Abstract
Acidovorax oryzae is a bacterium that has never before been reported as pathogenic in human subjects. Here we describe the first case of a successfully treated A. oryzae catheter-associated bloodstream infection in an immunocompetent patient prior to heart transplantation.
CASE REPORT
A 48-year-old man with severe heart failure was admitted to the coronary care unit. His symptoms and recent investigations indicated a significant decline in cardiac function, so admission to hospital for ionotropic support and further investigation was arranged.
His past medical history included nonischemic dilated cardiomyopathy, severe left ventricular systolic impairment, a biventricular pacemaker, hernia repair, gout, and anxiety with depression. The patient gave no history of foreign travel, gardening, farming, or contact with animals and enjoyed an unremarkable diet.
On admission, he was stabilized with ionotropes via a right internal jugular (IJ) catheter that was inserted without complication. He subsequently made a clinical improvement, and intracardiac pressure measurements indicated that he was suitable for transplantation. He was therefore listed for urgent transplant as an inpatient. There was no history of renal impairment, and his renal function remained within normal limits during his admission.
Two weeks into admission, it was noted that the levels of his inflammatory markers had begun to increase. The C-reactive protein (CRP) level peaked over 5 days to 25 mg/liter, although the levels of white cells and neutrophils remained within normal limits. He was asymptomatic, and a full examination did not reveal a source of infection. He was treated empirically with oral flucloxacillin at 500 mg four times a day for 2 days and then with intravenously (i.v.) administered piperacillin-tazobactam at 4.5 g three times a day for 6 days for a presumed catheter-related bloodstream infection. The CRP level subsequently decreased to within normal limits, and the patient's treatment regimen was deescalated to oral flucloxacillin (Fig. 1).
FIG 1.
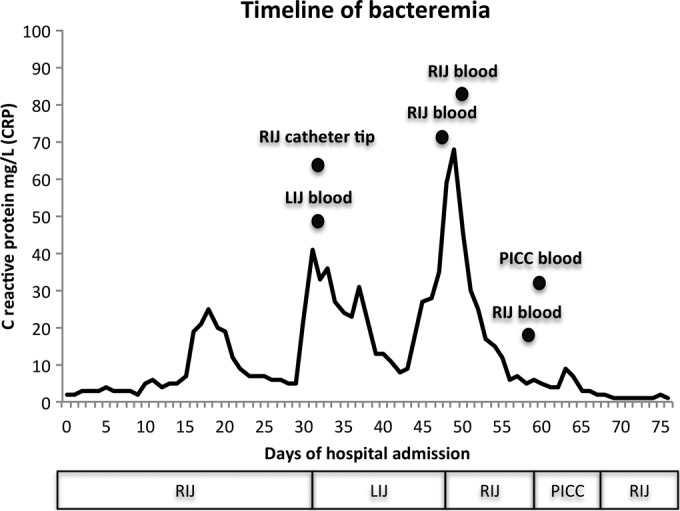
A timeline of the Acidovorax oryzae bacteremia. The CRP levels are displayed graphically as a line chart. The black circles indicate the times that the positive cultures were acquired and are labeled with the site of culture. Below the chart is a graphical representation of when the vascular catheters were implanted and explanted. The patient received the transplant on day 77. For clarity, only positive cultures are displayed; see Table 1 for a full list of positive and negative culture results. RIJ, right internal jugular catheter; LIJ, left internal jugular catheter; PICC, peripherally inserted central catheter.
A week later, the CRP level rose for a second time, reaching 42 mg/liter, without a detectable source of infection. Piperacillin-tazobactam treatment was recommenced, and the right IJ catheter was replaced with a left IJ catheter. Line replacement was technically difficult and was eventually achieved after four failed attempts. Central blood cultures were taken from the new left catheter at the time of insertion, and the explanted right catheter tip was sent for culture.
All central blood cultures were taken using an aseptic technique, placed into BacT/Alert culture bottles (bioMérieux Inc., Durham, NC), and incubated at 37°C routinely for 5 days. Positive cultures were then plated on Columbia blood agar (Oxoid/Thermo Scientific, Waltham, MA) and incubated aerobically at 37°C for 24 h and anaerobically on fastidious anaerobic agar (Oxoid/Thermo Scientific, Waltham, MA) at 37°C for 48 h. The catheter tip was cultured on Columbia blood agar at 37°C for 24 to 48 h, per Public Health England United Kingdom Standards for Microbiological Investigations (1).
After 48 h, the aerobic blood culture from the newly placed left IJ catheter had grown a Gram-negative organism. A single colony of the same organism had also grown from the explanted right IJ catheter tip. The results determined for all cultured blood and catheter tips are displayed in Table 1.
TABLE 1.
Tabulation of all blood cultures and catheter tip cultures taken during hospital admissiona
| Day of admission | Site of culture | Result | Time to positivity (h) |
|---|---|---|---|
| 33 | Right IJ catheter tip | A. orzyae | 48 |
| 33 | Left IJ blood | A. orzyae | 48 |
| 48 | Left IJ catheter tip | No growth | 48 |
| 48 | Right IJ blood | A. orzyae | 12 |
| 51 | Left IJ blood | A. orzyae | 12 |
| 59 | Left IJ blood | A. orzyae | 12 |
| 61 | PICC | A. orzyae | 48 |
| 62 | PICC | No growth | 48 |
| 63 | PICC | No growth | 48 |
| 64 | PICC | No growth | 48 |
| 66 | PICC | No growth | 48 |
| 67 | PICC | No growth | 48 |
| 70 | Right IJ blood | No growth | 48 |
The day of admission refers to the day that the cultures were acquired. IJ, internal jugular; PICC, peripherally inserted central catheter. All cultures are routinely held for 5 days.
The Gram-negative organism was evaluated using a Vitek2XL Gram-negative identification card (bioMérieux Inc., Durham, NC). This identified the organism as Acinetobacter ursingii with a 95% probability. The biocode was 4000000300500002, indicating that only 6 of 44 tests were positive. The positive test results were as follows: PyrA (l-pyrrolydonyl-arylamidase), 0.018 mg in well; TyrA (tyrose arylamidase), 0.0276 mg; URE (urease), 0.15 mg; ILATk (l-lactate alkalinization), 0.15 mg; SUCT (succinate alkalinization), 0.15 mg; and ELLM (Ellman), 0.03 mg. Per the local protocol, laboratory staff are trained to be suspicious of identifications with high probability with few positive tests. The sample was therefore sent to a reference laboratory for 16S RNA partial sequencing. Of note, only one representative sample was sent for identification at a reference laboratory for practical reasons. The rest of the positive cultures were matched to the representative sample via an identical Vitek biocode(s) and by matching colonial morphology.
To determine which antimicrobials would be effective against this unidentified organism, a Vitek 210 antibiotic susceptibility testing (AST) card (bioMérieux Inc., Durham, NC) was utilized but was not able to provide any susceptibility data for Acinetobacter ursingii because the organism was not listed in the Vitek database. While awaiting the results from the reference laboratory, our laboratory scientists needed to provide the treating clinicians with advice on which antimicrobials would be best to treat the organism. Therefore, noting that the organism was oxidase positive, the laboratory scientists prepared a manual Pseudomonas sensitivity preparation. This was achieved by inoculating a suspension of the test organism with a density equivalent to a McFarland 0.5 standard onto Mueller-Hinton agar (Oxoid) and incubating at 35°C ±1°C for 16 to 20 h. Sizes of zones of inhibition were calculated using a EUCAST (European Committee on Microbial Sensitivity Testing) disc diffusion method with a standard set of pseudomonal antimicrobials (Table 2). Zone sizes were compared to EUCAST standard zone sizes for Pseudomonas aeruginosa to define whether the organism was susceptible to each antimicrobial. Of note, the organism did grow on cysteine lactose electrolyte-deficient medium (CLED) agar, producing small non-lactose-fermenting colonies of approximately 1-mm diameter after 18 h of incubation.
TABLE 2.
Zones of inhibition of each antimicrobial tested against the cultured organisma
| Antimicrobial agent | Zone diam (mm) | S/I/R test result |
|---|---|---|
| Gentamicin | >15 | S |
| Imipenem | >20 | S |
| Ceftazidime | >16 | S |
| Ciprofloxacin | >25 | S |
| Meropenem | >24 | S |
| Piperacillin-tazobactam | >18 | S |
| Tobramycin | >16 | S |
Zones were calculated using a EUCAST (European Committee on Microbial Sensitivity Testing) disc diffusion method with a standard set of pseudomonal antimicrobials. Zone sizes were compared to EUCAST standard zone sizes for Pseudomonas aeruginosa to define whether the organism was susceptible to each antimicrobial. S, sensitive; I, intermediate; R, resistant.
Based on the zones of inhibition, the piperacillin-tazobactam treatment was continued for a further 10 days, after which the CRP level returned to 7 mg/liter and the treatment regimen was deescalated to oral flucloxacillin administered at 500 mg four times a day.
One week later, the condition of the patient deteriorated and the patient developed symptoms of general malaise and chills. He remained apyrexial, but the CRP level again began to rise, eventually reaching 68 mg/liter with a white cell count of 13.5 × 109/liter and neutrophils at 11.43 ×109/liter. At this time, his condition was reviewed by the infectious-diseases consultant. Further assessment and examination did not reveal a source of infection. A computed tomography scan of the chest, abdomen, and pelvis was unremarkable, an ultrasound scan of his pacemaker pocket revealed no collection, and a transesophageal ultrasound revealed no valvular vegetations. Oral flucloxacillin was discontinued, and i.v. administration of ciprofloxacin at 500 mg twice daily was commenced. Intravenous therapy was preferred as there was concern that significant cardiac failure might have prevented adequate absorption of the oral preparations.
With no other source of infection evident, the left IJ catheter was the presumed source of infection, so that catheter was removed and a replacement was inserted into the right IJ vein. Blood cultures were taken from the newly implanted catheter, and the explanted catheter tip was again sent for culture.
After 48 h of incubation, the explanted left catheter tip yielded no growth but blood cultures from the newly implanted (right) IJ catheter had grown the same unidentified organism. The levels of the inflammatory markers gradually declined, and the patient's symptoms resolved on i.v. ciprofloxacin treatment. However, two further blood cultures from the right IJ catheter were again positive 2 and 11 days after implantation despite continued i.v. ciprofloxacin treatment. Therefore, the remaining right IJ catheter was removed and a peripherally inserted central venous catheter (PICC) was simultaneously inserted into the left antecubital fossa in an attempt to clear the bacteremia. A blood culture taken at the time of implantation of the peripheral cannula proved positive for the same organism after 5 days of incubation.
Following the removal of all IJ catheters, five subsequent blood cultures taken peripherally were negative over the next 7 days. The dual lumen line was not ideal for the delivery of antibiotics and inotropes, so on day 8, the peripheral venous catheter was explanted and a right IJ catheter reinserted. Right IJ catheter cultures at the time of implantation were negative after 48 h of incubation. It was only following removal of all IJ catheters that the bacteremia resolved.
The patient remained on i.v. ciprofloxacin without symptoms and with normal inflammatory markers until his successful heart transplantation 77 days after admission. Posttransplantation, the patient remained on ciprofloxacin until all his lines were removed and he was discharged home without antibiotics 21 days posttransplantation. On the day of discharge, he was free of infective symptoms and the levels of his inflammatory markers were within normal limits. He remains well and has had no sequelae of his bacteremia. Histology of the explanted heart did not reveal any vegetation or infective source. The valves and coronary arteries were entirely unremarkable, and histological appearances supported a diagnosis of dilated cardiomyopathy.
The organism was eventually identified as Acidovorax oryzae by partial sequencing of the 16S rRNA gene. The sample was submitted to the national reference laboratory at the associated infection reference unit at Collingdale, London, United Kingdom. The sequence generated was a 100% match to A. oryzae with a 1,217/1,217-bp ratio, with the next best match A. avenae at 99.7% and 1,210/1,217 bp. The identification method used was first published by Edwards et al. in 2012 and consisted of block-based PCR and comparison with sequences in GenBank using the NCBI BLAST algorithm (2). The database used for comparison was BIBI, a bioinformatics bacterial identification database tool (3). The reference laboratory was also able to supply a full list of MICs for a variety of antimicrobials (listed in Table 3). MICS were calculated using the standardized British Society of Antimicrobial Chemotherapy (BSAC) disc susceptibility testing method (4).
TABLE 3.
MICs for antimicrobials tested against A. orzyaea
| Antimicrobial agent | MIC (mg/liter) |
|---|---|
| Amikacin | 2 |
| Gentamicin | 1 |
| Tobramycin | 1 |
| Aztreonam | 1 |
| Ceftazidime | 0.25 |
| Imipenem | 0.125 |
| Meropenem | 0.25 |
| Piperacillin | ≤1 |
| Piperacillin-tazobactam | ≤1 |
| Cotrimoxazole | 0.032 |
| Colistin | 32 |
| Ciprofloxacin | 0.25 |
| Minocycline | ≤0.125 |
MICS were calculated using the standardized British Society of Antimicrobial Chemotherapy (BSAC) disc susceptibility testing method.
A. oryzae is a member of the Comamonadaceae that is thought to be pathogenic only in plants. It is a Gram-negative, motile rod that infects a variety of crops, including corn, melons, oats, millet, orchids, sugar cane, and rice. A recent study using DNA sequencing and phenotypic examination demonstrated A. oryzae to be a newly defined organism distinct from A. avenae, which solely infects rice crops. It was therefore classified as a new taxon in 2009 (5).
There are very few reports of Comamonadaceae infection in humans. The predominant cases reported are ocular infections of patients with or without contact lenses who are topically or systemically immunosuppressed. Comamonas acidovorans, Comamonas testosteroni, and Delftia acidovorans have all been isolated in the setting of keratitis and corneal ulcers (6–8). However, most systemic infections have been associated with indwelling medical devices and immunosuppression (9).
A. avenae bacteremia has been reported on three prior occasions. Xu et al. processed 98 blood cultures in a study examining PCR and rRNA sequencing in blood from patients with hematological malignancies; one case was isolated. Shetty et al. have also isolated A. avenae in a patient with sarcoidosis treated with steroids for which ciprofloxacin was an effective treatment. Most recently, Malkan et al. cultured the same organism in addition to methicillin-sensitive Staphylococcus aureus from a patient with a longstanding central venous catheter. This organism was also susceptible to ciprofloxacin and a number of other antimicrobials (9–11).
To the best of our knowledge, this is the first reported case of A. oryzae bacteremia in humans. We have described a case of a bacteremia occurring in a patient pretransplantation who was successfully treated with ciprofloxacin and who subsequently underwent successful orthotopic heart transplantation. Infection with unusual organisms after transplantation in the setting of immunosuppression is well documented; in this case, however, the patient status was pretransplantation and, unlike the prior reports of A. avenae bacteremia, there was no history of immunosuppressive medication. There was also no coinfective organism isolated and no obvious source of infection identified other than the IJ catheter.
Currently, there is no information or guidance on the pathogenesis, infectivity, or management of Comamonadaceae infection in humans. Reviewing isolated case reports, it would appear that bacteremia is often associated with the presence of indwelling vascular devices. In cases where a catheter was implicated, most infections resolved following the removal of the implicated device and the application of appropriate antibiotic treatment.
The advent of modern laboratory techniques such as broad-range 16S rRNA PCR means that it might be anticipated that additional novel bacterial species will be identified in routine practice. In the past, it was likely that bacteremia such as that caused by A. oryzae would have been labeled as caused by “unidentified Gram negatives.” However, the addition of molecular diagnostic techniques such as 16S rRNA PCR enables the identification of bacteria that were previously difficult to characterize (12).
Ultimately, this case emphasizes the importance of the use of strict aseptic technique in handling medical devices, especially indwelling vascular catheters. It also highlights the importance of liaison with infectious-disease specialists and the utility of modern microbiology techniques that are available to identify an unusual bacterium.
Footnotes
Published ahead of print 1 October 2014
REFERENCES
- 1.Public Health England. 2014. Investigation of intravascular cannulae and associated specimens. Public Health England, London, United Kingdom: https://www.gov.uk/government/uploads/system/uploads/attachment_data/file/343933/B_20i5.2.pdf. [Google Scholar]
- 2.Edwards KJ, Logan JM, Langham S, Swift C, Gharbia SE. 2012. Utility of real-time amplification of selected 16S rRNA gene sequences as a tool for detection and identification of microbial signatures directly from clinical samples. J. Med. Microbiol. 61(Pt 5):645–652. 10.1099/jmm.0.041764-0. [DOI] [PubMed] [Google Scholar]
- 3.Devulder G, Perriere G, Baty F, Flandrois JP. 2003. BIBI, a bioinformatics bacterial identification tool. J. Clin. Microbiol. 41:1785–1787. 10.1128/JCM.41.4.1785-1787.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrews JM, Howe RA. 2011. BSAC standardized disc susceptibility testing method (version 10). J. Antimicrob. Chemother. 66:2726–2757. 10.1093/jac/dkr359. [DOI] [PubMed] [Google Scholar]
- 5.Schaad NW, Postnikova E, Sechler A, Claflin LE, Vidaver AK, Jones JB, Agarkova I, Ignatov A, Dickstein E, Ramundo BA. 2008. Reclassification of subspecies of Acidovorax avenae as A. Avenae (Manns 1905) emend., A. cattleyae (Pavarino, 1911) comb. nov., A. citrulli Schaad et al., 1978) comb. nov., and proposal of A. oryzae sp. nov. Syst. Appl. Microbiol. 31:434–446. 10.1016/j.syapm.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 6.Stonecipher KG, Jensen HG, Kastl PR, Faulkner A, Rowsey JJ. 1991. Ocular infections associated with Comamonas acidovorans. Am. J. Ophthalmol. 112:46–49. 10.1016/S0002-9394(14)76211-7. [DOI] [PubMed] [Google Scholar]
- 7.Reddy AK, Murthy SI, Jalali S, Gopinathan U. 2009. Post-operative endophthalmitis due to an unusual pathogen, Comamonas testosteroni. J. Med. Microbiol. 58(Pt 3):374–375. 10.1099/jmm.0.006072-0. [DOI] [PubMed] [Google Scholar]
- 8.Ray M, Lim DK. 2013. A rare polymicrobial keratitis involving Chryseobacterium meningosepticum and Delftia acidovorans in a cosmetic contact lens wearer. Eye Contact Lens 39:192–193. 10.1097/ICL.0b013e3182448881. [DOI] [PubMed] [Google Scholar]
- 9.Malkan AD, Strollo W, Scholand SJ, Dudrick SJ. 2009. Implanted-port-catheter-related sepsis caused by Acidovorax avenae and methicillin-sensitive Staphylococcus aureus. J. Clin. Microbiol. 47:3358–3361. 10.1128/JCM.01093-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu J, Moore JE, Millar BC, Alexander HD, McClurg R, Morris TC, Rooney PJ. 2004. Improved laboratory diagnosis of bacterial and fungal infections in patients with hematological malignancies using PCR and ribosomal RNA sequence analysis. Leuk. Lymphoma 45:1637–1641. 10.1080/10428190410001667695. [DOI] [PubMed] [Google Scholar]
- 11.Shetty A, Barnes RA, Healy B, Groves P. 2005. A case of sepsis caused by Acidovorax. J. Infect. 51:171–172. 10.1016/j.jinf.2004.12.014. [DOI] [PubMed] [Google Scholar]
- 12.Harris KA, Hartley JC. 2003. Development of broad-range 16S rDNA PCR for use in the routine diagnostic clinical microbiology service. J. Med. Microbiol. 52(Pt 8):685–691. 10.1099/jmm.0.05213-0. [DOI] [PubMed] [Google Scholar]


