Abstract
We compared paired enzyme immunoassay (EIA) and latex agglutination (LA) assay results with 185 blood and 164 cerebrospinal fluid (CSF) samples from 44 and 33 non-HIV cryptococcosis patients, respectively. The LA assay cutoff of 1:256 in the blood and 1:32 in the CSF was most highly predictive of a positive EIA result. The EIA missed 18.4% detected by the LA assay in the blood samples and 7.8% detected by the LA assay in the CSF samples. We note here the improved sensitivity of the LA assay over the EIA in non-HIV patients.
TEXT
Cryptococcus neoformans and Cryptococcus gattii are major causes of fungal meningoencephalitis in HIV-infected and -uninfected individuals. The polysaccharide capsule of these species is a virulence factor and is a major diagnostic tool used in clinical practice (1). Although there are many commercially available kits, the enzyme immunoassay (EIA) and latex agglutination (LA) test predominate. However, the LA assay requires the performance of a time-consuming and operator-variable visually assessed agglutination assay and requires repetitive dilutions for highly positive specimens, whereas the EIA utilizes an automated spectrophotometric method (2). Previous studies comparing various platforms have been performed on blood and cerebrospinal fluid (CSF) specimens, primarily those from AIDS patients. For example, in a study of 182 CSF and 90 blood samples from 49 C. neoformans culture-positive specimens, the sensitivity and specificity of the 4 LA assay and 1 EIA kits tested were 93 to 100% and 93 to 98%, respectively, for CSF samples. For blood samples, kits that did not pretreat with Pronase had reduced sensitivities (83%) but comparable specificities (93 to 100%). However, similar studies to determine the accuracy of the LA assay and EIA in diagnosing cryptococcosis in non-HIV patients have not been conducted, and the tests might be less sensitive due to lower antigen loads in this population. As such, we undertook a retrospective review of such patients with cryptococcosis (100% disease prevalence) to ascertain the relative sensitivities and specificities of the EIA and LA assay (2).
The subjects were participants in a National Institute for Allergy and Infectious Diseases (NIAID) institutional review board (IRB)-approved protocol on cryptococcosis in adults without known immunocompromising conditions. We included outside-referred patients from 2009 to 2013 and obtained the appropriate written informed consent.
Cryptococcal antigen (CrAg) EIA testing was performed on time-paired (same or close to the same date) sequential blood and CSF specimens from the same patient using the Premier cryptococcal antigen kit (Meridian Biosciences, Inc.), as per the manufacturer's instructions, with the exception of the following changes. Bloody CSF specimens were centrifuged at 3,000 × g for 10 min prior to testing, and nonbloody CSF specimens were tested directly. The blood specimens were allowed to clot for ≥10 min prior to processing, followed by centrifugation at 3,000 × g for 10 min and testing of the serum. The EIA results were read spectrophotometrically. A cryptococcal antigen LA assay was performed using the Cryptococcal antigen latex agglutination system (CALAS) kit (Meridian Biosciences, Inc.), as per the manufacturer's instructions, with the exception of the following changes. The CSF specimens were incubated at 56°C for 30 min and allowed to cool to room temperature. The serum specimens were digested with Pronase at a 1:1 ratio (200 μl each of serum and Pronase) for 15 min at 56°C, followed by 5 min at 100°C. Due to the added Pronase volume, the lowest dilution tested for the serum specimens was 1:2. The CSF specimens did not receive Pronase treatment, and the lowest dilution tested was 1:1.
We used logistic regression to ascertain how well the EIA and LA assay agreed for blood and CSF samples separately by treating the EIA as the binary outcome and base-2 logarithm of the corresponding LA assay as the independent covariate. We analyzed these longitudinally acquired data, accounting for correlated serial measurements per individual using cross-validation and bootstrapping. The resulting predicted probability of a positive EIA result and receiver operating characteristic sensitivity versus specificity curves were generated (3).
We identified 185 blood CrAg specimens from 44 patients with cryptococcosis and 164 CSF CrAg specimens from 33 patients with cryptococcosis (there were 106 from blood and 100 from CSF for which both EIA and LA assay measures were available [Table 1]). Figure 1 illustrates the probability of a positive EIA score as a function of the LA assay results among blood and CSF specimens. An LA assay score of 1:256 for a blood specimen was associated with a high probability (>90%) of a positive EIA result. An LA assay score of 1:4 was associated with the probability of a positive EIA result of approximately 10%. An LA assay score of ≥1:32 in the CSF highly predicted a correspondingly sampled positive EIA result. A receiver operating characteristic (ROC) curve of the relative sensitivity and specificity of the LA compared to those of the EIA (Fig. 2) demonstrated an area under the curve (AUC) for the assays for blood samples to be 0.90 [95% CI, 0.81 to 0.97], whereas the AUC for CSF samples was greater, at 0.96 [95% CI, 0.88 to 1.00]. The EIA missed 18.4% of the positives that were detected by the LA assay in blood (Table 2) and 7.8% of the positives that were detected by the LA assay in CSF (Table 2). These errors were decreased further in the cross-validation model. There were far fewer false-positive results in the CSF specimens than in the blood specimens.
TABLE 1.
Cross-tabulation of EIA and LA assay results from blood and CSF samples
| Sample type and latex agglutination dilution | No. with EIA result of: |
|
|---|---|---|
| Negative | Positive | |
| Blood | ||
| Undetectable | 28 | 1 |
| 1:2 | ||
| 1:4 | 6 | 0 |
| 1:8 | 3 | 1 |
| 1:16 | 6 | 7 |
| 1:32 | 10 | 7 |
| 1:64 | 4 | 8 |
| 1:128 | 0 | 4 |
| 1:256 | 0 | 7 |
| 1:512 | 0 | 9 |
| 1:1,024 | 0 | 5 |
| CSF | ||
| Undetectable | 36 | 3 |
| 1:2 | 4 | 2 |
| 1:4 | 1 | 5 |
| 1:8 | 0 | 6 |
| 1:16 | 0 | 5 |
| 1:32 | 1 | 9 |
| 1:64 | 0 | 5 |
| 1:128 | 0 | 6 |
| 1:256 | 0 | 3 |
| 1:512 | 0 | 4 |
| 1:1,024 | 1 | 6 |
| 1:2,048 | 0 | 3 |
FIG 1.
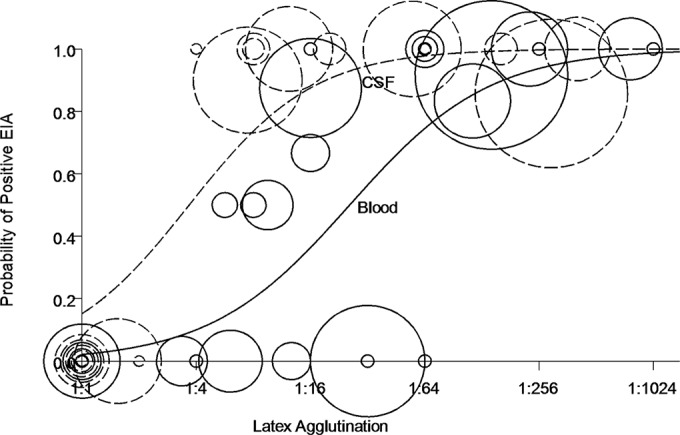
Predicted probability of a positive EIA result as a function of LA titer demonstrates reduced sensitivity of the EIA. The predicted probability of a positive enzyme immunoassay (EIA) result for cryptococcal antigen was determined as a function of the latex agglutination (LA) titer from the blood (solid line) and cerebrospinal fluid (CSF) (dashed line) among a cohort of patients with cryptococcosis without known immunocompromising conditions. The model was fit such that the probability of a positive EIA was exp(θ)/[1 + exp(θ)], where θ = −3.828 + 0.817 × log2(LA) in the blood and θ = −1.722 + 0.917 × log2(LA) in the CSF. The actual data are depicted as circles in which each circle represents one person. The size of the circle is based on the number of LA and EIA pairs for that person and specimen type, with the average LA value as a proportion of EIAs that were positive. Specimens testing negative by LA are represented by circles centered at the origin.
FIG 2.
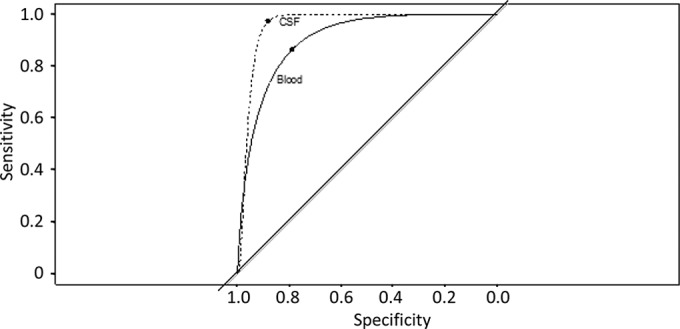
Receiver operating characteristic (ROC) curve demonstrates decreased relative sensitivity and specificity of EIA and LA tests from blood compared to those from CSF. The ROC curve of blood (solid line) was compared to that of CSF (dashed line). The upper left-hand corner of each curve signifies the values at which sensitivity and specificity were optimized (78.9% and 86.5% for blood and 88.3% and 97.3% for CSF, respectively); 1:10.6 and 1:2.7 were the LA threshold values for this optimal sensitivity and specificity from the blood and CSF, respectively. The area under the curve (AUC) for assays on the blood was 0.90 (95% confidence interval [CI], 0.81 to 0.97), whereas the AUC for CSF was greater, at 0.96 (95% CI, 0.88 to 1.00).
TABLE 2.
Relative sensitivity and specificity values of EIA and LA assaya
| Sample type and analysis | Sensitivity (%) | Specificity (%) |
|---|---|---|
| Blood | ||
| Applied to same data | 81.6 | 75.4 |
| Cross-validated | 91.8 | 59.6 |
| CSF | ||
| Applied to same data | 91.2 | 93.0 |
| Cross-validated | 91.2 | 83.7 |
The relative sensitivity and specificity values from the sample-derived model and cross-validated model were determined using an LA assay cutoff based on a 50% chance of a positive EIA result being lower in the blood than in the CSF. Using the same data set to generate a model and assess its performance is known to overestimate its predictive ability. To account for this, we used cross-validation; one at a time, we excluded a patient, fit the best model to the remaining patients, and then assessed its accuracy on the patient who was left out.
We demonstrated a high level of agreement between the EIA and LA assay in a less well-characterized cohort of previously healthy patients with cryptococcosis who had no known immune predisposition. The relative sensitivities and specificities of these platforms have not been described in this population. Since the LA assay cutoffs of 1:256 in the blood and 1:32 in the CSF were most highly predictive of a positive EIA result, the LA assay appears to have a lower limit of detection than the EIA in those without readily identifiable cellular immune deficits, such as AIDS, transplant recipients, cancer, and those receiving immunosuppressive medications, including corticosteroids. Although the results may have been altered with the use of antifungal therapy in the treated patients, there were no data to suggest this occurred, and all patients in the cohort were treated similarly, suggesting nondifferential therapeutic interventions. The C-statistic (AUC of the ROC) ranged from 0.90 to 0.96, indicating that the fitted model highly discerned the relative true positives from true negatives. The proportion of false negatives in the EIA was higher in the blood specimens than in the CSF specimens. Though more labor-intensive and requiring paired comparisons to previous samples, the LA assay appears to be more sensitive than the EIA in patients with cryptococcosis without known immunocompromising conditions.
ACKNOWLEDGMENTS
This research was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases (NIAID) at the National Institutes of Health (NIH).
The views herein do not reflect the official opinions of the Uniformed Services University or the Department of Defense.
Footnotes
Published ahead of print 24 September 2014
REFERENCES
- 1. Olszewski MA, Zhang Y, Huffnagle GB. 2010. Mechanisms of cryptococcal virulence and persistence. Future Microbiol. 5:1269–1288. 10.2217/fmb.10.93. [DOI] [PubMed] [Google Scholar]
- 2. Tanner DC, Weinstein MP, Fedorciw B, Joho KL, Thorpe JJ, Reller L. 1994. Comparison of commercial kits for detection of cryptococcal antigen. J. Clin. Microbiol. 32:1680–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Steyerberg EW, Vickers AJ, Cook NR, Gerds T, Gonen M, Obuchowski N, Pencina MJ, Kattan MW. 2010. Assessing the performance of prediction models: a framework for traditional and novel measures. Epidemiology 21:128–138. 10.1097/EDE.0b013e3181c30fb2. [DOI] [PMC free article] [PubMed] [Google Scholar]


