Abstract
No simple diagnostic tool is available to confirm Mycobacterium ulcerans infection, which is an emerging disease reported in many rural areas of Africa. Here, we report the 1-year results of a hospital laboratory that was created in an area of endemicity of Benin to facilitate the diagnosis of M. ulcerans infection.
TEXT
Buruli ulcer is a neglected tropical disease occurring mainly in poor, rural communities in West Africa and is caused by the environmental pathogen Mycobacterium ulcerans. This bacillus produces a unique toxin called mycolactone, which has cytotoxic and immunomodulatory activity and often causes severe skin ulcerations, disfigurement, and disability, with children the most affected. A combination of rifampin and streptomycin or rifampin and clarithromycin has been recommended by the World Health Organization (WHO) since 2004 as the first-line treatment (1, 2). M. ulcerans infection is confirmed by laboratory examination, including Ziehl-Neelsen staining, PCR, histology, and/or culture (3). A fine-needle aspiration (FNA) sample may be collected from nonulcerative lesions (nodule, plaque, or edema), and swabs are taken from the undermined edges of ulcerative lesions (4, 5). Biopsy specimens can be obtained from a punch biopsy sample or from excised necrotic tissue. Rapid diagnosis is essential for the management of patients. Since November 2012, the field laboratory established at the Buruli ulcer treatment center in Pobè, Benin, has been fully equipped to carry out M. ulcerans infection diagnosis and has three separate rooms to avoid cross-contamination. Technicians have been trained to perform quantitative PCR (qPCR) and culture in addition to smear examination. Here, we evaluated the 1-year results of this laboratory, which is the first one associated with a specialized treatment center located in a rural area of endemicity and able to carry out qPCR, culture, and direct smear examination (DSE) of a given sample. Detection and quantification of M. ulcerans by qPCR were compared with culture and Ziehl-Neelsen staining results, and the beneficial effects of this laboratory approach on patient management were evaluated.
Among 394 specimens analyzed, 203 (51%) were taken by swab, 101 (26%) by FNA, and 90 (23%) by biopsy. The samples were analyzed by molecular tests (qPCR) and microbiological tests (culture and Ziehl-Neelsen staining). Briefly, swabs were rehydrated and biopsy specimens were minced in 2 ml of sterile water. The Kubica method was used to decontaminate 400 μl of swab and skin biopsy specimens to isolate M. ulcerans strains (6). FNA specimens were not decontaminated, because the sampling procedure is considered sterile. After inoculation onto Lowenstein-Jensen medium, growth was monitored weekly for 5 months. Ziehl-Neelsen staining was performed as described previously (3). To prepare DNA, 400 μl of suspension was centrifuged, resuspended in 50 mM NaOH solution, and heated at 95°C for 10 min. DNA was then purified with a QIAquick PCR purification kit (Qiagen), according to the manufacturer's instructions. Negative and positive controls were systematically included. IS2404 primers and probe were selected from Rondini et al., and amplification and analysis were performed with a Step One real-time PCR system (Applied Biosystems) (4, 7). Sample DNA was quantified from an external standard curve made with a series of six 10-fold serial dilutions of M. ulcerans (strain 1G897) DNA (Fig. 1A).
FIG 1.
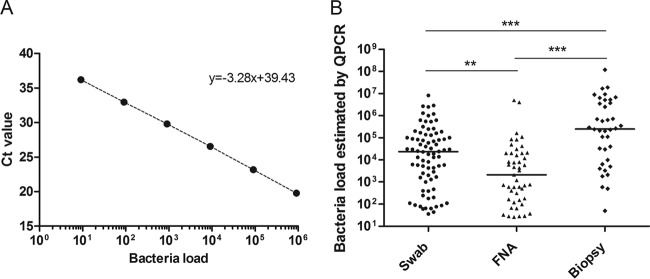
Quantification of M. ulcerans DNA by qPCR analysis of swabs, FNA samples, and biopsy specimens. (A) Representative external standard curve made from a series of six 10-fold serial dilutions of M. ulcerans DNA. Ct, threshold cycle. (B) Estimation of bacterial loads in swabs, FNA samples, and biopsy specimens (n = 163). **, P < 0.01; ***, P < 0.0001. Bacterial load data show the number of bacteria per ml detected by qPCR.
Of the 394 samples, 163 (41%) were qPCR positive, confirming the diagnosis of Buruli ulcer. The proportions of samples identified as positive for M. ulcerans by PCR were similar for FNA specimens, swabs, and skin biopsy specimens (P = 0.68). Indeed, M. ulcerans DNA was detected in 78 swabs (38%), 47 FNA specimens (47%), and 38 skin biopsy specimens (42%). The bacterial loads estimated by qPCR differed according to the type of sampling (with median loads of 24 × 103, 21 × 102, and 25 × 104 bacteria/ml in swabs, FNA specimens, and skin biopsy specimens, respectively) (Table 1). Skin biopsy specimens contained the largest amount of M. ulcerans DNA (P < 0.0001), followed by swabs (P < 0.01) (Fig. 1B). The amount of bacterial DNA detected did not depend on the WHO category of the lesion (1), and this was true for all types of sampling methods (data not shown).
TABLE 1.
Comparison of qPCR, DSE, and culture results with specimens obtained by swab, FNA, and biopsy
| Parameter | Value(s) |
|||
|---|---|---|---|---|
| Swab | FNA specimen | Biopsy specimen | Total | |
| % qPCR-positive specimens (no. of qPCR-positive specimens/total no. of specimens) | 38 (78/203) | 47 (47/101) | 42 (38/90) | 42% |
| Median bacterial DNA load estimated by qPCR (no. of bacteria/ml) | 23,500 | 2,100 | 250,000 | 91,900 |
| % positive DSEa (no. of qPCR-positive specimens/total no. of specimens) | 52 (41/78) | 13 (6/47) | 82 (31/38) | 47% |
| % positive cultureb (no. of qPCR-positive specimens/total no. of specimens) | 61 (41/67) | 63 (24/38) | 62 (10/16) | 62% |
DSE, direct smear examination.
Data include only samples taken before the beginning of antibiotic treatment.
For culture experiments, we analyzed only qPCR-positive specimens that were taken from qPCR-positive patients who had not started antibiotic treatment. Despite the relationship between the bacterial load estimated by qPCR and the type of sample, the positive culture rates were approximately the same for all three sampling methods: 61% (41/67) for swabs, 63% (24/38) for FNA specimens, and 62% (10/16) for skin biopsy specimens (Table 1). Three cultures (2.5%) showed fungal contamination, and the median incubation time required to obtain a positive culture was 45 days (minimum, 30; maximum, 120). Bacterial load estimated by qPCR was associated with a positive culture rate for swabs and FNA specimens. Indeed, the bacterial load was significantly higher in culture-positive specimens than in culture-negative specimens (Fig. 2A). However, the positive culture rate reached a plateau at around 80%, at which point an increase in bacterial load had no effect on the proportion of positive cultures (Fig. 2B). DSE results after Ziehl-Neelsen staining were positive in 47% of qPCR-positive specimens. The levels of sensitivity of DSE were highly dependent on the type of sampling and were 13% in FNA specimens, 52% in swabs, and 82% in skin biopsy specimens (Table 1), which is consistent with the results of a previous study (4).
FIG 2.
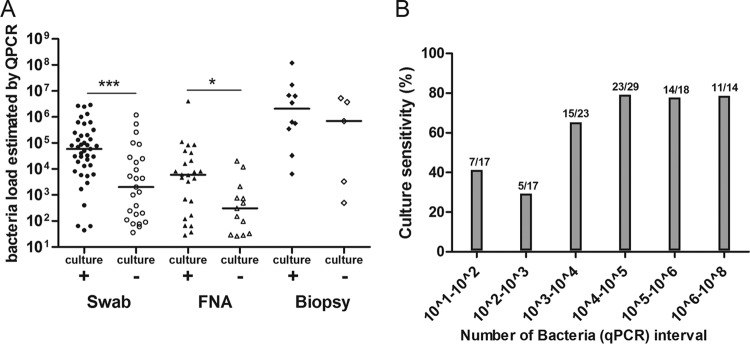
Sensitivity of culture in qPCR-positive specimens. (A) Bacterial load detected by qPCR in swabs, FNA samples, or biopsy specimens divided according to whether they gave positive or negative culture results. *, P < 0.1; ***, P < 0.001. (B) Positive culture rate as a function of bacterial load detected by qPCR in specimens (n = 118). Bacterial load data show the number of bacteria per ml determined by qPCR.
The Raoul Follereau hospital in Pobè, Benin, was established in 2004 and is located in the middle of an area of endemicity in southeastern Benin, close to the Nigerian border. This center treats around 150 Buruli ulcer patients per year. The creation of a molecular laboratory inside the hospital has enabled the direct processing of samples. We chose to use a qPCR method rather than a gel-based PCR method because this method minimizes cross-contamination from amplicons, is faster and more sensitive than gel-based PCR, and involves less toxic waste. Here, we describe for the first time the bacterial load of M. ulcerans estimated by qPCR in a large number of specimens and examine its relationship with the sensitivity of culture. The median bacterial load detected by qPCR in FNA specimens was lower than in swabs and in skin biopsy specimens, although the positive culture rates were equivalent for all sampling methods. The absence of the decontamination step for FNA specimens may explain this result, because this procedure negatively affects the population of cultivable mycobacteria. It is also possible that the number of dead bacteria in open lesions (collected using swabs) is higher than in closed lesions (collected using FNA). The positive culture rate among all qPCR-positive specimens was 62%, which is higher than those reported in previous studies (positive culture rates between 20% and 50%) (8–11). Moreover, the median incubation time required for the growth of M. ulcerans in culture was 1.7 times lower than that described previously (6 to 7 weeks versus 10 to 11 weeks, respectively) (8). Immediate processing of the specimens probably explains the high yield and short incubation period.
The recent establishment of the molecular laboratory in the peripheral hospital in Pobè is very beneficial for medical staff and patients. Results can be delivered quickly (Ziehl-Neelsen in 1 day, qPCR in 1 week), enabling rapid retesting if a first negative result does not correspond with clinical features. Indeed, second or third samples are needed to confirm Buruli ulcer in around 15% of patients. Some clinical forms of Buruli ulcer are difficult to diagnose clinically, even for experienced medical staff, and need biological confirmation (12). Furthermore, laboratory results are particularly valuable in cases of paradoxical reaction or suspected relapse (13–16). For example, during this investigation, 17 specimens were carefully evaluated because they came from patients with suspected paradoxical reactions. All these specimens were M. ulcerans-PCR positive, and none were culture positive, supporting the idea that the paradoxical reactions were not due to a failure in chemotherapy. Culture is also useful to monitor both the emergence of resistance to antibiotics and relapses and is essential for research on environmental dissemination and transmission.
ACKNOWLEDGMENTS
This study was supported by the Fondation Raoul Follereau, by the Institut National de la Santé et de la Recherche Médicale (Inserm, Programme Inserm Avenir), and by the Agence Nationale de Recherche sur le SIDA et les Hépatites.
Footnotes
Published ahead of print 15 October 2014
REFERENCES
- 1.World Health Organization. 2012. Treatment of Mycobacterium ulcerans (Buruli ulcer): guidance for health workers, p 73 World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Converse PJ, Nuermberger EL, Almeida DV, Grosset JH. 2011. Treating Mycobacterium ulcerans disease (Buruli ulcer): from surgery to antibiotics, is the pill mightier than the knife? Future Microbiol. 6:1185–1198. 10.2217/fmb.11.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Portaels F, Johnson P, Meyers WM. 2001. Buruli ulcer, diagnosis of M. ulcerans disease. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 4.Cassisa V, Chauty A, Marion E, Ardant MF, Eyangoh S, Cottin J, Aubry J, Koussemou H, Lelievre B, Ferec S, Tekaia F, Johnson C, Marsollier L. 2010. Use of fine-needle aspiration for diagnosis of Mycobacterium ulcerans infection. J. Clin. Microbiol. 48:2263–2264. 10.1128/JCM.00558-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eddyani M, Fraga AG, Schmitt F, Uwizeye C, Fissette K, Johnson C, Aguiar J, Sopoh G, Barogui Y, Meyers WM, Pedrosa J, Portaels F. 2009. Fine-needle aspiration, an efficient sampling technique for bacteriological diagnosis of nonulcerative Buruli ulcer. J. Clin. Microbiol. 47:1700–1704. 10.1128/JCM.00197-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kubica GP, Dye WE, Cohn ML, Middlebrook G. 1963. Sputum digestion and decontamination with N-acetyl-L-cysteine-sodium hydroxide for culture of mycobacteria. Am. Rev. Respir. Dis. 87:775–779. [DOI] [PubMed] [Google Scholar]
- 7.Rondini S, Mensah-Quainoo E, Troll H, Bodmer T, Pluschke G. 2003. Development and application of real-time PCR assay for quantification of Mycobacterium ulcerans DNA. J. Clin. Microbiol. 41:4231–4237. 10.1128/JCM.41.9.4231-4237.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eddyani M, Debacker M, Martin A, Aguiar J, Johnson CR, Uwizeye C, Fissette K, Portaels F. 2008. Primary culture of Mycobacterium ulcerans from human tissue specimens after storage in semisolid transport medium. J. Clin. Microbiol. 46:69–72. 10.1128/JCM.00301-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Phillips RO, Sarfo FS, Osei-Sarpong F, Boateng A, Tetteh I, Lartey A, Adentwe E, Opare W, Asiedu KB, Wansbrough-Jones M. 2009. Sensitivity of PCR targeting Mycobacterium ulcerans by use of fine-needle aspirates for diagnosis of Buruli ulcer. J. Clin. Microbiol. 47:924–926. 10.1128/JCM.01842-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reference deleted.
- 11.Yeboah-Manu D, Danso E, Ampah K, Asante-Poku A, Nakobu Z, Pluschke G. 2011. Isolation of Mycobacterium ulcerans from swab and fine-needle-aspiration specimens. J. Clin. Microbiol. 49:1997–1999. 10.1128/JCM.02279-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pommelet V, Vincent Q, Ardant MF, Adeye A, Tanase A, Tondeur L, Rega A, Landier J, Marion E, Alcais A, Marsollier L, Fontanet A, Chauty A. 21 July 2014, posting date Findings in patients from Benin with osteomyelitis and polymerase chain reaction-confirmed Mycobacterium ulcerans infection. Clin. Infect. Dis. 59:1256–1264. 10.1093/cid/ciu584. [DOI] [PubMed] [Google Scholar]
- 13.Kibadi K, Boelaert M, Fraga AG, Kayinua M, Longatto-Filho A, Minuku JB, Mputu-Yamba JB, Muyembe-Tamfum JJ, Pedrosa J, Roux JJ, Meyers WM, Portaels F. 2010. Response to treatment in a prospective cohort of patients with large ulcerated lesions suspected to be Buruli ulcer (Mycobacterium ulcerans disease). PLoS Negl. Trop. Dis. 4:e736. 10.1371/journal.pntd.0000736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nienhuis WA, Stienstra Y, Abass KM, Tuah W, Thompson WA, Awuah PC, Awuah-Boateng NY, Adjei O, Bretzel G, Schouten JP, van der Werf TS. 2012. Paradoxical responses after start of antimicrobial treatment in Mycobacterium ulcerans infection. Clin. Infect. Dis. 54:519–526. 10.1093/cid/cir856. [DOI] [PubMed] [Google Scholar]
- 15.O'Brien DP, Robson ME, Callan PP, McDonald AH. 2009. “Paradoxical” immune-mediated reactions to Mycobacterium ulcerans during antibiotic treatment: a result of treatment success, not failure. Med. J. Aust. 191:564–566. [DOI] [PubMed] [Google Scholar]
- 16.Ruf MT, Chauty A, Adeye A, Ardant MF, Koussemou H, Johnson RC, Pluschke G. 2011. Secondary Buruli ulcer skin lesions emerging several months after completion of chemotherapy: paradoxical reaction or evidence for immune protection? PLoS Negl. Trop. Dis. 5:e1252. 10.1371/journal.pntd.0001252. [DOI] [PMC free article] [PubMed] [Google Scholar]


