic resistance in bacterial pathogens presents a great threat to human health, and the prevention of the spread of such pathogens is important (1). The ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp. [2]) pathogens are especially problematic because of their antibiotic resistance mechanisms and nosocomial spread (2, 3). By using new strategies for the rapid typing of isolates, the possibility of preventing the uncontrolled spread of these pathogens is greatly increased. Pulsed-field gel electrophoresis (PFGE) has been considered the gold standard for epidemiological typing (4, 5). However, next-generation sequencing with whole-genome sequencing (WGS) is likely to become the new standard (6–10). The disadvantages of these methods are that they require both special equipment and standard protocols and training (4–6, 8–10). Thus, there is a need for a simple and fast typing method enabling real-time monitoring of nosocomial outbreaks. Most of the already-existing molecular typing methods, such as multilocus sequence typing (MLST), repetitive-sequence-based PCR (rep-PCR) (such as the commercial platform DiversiLab), and comparative genomic hybridization, although more rapid than PFGE still are expensive and species specific, and they exhibit insufficient resolution (11).
In this study, we further modified ligation-mediated PCR (LM PCR) with polyacrylamide gel analysis (12, 13) for use in a single-tube real-time PCR platform with high-resolution melt analysis (LMqPCR HRMA) for rapid epidemiological typing of the multiresistant ESKAPE pathogens. Following optimization, the method was evaluated blindly using a selection of ESKAPE pathogens that were previously defined by PFGE analysis as being identical, closely related, or unrelated (see the supplemental material).
Briefly, LMqPCR HRMA is based on fragmentation by a restriction enzyme (HindIII), followed by ligation of adaptor oligonucleotides to create amplicons. Analysis is done by HRM analysis, resulting in specific melt patterns depending on the number of amplified oligonucleotides, DNA sequence, G+C content, and length. By altering the denaturation temperature (TD) during PCR, the method was optimized for ESKAPE pathogens and performed in duplicate in three separate experiments. For a detailed description of the method, see the supplemental Materials and Methods. A complete list of the isolates, their origins, and resistance genes is presented in Table 1. All isolates had been analyzed with PFGE at the Public Health Agency of Sweden (see the supplemental Materials and Methods). A standardized protocol was developed based on several parameters of the melting curve shape to determine if two or more isolates were different within the same run (see the supplemental Materials and Methods).
TABLE 1.
Complete list of isolates included in the study
| Species and isolate | Origina | PFGE resultsb | LMqPCR HRMA resultsc | Resistance mechanism(s)d |
|---|---|---|---|---|
| E. faecium | ||||
| Ef01 | 1 | A | Ef-I | vanB |
| Ef02 | 1 | A1 | Ef-If | vanB |
| Ef03 | 1 | A | Ef-If | vanB |
| Ef04 | 1 | B | Ef-II | vanA |
| Ef05 | 1 | B | Ef-II | vanA |
| Ef06 | 1 | B1 | Ef-IIf | vanA |
| Ef07 | 1 | C | Ef-III | vanA |
| Ef08 | 1 | C | Ef-III | vanA |
| Ef09 | 1 | Unique | Similar to Ef-IIe,f | vanA |
| Ef10 | 1 | Unique | Unique | vanA |
| S. aureus | ||||
| Sa01 | 3 | A1 | Sa-If | mecA |
| Sa02 | 3 | B | Sa-II | mecA |
| Sa03 | 3 | B | Sa-II | mecA |
| Sa04 | 3 | Unique | Unique | mecA |
| Sa05 | 3 | C1 | Sa-III | mecA |
| Sa06 | 3 | C2 | Sa-IIIf | mecA |
| Sa07 | 3 | A2 | Sa-I | mecA |
| Sa08 | 3 | D | Sa-IV | mecA |
| Sa09 | 3 | D | Sa-IV | mecA |
| Sa10 | 3 | A2 | Sa-I | mecA |
| Sa11 | 3 | A2 | Sa-I | mecA |
| Sa12 | 3 | A3 | Sa-If | mecA |
| K. pneumoniae | ||||
| Kp01 | 1 | A | Kp-I | CTX-M-1 |
| Kp02 | 1 | A | Kp-I | CTX-M-1 |
| Kp03 | 1 | A | Kp-I | CTX-M-1 |
| Kp04 | 1 | B | Kp-II | CTX-M-1 |
| Kp05 | 1 | B | Kp-II | CTX-M-1 |
| Kp06 | 1 | B | Kp-II | CTX-M-1 |
| Kp07 | 1 | C | Kp-III | CTX-M-1 |
| Kp08 | 1 | C | Kp-III | CTX-M-1 |
| Kp09 | 1 | Unique | Unique | CTX-M-9 |
| Kp10 | 1 | Unique | Unique | CTX-M-1 |
| A. baumannii | ||||
| Ab01 | 2 | Unique | Unique | OXA-23, OXA-51 |
| Ab02 | 2 | A1 | Ab-Ie | OXA-23, OXA-51 |
| Ab03 | 2 | A2 | Ab-Ie,f | OXA-23, OXA-51 |
| Ab04 | 3 | B | Ab-II | OXA-23, OXA-51 |
| Ab05 | 2 | B | Ab-II | OXA-23, OXA-51 |
| Ab06 | 2 | B | Ab-II | OXA-23, OXA-51 |
| Ab07 | 1 | Unique | Unique | OXA-51 |
| Ab08 | 1 | A | Ab-III | OXA-23, OXA-51 |
| Ab09 | 1 | A | Ab-III | OXA-23, OXA-51 |
| Ab10 | 1 | Unique | Unique | No OXA |
| P. aeruginosa | ||||
| Pa01 | 1 | A | Pa-I | IMP |
| Pa02 | 1 | A | Pa-I | IMP |
| Pa03 | 1 | A | Pa-I | IMP |
| Pa04 | 1 | B | Pa-II | No MBL |
| Pa05 | 1 | B | Pa-II | No MBL |
| Pa06 | 1 | C | Pa-III | VIM |
| Pa07 | 1 | C1 | Pa-IIIf | VIM |
| Pa08 | 1 | C2 | Pa-IIIf | VIM |
| Pa09 | 1 | Unique | Unique | No MBL |
| Pa10 | 1 | Unique | Unique | No MBL |
| E. cloacae | ||||
| EnC01 | 1 | A | EnC-I | cAmpC |
| EnC02 | 1 | A | EnC-I | cAmpC |
| EnC03 | 1 | Unique | Unique | cAmpC |
| EnC04 | 1 | B | EnC-II | cAmpC |
| EnC05 | 1 | B | EnC-II | cAmpC |
| EnC06 | 1 | B | EnC-II | cAmpC |
| EnC07 | 1 | Unique | Unique | cAmpC |
| EnC08 | 1 | Unique | Unique | cAmpC |
| EnC09 | 1 | Unique | Unique | cAmpC |
| EnC10 | 1 | Unique | Unique | cAmpC |
Origins 1 to 3 are defined as follows: 1, the Public Health Agency of Sweden, Solna, Sweden; 2, Department of Clinical Microbiology, County Council of Östergötland, Linköping, Sweden; and 3, Department of Clinical Microbiology, Kalmar County Hospital, Kalmar, Sweden.
Arbitrary classification of PFGE cluster within each species are as follows: letters denote isolates within a cluster with >90 to 97% similarity, numbers denote closely related patterns with >97% similarity.
Classification of HRMA clusters I, II, III, etc. Ef, E. faecium; Sa, S. aureus; Kp, K. pneumonia; Ab, A. baumannii; Pa, P. aeruginosa; EnC, E. cloacae.
vanA and vanB indicate vancomycin resistance; mecA indicates methicillin resistance; CTX-M phylogenetic groups 1 and 9, species-specific OXA enzymes, and metallo-β-lactamases (MBL) IMP and VIM indicate β-lactamases; cAmpC indicates chromosomal β-lactamases.
Isolates for which the results from PFGE (with 90 to 97% similarity) and HRMA do not correspond.
Isolates for which the results from PFGE (with >97% similarity) and HRMA do not correspond.
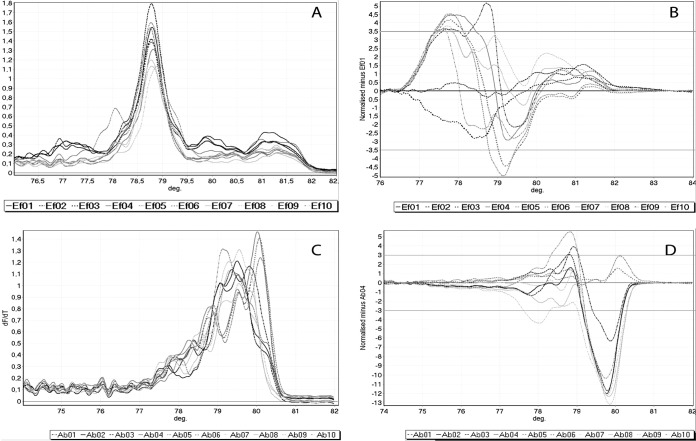
Overall, 59 out of 62 of the ESKAPE pathogen isolates were identified with full agreement between LMqPCR HRMA and PFGE (≥90% as the cutoff for similarity) (Table 1). The LMqPCR HRMA and PFGE results did not correlate for E. faecium strain Ef09 and A. baumannii strains Ab02 and Ab03 (Fig. 1). Ef09 clustered with other isolates using LMqPCR HRMA but was defined as unique by PFGE. When adding agarose gel electrophoresis analysis after HRMA, 60 out of 62 isolates (97%) were identified with full agreement with PFGE, with Ef09 being classified as unique. In contrast to PFGE, for which Ab02 and Ab03 were defined as being similar to A. baumannii strains Ab08 and Ab09, these two groups of isolates were not defined as similar by LMqPCR HRMA.
FIG 1.
LMqPCR HRMA results for E. faecium (A and B) and A. baumannii (C and D). (A and C) Complete melting curves for all isolates are visualized plotting df/dt against temperature (°C). The algorithm from Woksepp et al. (14) was used on the complete melting curves visualized in panels A and C, together with a subjective evaluation of the discrepancies between the melting curves for clustering analysis of the isolates. (B) Isolate E. faecium Ef01 is set as reference, plotting the difference of all other isolates to Ef01. The isolates within ±3.5 U of the y axis (Ef02 and Ef03) are considered to cluster with Ef01. (D) Isolate A. baumannii Ab04 is set as reference, plotting the difference of all other isolates to Ab04. Isolates within ±3 U of the y axis (Ab05 and Ab06) are considered to cluster with Ab04. For E. faecium, Ef01, Ef02, and Ef03 clustered together, Ef04, Ef05, and Ef06 clustered together, and Ef07 and Ef08 clustered together, with Ef09 being similar to the isolates Ef04, Ef05, and Ef06. Ef10 was unique. For A. baumannii, Ab02 and Ab03 clustered together, Ab04, Ab05, and Ab06 clustered together, and Ab08 and Ab09 clustered together. Ab01, Ab07, and Ab10 were unique. deg, degrees Celsius.
Using >97% as the similarity cutoff for the PFGE results, the methods were in agreement for 53 out of 62 isolates (85%), with discrepancies for E. faecium strains Ef02, Ef06, and Ef09, S. aureus strains Sa01, Sa06, and Sa12, A. baumannii Ab03, and P. aeruginosa strains Pa07 and Pa08 (Table 1). No discrepancies were found for K. pneumoniae and Enterobacter cloacae strains. None of the isolates determined as being unique according to LMqPCR HRMA were evaluated as being similar in the PFGE analysis (cutoff for PFGE, >97%). The HRMA method is therefore suitable for confirming that isolates in suspected nosocomial outbreaks are not related. The complete list of results is presented in Table 1.
We describe a rapid method based on restriction cleavage and ligation-mediated real-time PCR (qPCR) with high-resolution melt analysis, which can be used as a primary method for epidemiological typing in the investigation of suspected nosocomial outbreaks caused by pathogens from the ESKAPE group and as a definitive method for excluding unrelated isolates. This method has already been shown to identify a nosocomial outbreak of O25b-sequence type 131 (ST131)-associated extended-spectrum-β-lactamase (ESBL)-producing Escherichia coli isolates with a resolution comparable to that found with PFGE (14). Hence, in the evaluation of two or more isolates from a suspected outbreak in the routine microbiological laboratory, this method can confirm whether such isolates are different and to a very high certainty if they are similar. LMqPCR HRMA does not require any prior knowledge of the DNA sequence, although the typeability is dependent on the G+C content of the species. In order to determine the similarities and differences between isolates, it is possible to use the Rotor-Gene software and the algorithm previously developed by Woksepp et al. (14). Among the ESKAPE pathogens, the HRM patterns from the Gram-negative bacteria were more straightforward to evaluate than were the Gram-positive bacteria. When analyzing the HRMA results for Gram-negative bacteria, a visual inspection of the HRM curves was enough for all but A. baumannii to determine discrepancies between the isolates (see Fig. S1A in the supplemental material). For Gram-positive bacteria, our results show that additional agarose electrophoresis might be of benefit, especially in cases where the isolates are similar by HRM analysis only. The species that gave the highest resolution in LMqPCR HRMA were K. pneumoniae, P. aeruginosa, E. coli (14), and E. cloacae. All of these species have G+C contents between 51 and 66%, in contrast to E. faecium, S. aureus, and A. baumannii, which have G+C contents between 33% and 39% (http://www.ncbi.nlm.nih.gov/genome).
Our study has limitations, one being that it was performed on a limited number of isolates for each of the chosen species, although the isolates were carefully selected to represent identical, closely related, and unrelated types, based on the PFGE results. Another limitation is that despite the algorithm used, the HRMA data interpretation has a subjective component that is shared with other methods for epidemiological typing.
The LMqPCR HRMA method was primarily developed for the rapid investigation of clonal dissemination. For long-term surveillance, other methods, such as PFGE or WGS, should be used. However, the results confirm the usefulness of this rapid method, both as a screening method to exclude a potential nosocomial outbreak of ESKAPE pathogens but also to indicate relatedness to a high degree of certainty. It is important, however, to remember that typing results should be correlated to clinical and epidemiological data.
In conclusion, LMqPCR HRMA is a rapid and simple method for investigating nosocomial outbreaks of resistant ESKAPE pathogens. Results comparable to those from PFGE can be achieved within 1 day in a single-tube system.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by grants from Marianne and Marcus Wallenberg Foundation and FORSS (The Research Council of Southeast Sweden).
Footnotes
Published ahead of print 17 September 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.02537-14.
REFERENCES
- 1. Spellberg B, Bartlett JG, Gilbert DN. 2013. The future of antibiotics and resistance. N. Engl. J. Med. 368:299–302. 10.1056/NEJMp1215093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rice LB. 2008. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. J. Infect. Dis. 197:1079–1081. 10.1086/533452. [DOI] [PubMed] [Google Scholar]
- 3. Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. 2009. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 48:1–12. 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 4. van Belkum A. 1994. DNA fingerprinting of medically important microorganisms by use of PCR. Clin. Microbiol. Rev. 7:174–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. van Belkum A, Tassios PT, Dijkshoorn L, Haeggman S, Cookson B, Fry NK, Fussing V, Green J, Feil E, Gerner-Smidt P, Brisse S, Struelens M, European Society of Clinical Microbiology and Infectious Diseases (ESCMID) Study Group on Epidemiological Markers (ESGEM). 2007. Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clin. Microbiol. Infect. 13(Suppl 3):1–46. 10.1111/j.1469-0691.2007.01786.x. [DOI] [PubMed] [Google Scholar]
- 6. Bertelli C, Greub G. 2013. Rapid bacterial genome sequencing: methods and applications in clinical microbiology. Clin. Microbiol. Infect. 19:803–813. 10.1111/1469-0691.12217. [DOI] [PubMed] [Google Scholar]
- 7. Dark MJ. 2013. Whole-genome sequencing in bacteriology: state of the art. Infect. Drug Resist. 6:115–123. 10.2147/IDR.S35710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Köser CU, Ellington MJ, Cartwright EJ, Gillespie SH, Brown NM, Farrington M, Holden MT, Dougan G, Bentley SD, Parkhill J, Peacock SJ. 2012. Routine use of microbial whole genome sequencing in diagnostic and public health microbiology. PLoS Pathog. 8:e1002824. 10.1371/journal.ppat.1002824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Reuter S, Ellington MJ, Cartwright EJ, Köser CU, Török ME, Gouliouris T, Harris SR, Brown NM, Holden MT, Quail M, Parkhill J, Smith GP, Bentley SD, Peacock SJ. 2013. Rapid bacterial whole-genome sequencing to enhance diagnostic and public health microbiology. JAMA Intern. Med. 173:1397–1404. 10.1001/jamainternmed.2013.7734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Török ME, Peacock SJ. 2012. Rapid whole-genome sequencing of bacterial pathogens in the clinical microbiology laboratory–pipe dream or reality? J. Antimicrob. Chemother. 67:2307–2308. 10.1093/jac/dks247. [DOI] [PubMed] [Google Scholar]
- 11. Sabat AJ, Budimir A, Nashev D, Sá-Leão R, van Dijl J, Laurent F, Grundmann H, Friedrich AW. 2013. Overview of molecular typing methods for outbreak detection and epidemiological surveillance. Euro Surveill. 18:pii=20380 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20380. [DOI] [PubMed] [Google Scholar]
- 12. Masny A, Płucienniczak A. 2003. Ligation mediated PCR performed at low denaturation temperatures–PCR melting profiles. Nucleic Acids Res. 31:e114. 10.1093/nar/gng116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Krawczyk B, Samet A, Leibner J, Śledzińska A, Kur J. 2006. Evaluation of a PCR melting profile technique for bacterial strain differentiation. J. Clin. Microbiol. 44:2327–2332. 10.1128/JCM.00052-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Woksepp H, Jernberg C, Tärnberg M, Ryberg A, Brolund A, Nordvall M, Olsson-Liljequist B, Wisell KT, Monstein HJ, Nilsson LE, Schön T. 2011. High-resolution melting-curve analysis of ligation-mediated real-time PCR for rapid evaluation of an epidemiological outbreak of extended-spectrum-beta-lactamase-producing Escherichia coli. J. Clin. Microbiol. 49:4032–4039. 10.1128/JCM.01042-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.