Abstract
We present a case of tenosynovitis caused by a novel, slowly growing, nonchromogenic, nontuberculous mycobacterium (NTM). Originally misidentified as Mycobacterium tuberculosis complex, the NTM cross-reacts with the M. tuberculosis complex nucleic acid hybridization probe, a M. tuberculosis gamma interferon release assay, and is closely related to M. tuberculosis by 16S rRNA gene sequencing.
CASE REPORT
A58-year-old previously healthy female presented to her primary care physician with right thumb pain, swelling, warmth and erythema, and a blister at the base of the thumb. The lesion did not improve despite 2 weeks of amoxicillin treatment followed by 2 weeks of cephalexin treatment. An X-ray suggested osteoarthritis. After 2 months without improvement, an orthopedist injected a corticosteroid, with temporary improvement of the swelling and erythema. However, the swelling subsequently recurred, and magnetic resonance imaging (MRI) revealed moderate tenosynovitis of the flexor tendon. She underwent operative irrigation and debridement, and marked edema without purulence was noted. An operative specimen was negative by direct examination and culture for bacteria, fungi, and mycobacteria. Histologic examination revealed chronic inflammation, nonnecrotizing granulomas, fibrosis, and focal mild acute inflammation of a soft-tissue mass, as well as inflammation of the flexor tendon sheath. No further antimicrobial therapy was prescribed, and the patient noted improvement of the thumb.
Two weeks after surgery, new swelling was noted that was localized to her right wrist with focal symptoms of carpal tunnel syndrome, including a positive Tinel's sign result. The patient was treated with two injections of triamcinolone and lidocaine (Xylocaine) for presumed carpal tunnel syndrome without improvement. She underwent a second surgery for a carpal tunnel release and tenosynovectomy of the sublimis tendons. The operative report noted that the tendons were adherent and covered with a “thick, granulated-type tissue.” Direct smears and cultures were negative for bacteria, fungi, and mycobacteria. Evidence of granulomatous synovitis was seen by histologic examination of the tissue. The patient was referred to a rheumatologist, who initiated oral therapy with nonsteroidal anti-inflammatory agents and performed a corticosteroid injection into the interphalangeal (IP) joint. Extensive swelling and pain developed after the steroid injection with rapidly worsening erythema and painful swelling that progressed despite initiation of amoxicillin-clavulanate. The patient returned to the operating room for repeated debridement. The operative report noted “extensive chronic granulomatous-type tissue throughout the joint into the IP joint, under the extensor tendons and extending into the soft tissues.” Operative samples were obtained for culture. Throughout these episodes of waxing and waning swelling and erythema, the patient remained afebrile and felt systemically well; her only symptoms were the swelling and tenderness of her thumb and, subsequently, her wrist. She denied rash, involvement of other joints, cough, fatigue, weight loss, night sweats, or fevers.
On postoperative day 13, a mycobacterial broth culture from the third debridement was positive and contained a slowly growing, nonchromogenic mycobacteria. A DNA nucleic acid hybridization probe (Hologic GenProbe AccuProbe, San Diego, CA) identified the organism as Mycobacterium tuberculosis complex. A chest X-ray showed no evidence of active disease and no evidence of prior granulomatous disease. The result of a gamma interferon release assay (T-Spot.TB; Oxford Diagnostic Laboratories, Marlborough, MA) was also positive. She was started on M. tuberculosis antimicrobial therapy, which included rifampin (RIF), isoniazid (INH), pyrazinamide (PZA), and ethambutol (EMB) (RIPE therapy), with vitamin B6 supplementation.
Two days after initiating RIPE therapy, she developed worsening swelling and pain of the right thumb, as well as IP joint instability. Out of concern for possible bacterial superinfection, she was subjected to repeat bedside debridement with initiation of vancomycin and cefepime, in addition to ongoing RIPE therapy. All cultures showed no growth; parenteral antibiotics were discontinued, and she completed a 14-day course of amoxicillin-clavulanate treatment. Nineteen days after initiating RIPE therapy, she underwent repeat debridement, realignment, and arthrodesis with external fixation; cultures from this operative debridement grew the same Mycobacterium species. Fourteen days later, she had repeat operative debridement with bone graft, and cultures again showed no growth. From this time onward and while still on RIPE therapy, she showed sustained clinical improvement with ongoing reduction in swelling, erythema, and pain.
The patient had lived her entire life in the New England area, most recently residing in the Boston area. She had no known tuberculosis exposure history and did not recall prior tuberculosis testing. She had been vacationing in the U.S. Virgin Islands, where she had been gardening and swimming in tidal pools, 6 weeks prior to the appearance of the blister. Two weeks prior to the appearance of the blister, she also recalled sustaining a small cut on her hand in the same location.
Since the patient did not have risk factors for or clinical signs consistent with M. tuberculosis, the mycobacterial isolate was referred to a reference laboratory to confirm its identification. As discussed below, after extensive laboratory investigations, it was reported that the isolate was a novel nontuberculous species that closely resembled M. tuberculosis. Given the development of a pruritic, macular rash on her abdomen, PZA treatment was discontinued at month 4, and azithromycin was added to the remaining antimycobacterials (INH, RIF, and EMB). At month 6, drug susceptibility testing was available (Table 1), and her antimicrobial regimen was changed to INH, moxifloxacin, and azithromycin. At month 13, azithromycin was discontinued because the patient complained of tinnitus; she continued on moxifloxacin and INH to complete a total antimicrobial course of 18 months. Following discontinuation of antibiotics, she reported excellent thumb function, with no erythema, warmth, or swelling.
TABLE 1.
MICs for the novel NTM
| Agent | MIC(s) (μg/ml) |
|---|---|
| Amikacin | ≤1 |
| Ciprofloxacin | 4 |
| Clarithromycin | 4 |
| Doxycycline | 0.5 |
| Ethambutol | 4 |
| Linezolid | ≤1 |
| Moxifloxacin | 0.5 |
| Rifabutin | ≤0.25 |
| Rifampin | 2 |
| Trimethoprim-sulfamethoxazole | 0.25/4.75 |
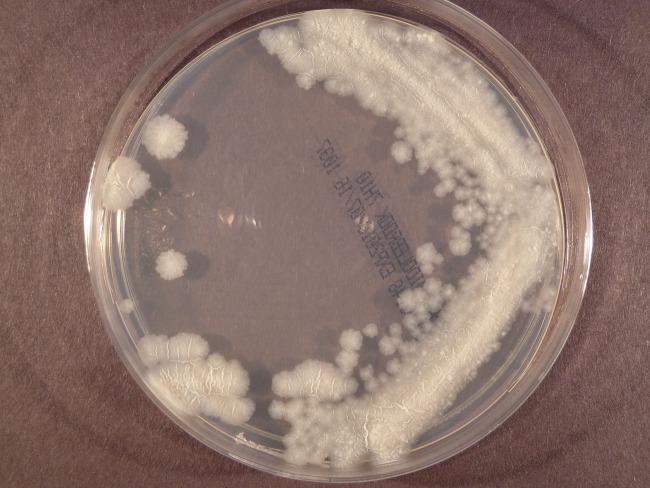
This novel slowly growing, nonchromogenic, nontuberculous mycobacterium species isolated from surgical specimen cultures was originally misidentified as M. tuberculosis complex both at the primary laboratory and again at a reference laboratory. The organism stained acid fast with the Kinyoun stain. The M. tuberculosis complex DNA AccuProbe (Hologic Gen-Probe) test was repeated at the reference laboratory and was positive at 50,094 relative light units (RLU; positivity cutoff, ≥30,000 RLU) when tested directly from mycobacterial growth indicator tube (MGIT; Becton Dickinson, Franklin Lakes, NJ) broth. A Middlebrook 7H11 subculture plate of the organism from broth displayed a colony morphology consistent with M. tuberculosis that was described as a dry, wrinkled, flat, and crystalline nonchromogen (Fig. 1). Because of the unusual clinical presentation, sequencing was performed to confirm the M. tuberculosis complex probe result. The first 500-bp sequence of the 16S rRNA gene resulted in a 99.09% match to an M. tuberculosis type strain in the reference laboratory's 16S rRNA gene library (1). According to Clinical and Laboratory Standards Institute (CLSI) guidelines, a sequencing score for of 99.00% to 99.99% for mycobacteria is not sufficient to identify to the species level and organisms with such a sequencing score should be reported as “Mycobacterium, most closely related to M. tuberculosis” (2). Antimicrobial susceptibility testing using INH, RIF, EMB, and PZA was performed using an FDA-approved VersaTREK platform (Trek/Thermo Scientific, Oakwood Village, OH) and a broth macrodilution method according to the Clinical and Laboratory Standards Institute methodology (3). Using interpretive criteria for M. tuberculosis, the results indicated the isolate was susceptible to RIF (1 μg/ml), high-level INH (0.4 μg/ml), and EMB (5 μg/ml) but resistant to low-level INH (0.1 μg/ml) and PZA (300 μg/ml). A laboratory-developed M. tuberculosis-specific PCR assay which targets the katG gene was performed and gave a negative result (4). In order to confirm the PZA resistance reported by the phenotypic broth method, the reference laboratory attempted to sequence the pyrazinamidase gene (pncA) (5–7). However, despite repeated attempts, pncA sequencing of the patient's isolate failed. Failure of an M. tuberculosis isolate to produce a pncA sequence, a rare event in the experience of the laboratory, in addition to the inability to detect the katG gene, the lack of 100% identity with M. tuberculosis by 16S rRNA gene sequencing, and the unusual clinical picture, raised suspicion that the isolate was not M. tuberculosis.
FIG 1.
Image of a Middlebrook 7H10 plate demonstrating the colony morphology of the novel NTM that was consistent with M. tuberculosis morphology and that was described as a dry, wrinkled, flat, and crystalline nonchromogen.
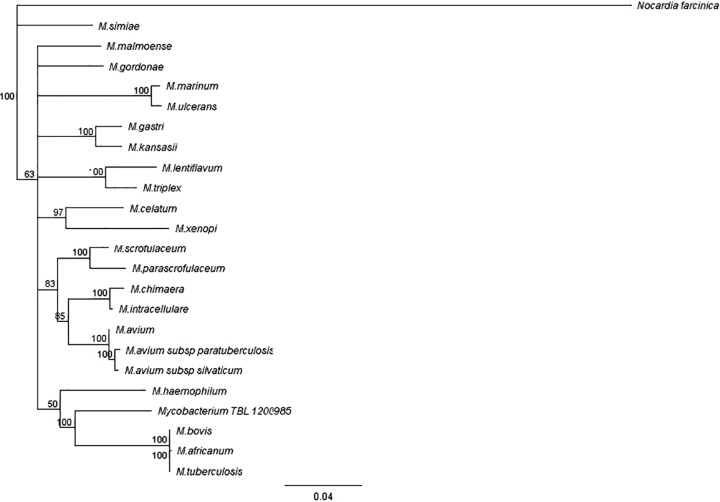
Given these inconsistencies, the isolate was then forwarded to the Mycobacteria/Nocardia Laboratory at the University of Texas Health Science Center at Tyler and the Division of Tuberculosis Elimination at the Centers for Disease Control (CDC) for further phylogenic analysis and broth microdilution MIC testing. Although sequencing of the full-length 16S rRNA gene indicated that the isolate was most closely related to M. tuberculosis complex, spoligotyping confirmed that the isolate did not belong to the M. tuberculosis complex and suggested the presence of a novel NTM (8, 9). The first 500-bp sequence of the 16S rRNA gene differed by 4 bp from that of M. tuberculosis but belonged to the same clade as M. tuberculosis, which includes M. marinum, M. ulcerans, and M. shottsii (all of which typically cause cutaneous disease) (Fig. 2) (10). Interestingly, 2 of the 4 nucleotide differences within the 500-bp sequence occurred within the M. tuberculosis complex nucleic acid hybridization probe binding area of the 16S rRNA between nucleotides 188 and 207 (equivalent to nucleotide positions 161 to 180; GenBank accession no. KF683289) but still allowed annealing of the probe (Fig. 3) (11, 12). M. celatum type 1 (one nucleotide substitution) (11, 12), M. holsaticum (two nucleotide substitutions) (13), and M. terrae (one nucleotide substitution) (11) have all been reported to cross-react with the M. tuberculosis complex probe. M. marinum has two nucleotide substitutions in the probe binding region similar to that of the novel NTM but at different locations and does not cross-react with the M. tuberculosis complex probe. Therefore, the number and location of nucleotide substitutions play a role in the ability of the M. tuberculosis complex probe to cross-react with other Mycobacterium species. By full 16S rRNA gene sequencing, the patient isolate was found to be more closely related to M. tuberculosis than to any other species. The isolate has a 7-bp mismatch and 2 gap differences with M. tuberculosis. M. marinum and other species have at least 14 differences, including gaps and base pair mismatches, with the patient strain. According to the results of sequencing of region V of the rpoB gene and of secA1, the patient's isolate was related only remotely (90.2% and 93.3%, respectively) to any validated species, with the most closely related being M. riyadhense and M. kansasii, respectively (14, 15). It had only 91% identity to M. tuberculosis by secA1 gene analysis. According to the results of hsp65 partial gene sequencing, it was related only remotely (95.5%) to any validated species, with the closest being M. intracellulare and other members of the M. avium complex (19-bp difference in 426 bp) (16). M. tuberculosis had only 94.6% identity (23-bp difference in 426 bp) to the patient's isolate by hsp65 sequencing. Molecular analysis suggests that this isolate represents a new NTM species (Fig. 4).
FIG 2.

Phylogenetic relationship of the novel NTM isolate (TBL 1200985) and related species of Mycobacterium based on 16S rRNA gene sequences. All sequences used, except for the novel NTM, are type strains. Sequences were aligned with MUSCLE (17) and the tree constructed by the neighbor-joining method using the Tamura-Nei distance model. All steps were performed using Geneious software. Mycobacterium abscessus CIP 104536 was used as an outgroup (GenBank accession no. AY457071). Branch support is recorded at the nodes as a percentage of 100 bootstrap iterations.
FIG 3.

Alignment of partial 16S rRNA gene sequence (positions 180 to 227) for M. tuberculosis (1), the patient's novel NTM isolate (2), M. celatum type 1 (3), M. terrae strain (4), and M. marinum (5). The grayed sequence is the target for the commercial M. tuberculosis complex DNA probe (Gen-Probe) between positions 188 and 207 of the 16S rRNA sequence. Nucleotides highlighted in yellow are differences in the 16S rRNA sequence of the NTM species (2 to 5) from that of M. tuberculosis (1). Dashes indicate deletions.
FIG 4.
Phylogenetic relationship of the novel NTM isolate (TBL 1200985) and related species of Mycobacterium, based on the concatenation of hsp65, rpoB, and secA1 sequences. All sequences used, except for that of the novel NTM, are type strains. Sequences were aligned with MUSCLE (17) and the tree constructed by the neighbor-joining method using the Tamura-Nei distance model. All steps were performed using Geneious software. Nocardia farcinica ATCC 3318T was used as an outgroup (GenBank accession no. AY756523, AB243742, and AY781800). Branch support is recorded at the nodes as a percentage of 100 bootstrap iterations.
In contrast to M. tuberculosis, the novel NTM was unable to accumulate niacin and could not reduce nitrate to nitrite. Levels of growth at 30°C and 37°C were equivalent. Addition of X-factor to the medium did not enhance growth of the isolate. After identification of the isolate as a slowly growing, nonchromogenic NTM, susceptibility testing was performed using the CLSI-recommended broth microdilution method for slowly growing mycobacteria using a Slomyco panel (TREK/Thermo Scientific), and, using interpretive criteria for rifampin-resistant M. kansasii and other slowly growing NTM, the isolate was found to be susceptible to clarithromycin, rifabutin, moxifloxacin, trimethoprim-sulfamethoxazole (TMP-SMX), amikacin, linezolid, and doxycycline (Table 1) (3). The isolate demonstrated intermediate susceptibility to EMB and was resistant to RIF and ciprofloxacin.
This case of granulomatous tenosynovitis was caused by a novel, slowly growing, nonchromogenic NTM which most closely resembled M. tuberculosis by 16S rRNA gene sequencing and which produced a false-positive cross-reaction with the M. tuberculosis complex nucleic acid hybridization probe and a gamma interferon release assay for M. tuberculosis. Similarly to the way in which M. marinum is commonly acquired, the most likely source of the mycobacteria was water or soil exposure while the patient was either in the U.S. Virgin Islands or at home in New England. Laboratories and clinicians should consider this novel NTM when the M. tuberculosis complex AccuProbe is positive but the clinical history makes tuberculosis unlikely. 16S rRNA gene sequencing can help to distinguish this novel species, since the first 500-bp sequence differs from M. tuberculosis by 4 nucleotides and the full 16S rRNA sequence differs by 7-bp with 2 gaps.
Nucleotide sequence accession numbers.
The gene sequences for the novel NTM isolate have been deposited in the NCBI GenBank database (for the 16S rRNA gene, accession no. KF683289; for the rpoB gene, accession no. KJ371034; for the hsp65 partial gene sequence, accession no. KJ371035; and for the secA1 gene, accession no. KJ371036).
Supplementary Material
ACKNOWLEDGMENTS
This study was supported in part by a grant from the Carter Foundation to the Mycobacteria/Nocardia Laboratory at the University of Texas Health Science Center.
We declare no conflicts of interest. The findings and conclusions in this report are ours and do not necessarily represent the official position of the Centers for Disease Control and Prevention. Use of trade names and commercial sources is for identification purposes and does not constitute endorsement by the Public Health Service or the U.S. Department of Health and Human Services.
Footnotes
Published ahead of print 24 September 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.00967-14.
REFERENCES
- 1.Hall L, Doerr KA, Wohlfiel SL, Roberts GD. 2003. Evaluation of the MicroSeq system for identification of mycobacteria by 16S ribosomal DNA sequencing and its integration into a routine clinical mycobacteriology laboratory. J. Clin. Microbiol. 41:1447–1453. 10.1128/JCM.41.4.1447-1453.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clinical and Laboratory Standards Institute (CLSI). 2008. Interpretive criteria for identification of bacteria and fungi by DNA target sequencing; approved guideline. CLSI document MM18-A (ISBN 1-56238-664-6). CLSI, Wayne, PA. [Google Scholar]
- 3.Clinical and Laboratory Standards Institute (CLSI). 2008. Susceptibility testing of mycobacteria, nocardiae, and other aerobic actinomycetes; approved standard—2nd ed. CLSI document MM18-A (ISBN 1-56238-664-6). CLSI, Wayne, PA. [PubMed] [Google Scholar]
- 4.Buckwalter SP, Buschur L, Clark S, Ruth C, Short G, Vandorp J, Wohlfiel SL, Wengenack NL. 2008. Comparison of culture with auramine rhodamine stain, the amplified direct test and an M. tuberculosis real-time PCR assay for respiratory specimens, abstr C-204. Abstr. 108th Gen. Meet. Am. Soc. Microbiol., Boston, MA, June 1 to 5 American Society for Microbiology, Washington, DC. [Google Scholar]
- 5.Shenai S, Rodrigues C, Sadani M, Sukhadia N, Mehta A. 2009. Comparison of phenotypic and genotypic methods for pyrazinamide susceptibility testing. Indian J. Tuberc. 56:82–90. [PubMed] [Google Scholar]
- 6.Simons SO, van Ingen J, van der Laan T, Mulder A, Dekhuijzen PN, Boeree MJ, van Soolingen D. 2012. Validation of pncA gene sequencing in combination with the mycobacterial growth indicator tube method to test susceptibility of Mycobacterium tuberculosis to pyrazinamide. J. Clin. Microbiol. 50:428–434. 10.1128/JCM.05435-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Somoskovi A, Dormandy J, Parsons LM, Kaswa M, Goh KS, Rastogi N, Salfinger M. 2007. Sequencing of the pncA gene in members of the Mycobacterium tuberculosis complex has important diagnostic applications: identification of a species-specific pncA mutation in “Mycobacterium canettii” and the reliable and rapid predictor of pyrazinamide resistance. J. Clin. Microbiol. 45:595–599. 10.1128/JCM.01454-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cowan LS, Diem L, Brake MC, Crawford JT. 2004. Transfer of a Mycobacterium tuberculosis genotyping method, spoligotyping, from a reverse line-blot hybridization, membrane-based assay to the Luminex multianalyte profiling system. J. Clin. Microbiol. 42:474–477. 10.1128/JCM.42.1.474-477.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, Bunschoten A, Molhuizen H, Shaw R, Goyal M, van Embden J. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Devulder G, Perouse de Montclos M, Flandrois JP. 2005. A multigene approach to phylogenetic analysis using the genus Mycobacterium as a model. Int. J. Syst. Evol. Microbiol. 55:293–302. 10.1099/ijs.0.63222-0. [DOI] [PubMed] [Google Scholar]
- 11.Emler S, Ninet B, Rohner P, Auckenthaler R, Jager D, Hirschel B. 1995. Molecular basis for cross-reactivity between a strain of Mycobacterium terrae and DNA probes for Mycobacterium tuberculosis complex. Eur. J. Clin. Microbiol. Infect. Dis. 14:627–629. 10.1007/BF01690741. [DOI] [PubMed] [Google Scholar]
- 12.Somoskövi A, Hotaling JE, Fitzgerald M, Jonas V, Stasik D, Parsons LM, Salfinger M. 2000. False-positive results for Mycobacterium celatum with the AccuProbe Mycobacterium tuberculosis complex assay. J. Clin. Microbiol. 38:2743–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tortoli E, Pecorari M, Fabio G, Messino M, Fabio A. 2010. Commercial DNA probes for mycobacteria incorrectly identify a number of less frequently encountered species. J. Clin. Microbiol. 48:307–310. 10.1128/JCM.01536-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adékambi T, Colson P, Drancourt M. 2003. rpoB-based identification of nonpigmented and late-pigmenting rapidly growing mycobacteria. J. Clin. Microbiol. 41:5699–5708. 10.1128/JCM.41.12.5699-5708.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zelazny AM, Calhoun LB, Li L, Shea YR, Fischer SH. 2005. Identification of Mycobacterium species by secA1 sequences. J. Clin. Microbiol. 43:1051–1058. 10.1128/JCM.43.3.1051-1058.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Telenti A, Marchesi F, Balz M, Bally F, Bottger EC, Bodmer T. 1993. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J. Clin. Microbiol. 31:175–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.