LETTER
The blaKPC-2 gene, encoding the KPC-2 carbapenemase, was first identified in the United States in 1996, and blaKPC variants now appear to be endemic in several countries, including the United States, Israel, Greece, and China, with sporadic reports in other locations (1). blaKPC genes have generally been found flanked by the insertion sequences ISKpn7 and ISKpn6 within the Tn3-family transposon Tn4401 (2) or truncated versions of this structure. Tn4401 has been detected in plasmids from several incompatibility groups, including IncFIIK plasmids related to pKpQIL (3), IncN, ColE (4), IncI2 (5), and IncX3 (6) plasmids.
A recent paper linked an ∼11,109-Da peak detected by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) with the presence of blaKPC and used it to track pKpQIL-like plasmids (7). The peak corresponds to the cleavage product of a hypothetical protein designated p019 in the pKpQIL sequence (GenBank accession no. NC_014016.1). The authors noted that although p019 “appears to be fairly closely linked to the blaKPC gene,” “genetic events in the plasmid may change the association of the pKpQIL_019 MALDI-TOF MS peak with the presence of functional carbapenemase” (7).
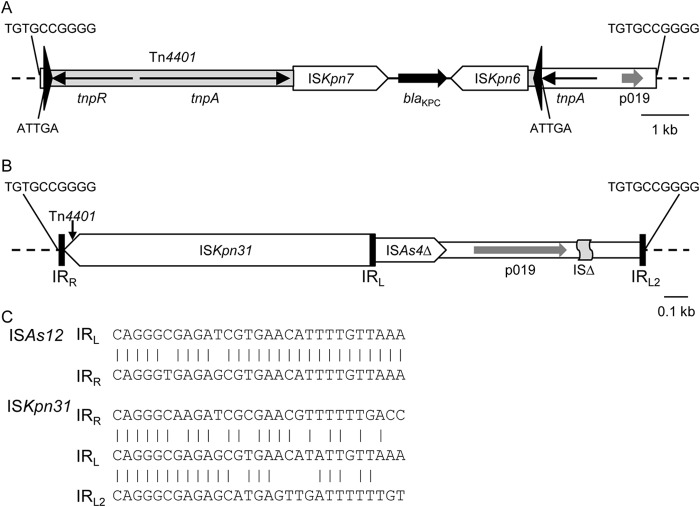
Detailed analysis of the pKpQIL sequence suggests that the p019 gene is part of an insertion into which Tn4401 has then been inserted. This insertion is flanked by 10-bp direct repeats (DR) and is 2,328 bp in length (excluding one 5-bp DR flanking Tn4401) (Fig. 1A). As noted previously (8), part of this region is related to the insertion sequence ISAs12 (∼91% nucleotide identity), which creates 10 bp (ISfinder, https://www-is.biotoul.fr/) (9). The ends of this putative IS, designated ISKpn31 by ISfinder, resemble the terminal inverted repeats (IR) of ISAs12. However, the adjacent region containing the p019 gene and other IS fragments, which has no equivalent in ISAs12, also ends in a similar sequence (Fig. 1C). It is possible that this region had previously become incorporated as an internal part of an ISKpn31-derived mobile element or that it was mobilized in a way similar to that of ISEcp1-mediated capture of regions adjacent to the right IR (IRR) (10), except that it lies adjacent to the left end of ISKpn31.
FIG 1.
Insertion in pKpQIL-like plasmids. (A) Complete insertion (positions 8823 to 21062 in GenBank accession no. NC_014016.1). (B) Expanded diagram showing details of the inserted region with Tn4401 and one 5-bp DR removed (positions 8823 to 8881 and 18794 to 21062). Inverted repeats (IR) of Tn4401 are shown as tall black triangles, and IR of ISKpn31 are indicated by black bars. IS are shown by boxes, with the pointed end indicating IRR. Labeled arrows indicate the directions and extents of selected genes. The sequences of DR flanking insertions are shown. ISΔ is a fragment of an IS21-like IS. (C) Comparison of IR sequences. (ISAs12 IR are from the 16 copies in the Aeromonas media chromosome in GenBank accession no. CP007567).
Although BLAST searches indicate that the insertion containing p019 has been found only in plasmids carrying blaKPC to date, these two genes are part of different mobile elements. Independent movement is already evident for Tn4401, which has been found in different plasmid backbones flanked by 5-bp DR, indicative of direct insertion, e.g., in p15S (GenBank accession no. FJ223606) (4) and pCOL-1 (KC609323) (11). The presence of p019 and blaKPC on IncFIIK plasmids less closely related to pKpQIL (e.g., pKPN-101-IT) (12) or those that belong to other Inc groups, e.g., IncI2 (pBK15692) (5) or IncX3 (pKPC-NY79) (6), also means that detecting p019 does not always indicate closely related or even similar plasmids. Thus, caution needs to be used in correlating the presence of p019 with the presence of blaKPC or a particular plasmid, and understanding the genetic contexts of markers apparently linked to resistance genes is important.
Footnotes
For the author reply, see doi:10.1128/JCM.02614-14.
REFERENCES
- 1.Nordmann P, Naas T, Poirel L. 2011. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 17:1791–1798. 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Naas T, Cuzon G, Villegas MV, Lartigue MF, Quinn JP, Nordmann P. 2008. Genetic structures at the origin of acquisition of the β-lactamase blaKPC gene. Antimicrob. Agents Chemother. 52:1257–1263. 10.1128/AAC.01451-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leavitt A, Chmelnitsky I, Carmeli Y, Navon-Venezia S. 2010. Complete nucleotide sequence of KPC-3-encoding plasmid pKpQIL in the epidemic Klebsiella pneumoniae sequence type 258. Antimicrob. Agents Chemother. 54:4493–4496. 10.1128/AAC.00175-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gootz TD, Lescoe MK, Dib-Hajj F, Dougherty BA, He W, Della-Latta P, Huard RC. 2009. Genetic organization of transposase regions surrounding blaKPC carbapenemase genes on plasmids from Klebsiella strains isolated in a New York City hospital. Antimicrob. Agents Chemother. 53:1998–2004. 10.1128/AAC.01355-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen L, Chavda KD, Al Laham N, Melano RG, Jacobs MR, Bonomo RA, Kreiswirth BN. 2013. Complete nucleotide sequence of a blaKPC-harboring IncI2 plasmid and its dissemination in New Jersey and New York hospitals: a hidden threat. Antimicrob. Agents Chemother. 57:5019–5025. 10.1128/AAC.01397-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ho PL, Cheung YY, Lo WU, Li Z, Chow KH, Lin CH, Chan JF, Cheng VC. 2013. Molecular characterization of an atypical IncX3 plasmid pKPC-NY79 carrying blaKPC-2 in a Klebsiella pneumoniae. Curr. Microbiol. 67:493–498. 10.1007/s00284-013-0398-2. [DOI] [PubMed] [Google Scholar]
- 7.Lau AF, Wang H, Weingarten RA, Drake SK, Suffredini AF, Garfield MK, Chen Y, Gucek M, Youn JH, Stock F, Tso H, DeLeo J, Cimino JJ, Frank KM, Dekker JP. 2014. A rapid matrix-assisted laser desorption ionization-time of flight mass spectrometry-based method for single-plasmid tracking in an outbreak of carbapenem-resistant Enterobacteriaceae. J. Clin. Microbiol. 52:2804–2812. 10.1128/JCM.00694-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moura A, Pereira C, Henriques I, Correia A. 2012. Novel gene cassettes and integrons in antibiotic-resistant bacteria isolated from urban wastewaters. Res. Microbiol. 163:92–100. 10.1016/j.resmic.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 9.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34:D32–D36. 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poirel L, Lartigue MF, Decousser JW, Nordmann P. 2005. ISEcp1B-mediated transposition of blaCTX-M in Escherichia coli. Antimicrob. Agents Chemother. 49:447–450. 10.1128/AAC.49.1.447-450.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Naas T, Bonnin RA, Cuzon G, Villegas MV, Nordmann P. 2013. Complete sequence of two KPC-harbouring plasmids from Pseudomonas aeruginosa. J. Antimicrob. Chemother. 68:1757–1762. 10.1093/jac/dkt094. [DOI] [PubMed] [Google Scholar]
- 12.Frasson I, Lavezzo E, Franchin E, Toppo S, Barzon L, Cavallaro A, Richter SN, Palu G. 2012. Antimicrobial treatment and containment measures for an extremely drug-resistant Klebsiella pneumoniae ST101 isolate carrying pKPN101-IT, a novel fully sequenced blaKPC-2 plasmid. J. Clin. Microbiol. 50:3768–3772. 10.1128/JCM.01892-12. [DOI] [PMC free article] [PubMed] [Google Scholar]