LETTER
The 24-locus mycobacterial interspersed repetitive-unit–variable-number tandem-repeat (MIRU-VNTR) typing method is considered the “gold standard” for the study of tuberculosis (TB) epidemiology. The protocols and the nomenclature for this technology have been fully standardized (1).
Both the 580 locus (also known as exact tandem repeat D [ETRD] or MIRU 4) and 4348 locus (also known as MIRU 39) are included in the standard 24 loci used for MIRU-VNTR typing of Mycobacterium tuberculosis complex (MTC) strains and consist of a variable number of repeat units of 77 and 53 bp, respectively. While locus 580 is included in the subset of 15 MIRU loci with the highest discriminatory power, locus 4348 is more conserved (2).
This study describes large mutations found in these two loci that prevented us from correctly classifying them using the current standard nomenclature.
A total of 655 MTC isolates collected from patients in Albania over 4-year period (from 2007 to 2010) within the framework of the European Union-funded TM-REST project (FP7-202145) were typed by spoligotyping (3) and by 24-locus MIRU-VNTR using the quadruplex version of the GenoScreen MIRU typing kit (GenoScreen, Lille, France) and GeneMapper version 3.7 (Applied Biosystems, CA). The loci that we were not able to assign to a specific allele were amplified by single PCR and run on a 3% agarose gel. To determine the sequences of these alleles, we sequenced the corresponding genome regions by the Sanger method using the same primers used for the MIRU-VNTR typing (2). Sequences were then aligned by BioEdit Sequence Alignment Editor (4).
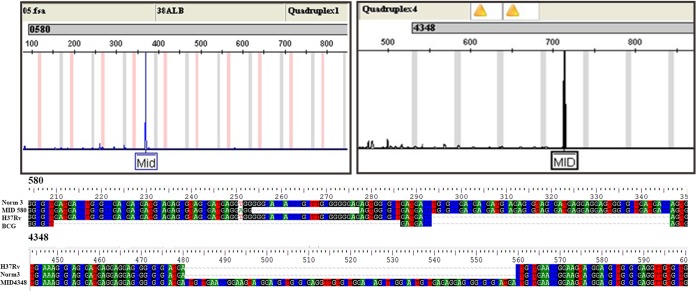
Out of 655 isolates typed, 25 isolates for locus 580 (3.8%) and 7 isolates for locus 4348 (1.1%) were found to carry an “out-of-bin” allele, in the middle (designated “MID”) of two standard markers (Fig. 1). Long-run gel electrophoresis excluded any possible mistake due to incorrect calibration of the sequencer or any possible marker deviation in the analysis performed using GeneMapper version 3.7 (Applied Biosystems).
FIG 1.
GeneMapper version 3.7 screenshot of alleles located in the middle (MID) of two bins (between 3 and 3s for MID 580 and between 3 and 4 for MID 4348) and nucleotide structures of alleles MID 580 and 4348 compared to the reference strains. The figure was created using GeneMapper version 3.7 software (Applied Biosystems, Foster City, CA).
These 32 isolates shared the same lineage, UgandaI, which represents 18.17% of our strain collection, but belonged to different spoligo-international types (SITs): SIT 613 for samples harboring locus MID 580 and SIT 52 for those with locus MID 4348. These two groups of strains showed the same or a very similar 24-locus MIRU-VNTR code.
The 25 samples carrying the MID 580 allele showed a 24-bp deletion in the third repetition preceded by a T→A substitution at position 251, whereas all 7 samples with the unclassified locus 4348 showed two full repeats plus a 79-bp insertion after the second repetition, resembling part of the 3′-end flanking region (Fig. 1).
In this study, we highlight two different uncommon MIRU-VNTR loci that cannot be assigned to any known allele included in the current standard nomenclature.
Large polymorphisms inside locus 580 have been previously reported, strongly suggesting that this locus is prone to mutation (5, 6). Smittipat et al. reported a 24-bp deletion affecting a slightly different part of the locus without nucleotide substitution at position 251 (5). Tafaj et al. suggested the presence of a large mutation in SIT 613 (6). To our knowledge, no large polymorphisms have been previously reported for locus 4348.
In conclusion, we report the first collection of MTC strains in which alleles unclassifiable with the current standard MIRU-VNTR nomenclature are not a rare event (3.8% and 1.1% of our collection). Despite the fact that they show the same or a very similar MIRU-VNTR code, in some of these strains, the recent transmission has been excluded by epidemiological investigation. It is possible that previous studies, especially when performed mainly by manual methodology, could have misclassified those loci, over- or underestimating transmission. A larger typing study on the UgandaI lineage could highlight the true representation of these alleles.
If these large mutations will be confirmed in other strain collections, they will have to be included in the current 24-locus MIRU-VNTR standardized nomenclature to ensure univocal genotypic characterization of these strains and to avoid improper cluster analysis, which would affect an efficient contact tracing strategy.
Footnotes
Published ahead of print 24 September 2014
REFERENCES
- 1.de Beer JL, Kremer K, Ködmön C, Supply P, van Soolingen D, Global Network for the Molecular Surveillance of Tuberculosis 2009. 2012. First worldwide proficiency study on variable-number-tandem-repeat typing of Mycobacterium tuberculosis complex strains. J. Clin. Microbiol. 50:662–669. 10.1128/JCM.00607-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Supply P, Allix C, Lesjean S, Cardoso-Oelemann M, Rüsch-Gerdes S, Willery E, Savine E, de Haas P, van Deutekom H, Roring S, Bifani P, Kurepina N, Kreiswirth B, Sola C, Rastogi N, Vatin V, Gutierrez MC, Fauville M, Niemann S, Skuce R, Kremer K, Locht C, van Soolingen D. 2006. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 44:4498–4510. 10.1128/jcm.01392-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, Bunschoten A, Molhuizen H, Shaw R, Goyal M, van Embden J. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic. Acids Symp. Ser. 41:95–98. [Google Scholar]
- 5.Smittipat N, Billamas P, Palittapongarnpim M, Thong-On A, Temu MM, Thanakijcharoen P, Karnkawinpong O, Palittapongarnpim P. 2005. Polymorphism of variable-number tandem repeats at multiple loci in Mycobacterium tuberculosis. J. Clin. Microbiol. 43:5034–5043. 10.1128/JCM.43.10.5034-5043.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tafaj S, Zhang J, Hauck Y, Pourcel C, Hafizi H, Zoraqi G, Sola C. 2009. First insight into genetic diversity of the Mycobacterium tuberculosis complex in Albania obtained by multilocus variable-number tandem-repeat analysis and spoligotyping reveals the presence of Beijing multidrug-resistant isolates. J. Clin. Microbiol. 47:1581–1584. 10.1128/JCM.02284-08. [DOI] [PMC free article] [PubMed] [Google Scholar]