PREFACE
The ability to sense and respond to fluctuations in environmental nutrient levels is a requisite for life. Nutrient scarcity is a selective pressure that has shaped the evolution of most cellular processes. Different pathways that detect intracellular and extracellular levels of sugars, amino acids and lipids, and surrogate metabolites, are then integrated and coordinated at the organismal level via hormonal signals. During food abundance, nutrient sensing pathways engage anabolism and storage, and scarcity triggers homeostatic mechanisms, like the mobilization of internal stores through mechanisms such as autophagy. Nutrient sensing pathways are commonly deregulated in human metabolic diseases.
Nutrients (also referred to as macronutrients) are simple organic compounds involved in biochemical reactions that produce energy or are constituents of cellular biomass. Glucose and related sugars, amino acids, and lipids (including cholesterol) are important cellular nutrients, and distinct mechanisms to sense their abundances operate in mammalian cells. Essentiality is not necessarily a hallmark of nutrients, as for certain amino acids, such as arginine, cysteine, glutamine, glycine, proline and tyrosine, their essentiality is context dependent. In healthy individuals, the de novo synthesis of these amino acids from other molecules meets organismal requirements, but under particular metabolic needs, as during the rapid growth of infants1,2, they must be also obtained from the environment. Nutrient scarcity has operated as a strong pressure for selecting efficient mechanisms of nutrient sensing in all organisms. Considering the importance of nutrient homeostasis for all living organisms, and for human health in particular, it is surprising that we know relatively little about direct nutrient sensing mechanisms.
The sensing of a particular nutrient may involve the direct binding of the sensed molecule to the sensor, or occur by an indirect mechanism relying on the detection of a surrogate molecule that reflects nutrient abundance. Regardless of the manner in which nutrient sensing occurs, in order to consider a sensor as such, the affinity constant must be within the range of physiological fluctuations of the concentration of the nutrient or its surrogate.
Unicellular organisms are directly exposed to environmental fluctuations of nutrients, and sense both intracellular and environmental nutrient levels. In contrast, most cells in multicellular eukaryotes are not directly exposed to changes in environmental nutrients, and homeostatic responses aimed to maintain circulating nutrient levels within a narrow range exist. Nevertheless, internal nutrient levels do fluctuate, and hence intracellular and extracellular nutrient sensing mechanisms exist also in mammals. In multicellular organisms, nutrients also trigger the release of hormones, which act as long-range signals with non-cell autonomous effects, to facilitate the coordination of coherent responses in the organism as a whole.
Here, we will discuss intracellular and extracellular glucose, amino acid, and lipid sensing mechanisms and signaling events in mammals; how these sensing mechanisms become deregulated in human disease; and also elaborate on how internal nutrient stores are mobilized during nutrient scarcity.
LIPID SENSING
Lipids are a large and diverse set of nutrients (e.g. fatty acids or cholesterol) characterized by hydrophobic carbon backbones that are used for energy storage and membrane biosynthesis, among other cellular processes. Due to their non-polar nature, lipids are either packaged into lipoproteins and chylomicrons or bound by albumin in the serum3, and are rarely found free in a soluble form the organism. Despite the morbidity caused by increased lipid ingestion and deregulated lipid storage, as occurring in obese states, our knowledge about lipid sensing mechanisms is, with some exceptions, quite limited.
Fatty acid signaling
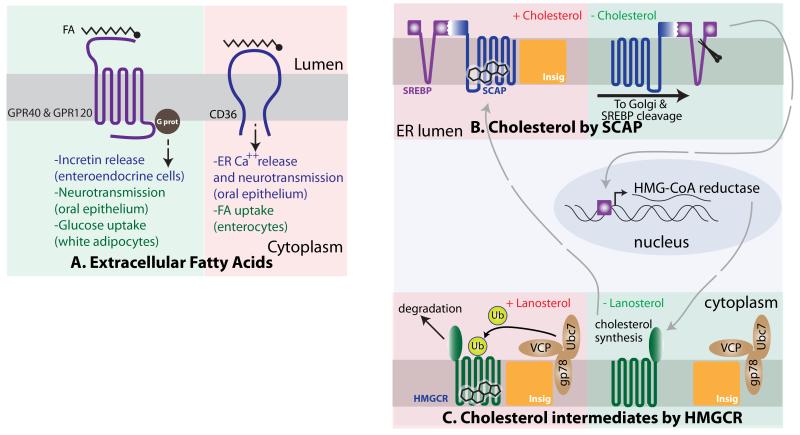
A family of G-protein coupled receptors, best characterized by GPR40 and GPR120, detect long chain unsaturated fatty acids (FAs). In mechanisms not fully understood, free FA stimulation of GPR40 at the plasma membrane of pancreatic beta cells augments glucose-stimulated insulin release4 (Figure 1A). GPR120 also mediates insulinotropic activity, albeit by an indirect mechanism, involving production of GLP1. GLP1 belongs to a group of gastrointestinal hormones called incretins that promote insulin release in beta cells5. These examples demonstrate how an increase in one particular nutrient (FAs) anticipates a response to the imminent increase in another nutrient (glucose), as food intake rarely provides solely one nutrient species. Additionally, activation of GPR120 at the plasma membrane of white adipocytes leads to a signal transduction cascade that promotes PI3K/AKT activation, leading to the cell-autonomous induction of glucose uptake6 (Figure 1A). Genetic mutations that disrupt GPR120 function occur in obese humans, and ablation of Gpr120 in mice contributes to diet induced-obesity, suggesting a key role for this signal transduction pathway in the systemic control of nutrient homeostasis7. Naturally, these findings have spurred interest toward the development of GPR120 agonists to control the onset of obesity8.
Figure 1. Lipid Sensing Mechanism.
A. Fatty Acid (FA) detection mechanisms by GPR40 and 120 (left) and CD36 (right). These GPR family members are expressed in several cell types including entero-endocrine cells, taste buds and white adipocytes. In the enteroendocrine cells, binding to FAs occurs in the luminal side, and the signal is transduced via G protein, leading to the release of incretins into the circulation. In taste buds, they trigger the release of neurotransmitters; in white adipocytes, activation of GPR120 indirectly promotes glucose uptake. Binding of CD36 to free FAs in the oral taste buds triggers Ca++ release and neurotransmission; in enterocytes, it directly promotes FA uptake. B. Cholesterol sensing by SCAP. In the presence of cholesterol, the SCAP/SREBP complex binds the INSIG proteins at the endoplasmic reticulum (ER) membrane and remains anchored in the ER. When cholesterol is absent and SCAP/SREBP do not bind INSIG, the complex traffics to the Golgi where the cytoplasmic tail of SREBP gets released by proteolytic cleavage, and triggers a cholesterol synthesis transcriptional program at the nucleus, including the synthesis of HMG-CoA reductase (HMGCR). C. The enzyme HMGCR catalyzes a rate-limiting step in cholesterol synthesis, and is synthesized when cholesterol levels are low. HMGCR is embedded in the ER membrane and also has cytoplasmic domains, which include its catalytic activity. In the presence of abundant intermediate species in the cholesterol biosynthetic pathway, HMGCR interacts with the INSIG proteins, constitutively bound to an ubiquitination complex. This leads to HMGCR ubiquitination and degradation and halts the synthesis of cholesterol in a rapid regulatory mechanism, key to the anticipation of an imminent increase in cholesterol levels.
In addition to GPR120, the FAT/CD36 receptor has been implicated in direct binding and uptake of intestinal luminal FAs9, and interestingly GPR40, GPR120, and CD36 have FA-sensing properties in cells within the oral epithelium involved in gustatory perception10-13 (Figure 1A).
Cholesterol sensing
Our limited knowledge about the sensing of other lipidic species contrasts with the profound understanding of the cholesterol sensing mechanism, deciphered by Brown and Goldstein14. Sterols, including cholesterol, are fundamental constituents of mammalian membranes that provide membrane fluidity and are also needed for the synthesis of steroid hormones. Cholesterol can be obtained from the diet and also synthesized de novo. Hence, adequate sensing of internal cholesterol levels allows for the control of the energetically demanding cholesterol biosynthetic pathway, so that is only active when external supply and internal levels of sterols are low. Cholesterol sensing occurs in close proximity to the regulation of the cholesterol biosynthetic pathway. The cholesterol sensing protein, and the transcription factor that induces the expression of enzymes involved in the cholesterol biosynthetic pathway, from a constitutively bound complex on the endoplasmic reticulum (ER). The cholesterol-sensing protein SCAP (SREBP1 cleavage activating protein) directly binds cholesterol via a region originally found to span its 5 transmembrane sterol sensing domains (SSD)15,16. The initial mapping observations were later refined to a loop in the ER side of the membrane, likely embedded in the lipid bilayer17 (Figure 1B). SCAP is constitutively bound to SREBP (Sterol Regulatory Element-Binding Protein), which transactivates genes critical for cholesterol synthesis. When cholesterol levels are high, cholesterol binding to SCAP triggers a conformational change that increases its affinity to the INSIG protein18, an anchor for SCAP and SREBP within ER membranes. Conversely, when cholesterol levels are low and SCAP is not bound to cholesterol, the SCAP/SREBP tandem dissociates from INSIG and shuttles to the Golgi apparatus19 (Figure 1B). This step is of crucial importance, because the presence of the SCAP/SREBP complex at the Golgi allows the cleavage and release of the cytoplasmic N-terminus of SREBP by proteases resident at the Golgi20,21. In turn, the cleaved cytoplasmic fragment of SREBP translocates to the nucleus and induces genes involved in lipid anabolism. Replete cholesterol levels then initiate a negative feedback by interacting with SCAP and inhibiting further cleavage of SREBP22.
Substantial evidence also supports an additional sterol-sensing event that occurs within the ER, involving the enzyme HMG-CoA reductase. HMG-CoA reductase catalyzes the rate-limiting step in de novo cholesterol synthesis and is a transcriptional target of SREBP in response to low cholesterol levels. The C-terminus of HMG-CoA reductase, containing its catalytic activity, is exposed to the cytoplasm, while several transmembrane domains, including the sterol-sensing domain reminiscent to that of SCAP, are embedded in the ER membrane23. High levels of intermediate lipid species in cholesterol synthesis, such as lanosterol, trigger the binding of HMG-CoA reductase to INSIG, which is also bound constitutively to an ubiquitination complex formed by VCP, GP78 and UBC7. This interaction promotes the ubiquitin-mediated degradation of HMG-CoA reductase24 (Figure 1C). As mentioned before, HMG-CoA reductase catalyzes an early (and rate limiting) step in cholesterol synthesis, but the levels of HMG-CoA reductase are regulated by a slow, transcriptional mechanism that is shutoff only after cholesterol levels have already been replenished. Hence, the interaction of HMG-CoA reductase with Insig, leading to its turnover by the proteasome, constitutes a faster regulatory loop that aims to put a brake in cholesterol synthesis when the presence of precursor molecules already warranties its imminent increase.
Sensors upstream of adipokines
Adipokines, hormones secreted by adipocytes, exert systemic effects that include the regulation of appetite, energy expenditure and other processes that contribute to nutrient homeostasis. Their levels do not necessarily reflect circulating lipid levels, but report on organismal lipid storage25, and some adipokines, as LEPTIN, can be considered a surrogate indicator of lipid storage abundance. Surprisingly, the identity of the sensor that connects high levels of stored lipids with LEPTIN production remains a mystery, despite the identification of regulatory elements in the promoter region of the LEPTIN gene26. We know significantly more regarding the systemic effects downstream of LEPTIN. LEPTIN receptor is expressed both in the central nervous system and in peripheral tissues and its activation coordinates food intake and organismal metabolism. In hypothalamic neurons that suppress appetite (anorexigenic), LEPTIN activity antagonizes the effect of appetite-stimulating neuropeptides and neurotransmitters. Lipid mobilization by adipocytes, as occurring in fasting states, results in decreased LEPTIN production, thereby stimulating appetite and promoting nutrient acquisition behavior. Indeed, mutations in the Leptin receptor gene were found in morbidly obese patients27, and mice harboring inactivating mutations in the Leptin28 or Leptin receptor29 genes are so hyper-fagic that they can double the mass of normal mice.
In addition to LEPTIN, adipocytes also synthesize the hormone Adiponectin (also known as ADIPOQ)30,31, though the regulation of its production is even less understood32. In contrast to LEPTIN, circulating ADIPOQ levels inversely correlate with lipid storage, and this adipokine exerts a multitude of systemic effects that include the promotion of energy expenditure, insulin sensitivity and loss of appetite33-35. Mutations and polymorphisms in the human ADIPOQ gene strongly correlate with obesity and the development of type 2 diabetes36-38.
AMINO ACID SENSING
Amino acids (AAs) are the building blocks for proteins, the most abundant macromolecule in cells. Protein synthesis is energetically expensive and complex; accordingly, cells sense extracellular and intracellular AAs to couple abundance to use. Under conditions of AA scarcity, proteins constitute reservoirs of AAs that catabolic programs such as proteasome-mediated degradation and autophagy mobilize. AAs are subsequently recycled and allocated for the synthesis of specific proteins required under nutrient limitation. Furthermore, under periods of prolonged starvation and hypoglycaemia, AAs are catabolized for the production of other forms of energy, as glucose and ketone bodies, required to fuel the particular needs of certain organs, like the brain. Hence, accurate sensing of AA levels is key for an efficient regulation of protein and AA synthesis and catabolism, and also for the control of food intake.
GCN2
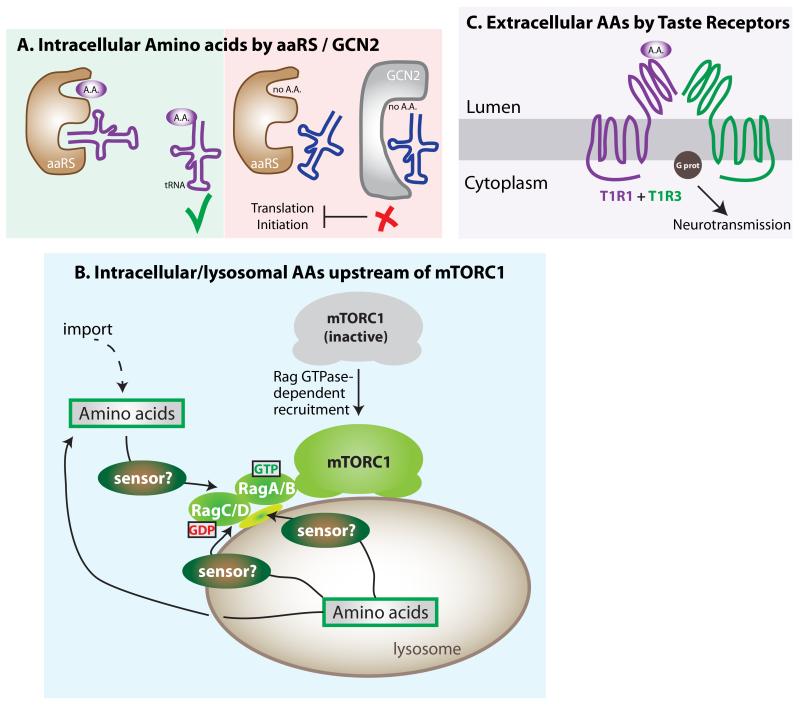
As no AA compensates for the absence of another in protein synthesis, the cell must be able to efficiently detect the lack of any AA in order to prevent potential failures in peptide chain synthesis. The structural unit of the protein synthesis machinery, the ribosome, incorporates AAs into a nascent peptide by the sequential binding of specific transfer RNAs (tRNAs) covalently linked to its cognate AA. Amino acid-specific aminoacyl tRNA synthetases (aaRS) execute the loading of AAs to their cognate tRNAs39, and uncharged tRNAs accumulate during low levels of free AAs. Failure to finish a peptide chain due to a stalled ribosome under AA scarcity is inefficient and energetically onerous, so cells anticipate this situation by preventing translation initiation. The mechanism involves a single protein that is able to detect any uncharged tRNA, regardless of its AA specificity, allowing for the detection of low levels of any AA in the context of abundance of the other 19. This protein is GCN2 (kinase general control nonderepresible 2), which has high affinity to all uncharged tRNAs40 (Figure 2A), and represents an elegant example of AA sensing by the detection of a surrogate molecule. Under low intracellular AA levels, the binding of GCN2 to a given uncharged tRNA triggers a conformational change that leads to kinase activation and inhibitory phosphorylation of a key early activator of translation initiation: the eukaryotic initiator factor 2 alpha (eIF2α)41. Mouse models have proven the importance of GCN2/eIF2a in mammalian responses to transient drops in AAs42,43 and, interestingly, this AA sensing pathway seems to play a key role in the central nervous system for the detection of imbalances in AA composition in food, independently of taste44-46.
Figure 2. Amino acid Sensing Mechanisms.
A. GCN2 detects insufficiencies of cellular amino acids (AAs). During low levels of any AA, its cognate aaRS fails to load the transfer RNA (tRNA), which is then detected by GCN2 kinase, halting translation initiation. B. mTORC1 is activated downstream of elevated intracellular AAs via its recruitment to the outer lysosomal surface through a Rag GTPase-mediated mechanism. Increases in intra-lysosomal levels of AAs control Rag GTPase function, which recruits mTORC1 to the outer lysosomal membrane, an essential step in its activation. The identities of the sensor for AAs remain unidentified, and several non-mutually exclusive possibilities exist: a) an intra-lysosomal sensor that transduces the signal through the membrane; b) a lysosomal transmembrane sensor that both detects and transduces the signal; and c) and a cytoplasmic sensor that operates downstream of AA export from the lysosome. C. Extra-organismal AA sensing by oral taste receptors. The heterodimeric receptor T1R1+T1R3 binds AAs at high concentrations only, and triggers a signal transduction cascade via G-protein. In the intestinal epithelium, it also leads to the localization of GLUT2 to the apical membrane, facilitating glucose import.
Inhibition of protein synthesis by GCN2/eIF2a occurs in concert with other cellular responses to AA depletion, such as the inhibition of the mTOR pathway (see below), restricting translation to those mRNAs encoding proteins required for cellular adaptation to nutrient starvation while impairing synthesis of most other proteins47. Minimizing translation also enables the use of AA as energetic sources.
mTORC1
The mechanistic target of rapamycin (mTOR) kinase, when part of mTOR complex 1 (mTORC1), controls cellular energetics by inducing numerous anabolic processes, including protein and lipid synthesis48. Growth factors activate mTORC1 via a well-understood signal transduction cascade initiated by the binding of a receptor at the plasma membrane, and culminating with the activation of the Rheb GTPase. Rheb directly binds mTORC1 and activates its kinase in a growth factor-dependent manner49-52. In addition to the regulation by protein hormones, intracellular AAs also activate mTORC1, so the complex integrates information on both systemic and cellular nutrient levels. In spite of the fact that mTORC1 activity is highly responsive to changes in AA levels, it is not an AA sensor. Indeed, mTORC1 activation is one of the several examples of a key sensing signaling process where, in spite of intense interest, actual nutrient sensors remain unidentified (Figure 2B). mTORC1 is not equally sensitive to all AAs, leucine being particularly important for its activation53. We can only speculate about the selective importance of leucine levels for mTORC1 activation, mentioning that is one of the most abundant AA in proteins, and hence, more likely to be limiting during protein synthesis. Intriguingly, GCN2-null mice fed a leucine deficient diet display a more severe phenotype than the same animals fed diets lacking tryptophan or glycine43. Thus, leucine seems critical for the organismal sensing of AA sufficiency and deprivation by different pathways. The molecular characterization of the AA-dependent activation of mTORC1 started only a few years ago with the identification of the Rag family of GTPases54,55, which regulate mTORC1 via a distinct mechanism to that of growth factors. Whereas growth factors regulate the kinase activity of mTORC1, the Rag GTPases recruit mTORC1 to the outer lysosomal surface, an essential step in its activation56. Because mTORC1 kinase activation by Rheb occurs at the outer lysosomal surface, it is only possible following Rag GTPase-dependent recruitment of mTORC1 (Figure 2B). Hence, AA abundance and the consequent recruitment of mTORC1 is a prerequisite for the activation of mTORC1 by growth factors (Figure 2). Although the sensors for AAs have not been identified as yet, a few pieces in the puzzle of AA-dependent regulation of mTORC1 have been added recently. Cell-based biochemical studies have identified the proteins responsible for tethering the Rags to the lysosomal surface56, guanine exchange factors (GEFs) and GTPase-activating proteins (GAPs), as well as other regulatory proteins operating upstream of the Rag GTPases57-63.
Although the reason for the recruitment of mTORC1 to the lysosomal surface may sound puzzling, independent pieces of evidence suggest that the lysosome plays a key role in AA homeostasis. The yeast vacuole, organelle equivalent to mammalian lysosome, accumulates nutrients, such as AAs64, and the mechanism of mTORC1 recruitment is conserved in yeast65. In addition, high intraluminal concentrations of certain AAs have been shown also in lysosomes66. Protists such as D. dyscoideum obtain energy via phagocytosis and lysosomal degradation67, which is followed by a transient increase in intralysosomal nutrient levels. Finally, both the lysosome and the vacuole are the organelles where AAs and other nutrients are scavenged from cellular components, via the catabolic process of autophagy (Figure 4). Hence, high levels of AAs within the lysosome/vacuole system seem to reflect to some extent cellular AA abundance, and so it is reasonable to couple its sensing with recruitment and activation of mTORC1, a critical regulator of most anabolic processes, including protein synthesis.
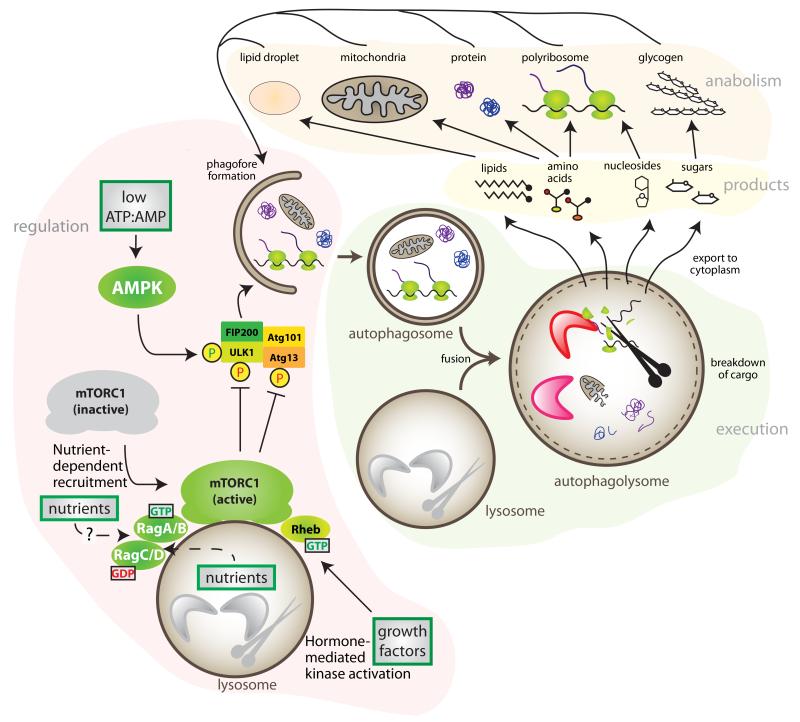
Figure 4. Nutrients and Autophagy.
Autophagy serves as an internal source of stored nutrients under conditions of nutrient limitation. Two main regulatory inputs for autophagy are AMPK and mTORC1. Autophagy initiation can be promoted by the activation of ULK1 via AMPK-dependent phosphorylation during low ATP:AMP ratio. mTORC1 is activated by growth factors at the outer lysosomal surface if cellular AAs and glucose have recruited mTORC1 via the action of the Rag GTPases. Once activated, mTORC1 inhibits ULK1 and Atg13 by phosphorylation. Hence, low nutrients promote autophagy by the inhibition of mTORC1. Autophagy starts with the engulfment of cellular constituents: glycogen, lipids from lipid droplets, soluble proteins, ribosomes or organelles in a double membrane structure that then fuses with lysosomes, where the enzymatic breakdown occurs. The products of autophagy, basic nutrients (sugars, lipids, amino acid, and nucleosides), are then exported into the cytoplasm, where may be used as a source of energy, or re-used for anabolism.
Germline and sporadic mutations in genes involved in the signal transduction of nutrient levels upstream of the Rag GTPases have been found in human syndromes characterized by growth defects, neurological disorders, skin and immunological problems, and tumors60,61,68-70.
Amino acid-sensing taste receptors
As strict heterotrophs, animals must obtain energy and nutrients from external organic sources, and predicting the nutritional value of food before digestion allows for accurate selection of food sources and for anticipation of increased nutrient abundance. Several mechanisms act synergistically, including experience and social rules in humans, but a fundamental nutrient sensing event occurs at the level of the oral taste buds. Nutrient sensing by taste receptors is not just a means of sensing extracellular nutrients, it is a mechanism of extra-organismal sensing that allows interrogation of prospective food sources. In humans, taste is divided into five categories: sweet, umami, bitter, sour and salty, and is generated by signals elicited in taste buds, groups of cells in the tongue, palatal, and esophageal epithelium. Within these cells, the taste receptors are logically exposed in the apical membrane oriented toward the environment71.
Taste receptors belong to the T1R and T2R families of G-protein coupled receptors, and are characterized by 7 trans-membrane domains with an extracellular N-terminus and an intracellular C-terminus. Molecular and genetic information regarding the different members of the taste receptors genes can be found elsewhere71. The T2R family is involved in the detection of bitter molecules, a category that includes potentially toxic compounds, and two T1R family members are responsible for sensing the presence of AAs (the umami taste). Although other taste receptors also exist71,72, elegant genetic studies employing heterologous expression experiments defined that the T1R1+T1R3 heterodimer senses AAs (Figure 2C). Human AA taste receptors have particularly high affinity to glutamate, but other L-amino acids also serve as ligands, while D-amino acids do not73. AA binding to a taste receptor triggers signal transduction through the plasma membrane, followed by G-protein activation and neurotransmitter release74, which is then integrated with other neurotransmission events at the central nervous system level.
In addition to the presence of taste buds in the oral epithelium, taste receptors also exist in endocrine cells in certain regions of the gut75. Intestinal taste receptors operate via G-protein activation in a similar manner to that of the oral epithelium, but instead of inducing the release of a neurotransmitter that activates an afferent signal to the brain, the cascade elicited by enteral taste receptors culminates with the release of incretins into blood circulation, serving as an anticipatory signal that prepares responses for the imminent digestion and systemic increase in nutrient abundance.
Interestingly, extracellular AA sensing at the plasma membrane by taste receptors can modulate mTORC1 activation without affecting intracellular AA levels76, a meaningful cross-talk that engages the anabolic machinery of the cell in anticipation to an elevation in intracellular AA levels, following import.
GLUCOSE SENSING
Mammals rely on multiple means of maintaining glucose levels within a narrow physiological range. Glucose intake, storage, mobilization and breakdown are tightly regulated at different levels, and multiple mechanisms of glucose sensing coexist: extraorganismal, extracellular and intracellular. In addition, and a network of hormone signals, exemplified by insulin and glucagon, aim to coordinate coherent responses to systemic glucose levels in distant organs. Deregulated glucose homeostasis mechanisms, from glucose sensing to import, storage and mobilization underlie the pathogenesis of type 2 diabetes and other human diseases.
GCK
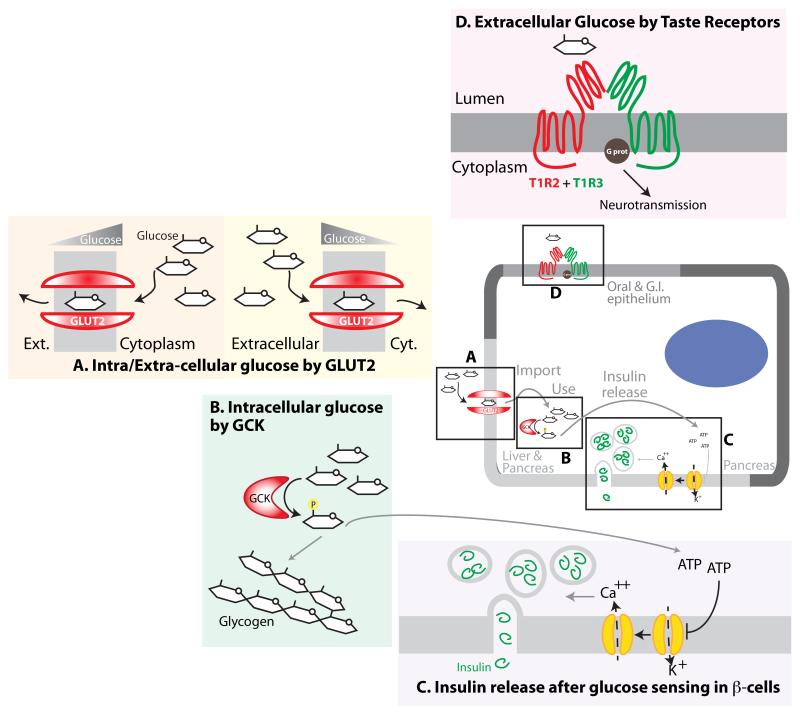
Glucokinase (GCK) catalyzes the first step in the storage and consumption of glucose, glycogen synthesis and glycolysis, and its function constitutes a simple, direct intracellular nutrient sensing mechanism that controls systemic glucose homeostasis. Like all hexokinases, GCK phosphorylates glucose to make glucose-6-phosphate (G6P), but unlike the other isozymes, only GCK functions as a glucose sensor77. This uniqueness occurs because, unlike the other hexokinases, which have Kms for glucose much below the minimum physiological level of glucose, GCK has a significantly lower affinity and is only active when glucose levels are relatively high (~120 mg/dl, or 7 mM, and greater). Hence, while the other hexokinases function as ‘phosphorylation machines’ regardless of the actual glucose levels, GCK is active only during glucose abundance, and it controls systemic glucose fate through its effects in the liver and pancreas (Figure 3B). The liver maintains glycaemia through gluconeogenesis and glycogen breakdown during periods of systemic glucose scarcity, or by storing glucose in the form of glycogen when it is in excess78. GCK is the most abundant hexokinase in liver and because is inactive under conditions of glucose limitation, it permits export of un-phosphorylated glucose from the liver in order to supply the energetic demands of the brain and muscles. When hepatic glucose levels are high, GCK-mediated conversion of glucose to the metabolic intermediate G6P allows its shunting into glycolysis (for energy production) or glycogen synthesis (for storage).
Figure 3. Glucose Sensing Mechanisms.
A. Glucose sensing by the GLUT2 transporter. Due to low affinity, this transporter actively imports glucose only during high glycaemic states (right). Due to its bidirectional properties, it can also export glucose from hepatocytes into the circulation under hypoglycaemic states if hepatic gluconeogenesis and glycogen breakdown raise the intrahepatic glucose levels (left) B. Intracellular glucose sensing by glucokinase (GCK) in hepatic and pancreatic cells. GCK has low affinity for glucose, and shunts glucose-6-phosphate into either glycolysis or glycogen synthesis only when glucose is abundant. C. Mechanism of insulin release downstream of glucose sensing in pancreatic beta-cells. A multi-step process that relies on glucose phosphorylation by GCK, subsequent ATP production, and ATP-mediated blockade of K+ channels. This leads to a Ca++ influx that facilitates insulin release from vesicles into the bloodstream. D. Extra-organismal glucose sensing by oral taste receptors. Dimeric receptors T1R2+T1R3 bind glucose, sucrose, fructose and artificial sweeteners at high concentration only, and trigger a signal transduction cascade via G-protein.
GCK is also expressed in beta cells (see below), and in neurons and glial cells in the hypothalamus, and although work remains to be done to understand the role of this glucose sensor in the brain, systemic effects, such as feeding responses and insulin release, are likely downstream of hypothalamic GCK activity79.
Dozens of germline mutations in GCK in patients with abnormal glycaemia and diabetes80, together with conditional deletion of the murine Gck gene in liver and pancreas81, support the fundamental role for GCK in maintaining organismal glucose homeostasis.
GLUT2
The glucose transporter GLUT2 (SLC2A2) is a sensor of extracellular glucose levels; like GCK, GLUT2 has a higher Km (20 mM) than other glucose transporters of the same family. The Km for GLUT1 is approximately 1 mM and that of GLUT4 is ~5 mM82, so they are close to saturation even during fasting glycaemia (~4 mM). The low affinity of GLUT2, in contrast, allows for efficient transport of glucose across the plasma membrane only when glycaemia is high, but not under the low concentrations that still saturate the other transporters. Accordingly, GLUT2 plays critical roles in directing organismal glucose handling following feeding. Hepatic glucose import mediated by GLUT2 is followed by GCK-dependent phosphorylation for storage and energy production, as described above. Importantly, during periods of low glycaemia, hepatic glycogenolysis and gluconeogenesis increase intrahepatic glucose levels. Because GLUT2 can transport glucose in a bi-directional manner, it now exports glucose to the circulation (Figure 3A). Hence, GLUT2-mediated import occurs only during transient hyperglycaemic states, and GLUT2-mediated export only happens when intrahepatic glucose levels are high, thus constituting a key controller of glucose homeostasis. Not surprisingly, inactivating mutations in GLUT2 lead to human metabolic disorders, such as the Fanconi-Bickel syndrome, which is characterized by deregulated glycogen accumulation, hepatomegaly and hypoglycaemia, among other symptoms of disrupted glycaemic homeostasis83.
Beta cells in the pancreas have a specialized role in sensing systemic glucose levels, and are responsible for the synthesis and secretion of insulin. Glucose is imported in beta cells and phosphorylated by the tandem of GLUT2 (or GLUT1) and GCK, respectively, and, as it is consumed, leads to an increased ATP:ADP ratio. This closes K+-channels at the plasma membrane, and causes the membrane to depolarize. Dissipation of membrane potential results in a transient increase of intracellular Ca++ that facilitates the fusion of insulin-containing vesicles with the plasma membrane, releasing its cargo into systemic circulation (Figure 3C). It is important to mention that whereas the predominant transporter in murine beta cells is GLUT2, the relative abundance of the GLUT2 transporter in human islets seems to be minor compared to that of the high affinity GLUT1 transporter, so that the relevance of GLUT2 for glucose transport in human beta cells is not clear84.
Elevated sugar intake and chronic hyperglycaemia deregulates normal glucose sensing via several mechanisms, including ER stress, elevated intracellular Ca++ levels, mitochondrial dysfunction, reactive oxygen species, and chronic inflammation, all of which seem to contribute to the corruption of insulin secretion in type 2 diabetes85.
Finally, although the other glucose transports, such as GLUT1 and GLUT4, do not behave as sensors, their activities and effects are regulated by different means, in order to meet particular requirements of glucose use and storage. GLUT4 is expressed in skeletal muscle and adipose tissue, two organs important for post-prandial glucose uptake and storage82, and although GLUT4 has a low Km, glucose uptake in these organs is a regulated process. Insulin triggers a PI3K-AKT dependent signal transduction cascade that results in GLUT4 localization to the plasma membrane, allowing glucose uptake in these tissues86. Because glucose import and storage are insulin-dependent, and thus secondary to direct glucose sensing mechanisms in liver and pancreas, they occurs only after the organism has reached a threshold of internal glucose abundance. GLUT1 is expressed in fetal tissues and its constant activity provides glucose to all tissues to sustain the rapid growth of the organism.
AMPK and ATP:AMP ratios
The AMP-activated protein kinase (AMPK) is a fundamental regulator of cellular metabolism and coordinates several metabolic responses in different cell types. It is exquisitely responsive to cellular energy levels, as a surrogate sensing mechanism for glucose abundance, increased levels of AMP and ADP directly activate the kinase. AMPK has been the subject of a number of excellent reviews addressing its activation, regulation, and downstream consequences can be consulted87,88, and will be briefly discussed herein in the context of the regulation of autophagy.
mTORC1 and the sensing of glucose
The regulation of mTORC1 through Rag GTPase-mediated recruitment is not restricted to AAs; cellular glucose levels also affect the activity of the Rag GTPases 89. Unlike our understanding of the activation of mTORC1 by cellular AAs, with some molecular players upstream of the Rag GTPases and downstream of AA already identified, less clear is the mechanism by which glucose regulates the Rags. Some aspects downstream of glucose and AA sensing are shared, such as the involvement of the lysosomal v-ATPase48,89,90, but additional players remain unidentified. Because the AA and glucose sensing mechanisms are generally independent phenomena, as herein illustrated, it is very likely that AAs and glucose sensing upstream of mTORC1 occur in parallel and converge upstream of the Rag GTPases, but precisely how this integration occurs is unresolved.
Glucose-sensing taste receptors
In a similar manner to AA sensing in taste buds by T1R1+T1R3, the heterodimer composed of T1R2+T1R3 constitutes the glucose taste receptor (Figure 3D). The extracellular N-terminal domains of both T1R1 and T1R2 are essential for determining the specificity for their natural ligands91. Millimolar concentration of the saccharides glucose, fructose or sucrose activate the T1R2+T1R3 receptor92; this concentration may seem high, but sucrose concentration in an apple is ~100-200 mM, and hence, this process is selective and efficient for the detection of highly energetic foods.
Glucose taste receptors are also expressed in the intestinal epithelium, and although the sensing process is identical to that of the oral epithelium, the signal transduction does not trigger an afferent signal to the brain, but results in the transient localization of the GLUT2 transporter to the apical membrane, leading to increased absorption of glucose from the intestinal lumen after feeding93,94.
In addition to natural ligands, glucose taste buds also respond to artificial sweeteners as saccharine, cyclamate and aspartame92. Activation of glucose taste receptors by artificial ligands has clinical implications for obesity and type 2 diabetes, as sweeteners may increase nutrient absorption and activate other nutrient-sensing signaling cascades at different levels, regardless of nutritional value. Indeed, some studies have shown that consumers of artificial sweeteners are at higher risk to develop metabolic disease95. The phenomenon of artificial activation of this nutrient sensing pathway is currently an active field of research.
AUTOPHAGY: ACCESSING INTERNAL NUTRIENT STORES
Because environmental nutrient availability can be intermittent, cells and organisms have evolved efficient ways of storing nutrients during periods of abundance. This occurs in unicellular organisms and is more obvious and prominent in animals, with the emergence of organs specialized in nutrient storage, such as the fat tissue, liver and skeletal muscle. Mammalian cells accumulate and store glucose in the form of glycogen, lipids within lipid droplets and internal membranes, and AAs in proteins and organelles;all of which can be mobilized and catabolized to endure periods of nutrient limitation. Cells exploit different means to obtain the basic nutrients from internal stores, including autophagy, the controlled process of recycling of cellular constituents confined within a double membrane structure. Autophagy starts with the de novo formation of a membrane structure termed phagophore, which engulfs its cargo and closes as a cytoplasmic double-membrane autophagosome. An autophagosome then fuses with a lysosome, which leads to the enzymatic breakdown of the autophagosomal cargo into its basic building blocks, then exported from the autophagolysosomes and further catabolized to produce energy, or used again in other anabolic reactions (Figure 4).
The process of autophagy is unique because it can target any cellular component and nutrient storage depot, and as a key internal source under scarcity, is highly regulated at multiple levels by nutrients and nutrient signaling96. AMPK kinase, directly activated by a low ATP:ADP ratio, phosphorylates and activates ULK1, a kinase that regulates autophagy initiation97,98. AMPK also activates the FoxO transcription factors, which transactivate ATG genes, responsible for the initiation and completion of autophagy99. Hence, AMPK regulates autophagy acutely, and also by means of a slower, transcriptional mechanism.
A critical regulator of autophagy, as shown in all eukaryotes using both cultured cells and model organisms, is mTORC1, through its inhibitory phosphorylation of ULK1 and Atg13100. mTORC1 appears to play a dominant role on the regulation autophagy, as mTORC1 inhibition is sufficient to induce it101, while its constitutive activation is sufficient to block it89. Nutrient depletion is perhaps the most potent inducer of autophagy, and the regulation of mTORC1 by the Rag GTPases downstream of nutrient scarcity appears to be essential for the regulation of autophagy. Mice with constitutive RagA activity, and hence, constitutive activation of mTORC1 regardless of nutrient levels, develop normally but succumb within the first day of life, similarly to mice lacking essential autophagy genes Atg5 and Atg789,102,103. Constitutive RagA activity in neonatal mice leads to a profound glucose and AA homeostasis defect secondary to an impairment in the detection of nutrient shortage after the trans-placental supply of nutrients is interrupted at birth, and the consequent inability to trigger autophagy.
In addition to the regulation of autophagy initiation, mTORC1 activity is required for autophagy termination104. Cellular free AAs, produced by autophagy, result in an increase in mTORC1 activity and the reformation of lysosomes. Systemic levels of nutrients also regulate autophagy via the effects of insulin105. The intracellular cascade of insulin activates AKT, a positive input for mTORC1, and also a negative regulator of the FoxO transcription factors. Hence, both local and systemic nutrients regulate the process. In addition to nutrients, hypoxia, ER stress, DNA damage, among others forms of stress, also regulate autophagy106.
Several studies that generated autophagy-deficient tissues in a temporal specific manner have determined the importance of autophagy in mammalian physiology. Besides the aforementioned role of autophagy in the early neonatal starvation period102,103, autophagy is essential for the survival of embryos in the pre-implantation stage107. Whole-body acute deletion of autophagy genes in adult mice eventually culminates in neurodegeneration and death, presumably due to the accumulation of harmful organelles and proteins, which likely cause neuronal toxicity108,109. Liver-specific impairment in autophagy results in accumulation of abnormal cellular endomembranes, mitochondria and ubiquitinated proteins103, and impaired lipid mobilization110. An impaired autophagy seems to preferentially affect cells specialized in vesicle trafficking, such as lymphocytes, beta cells, and others111, but some of these effects may be due to a deranged endomembrane trafficking system, rather than a direct consequence of a nutrient homeostasis defect.
CONCLUDING REMARKS
In spite of intense research, our understanding of nutrient sensing mechanisms is far from complete. For instance, we have not yet deciphered what links lipid storage levels with LEPTIN synthesis and release. Equally unclear is what the glucose and AA sensors upstream of mTORC1 are. Toward the identification of nutrient sensors upstream of mTORC1, the lysosome appears to be a key organelle in nutrient sensing; yet we still need to determine what and how is sensed at the lysosome. Besides these and other fundamental unanswered questions of direct nutrient sensing, the mechanisms discussed herein were outlined mostly in a modular manner. This reflects that we still lack an integrative view of the nutrient sensing pathways; connecting different aspects of nutrient sensing is one of the challenges of future research. We know that mTORC1 is a node where hormone and nutrient inputs converge, but we ignore whether these signaling cascades cross talk upstream of mTORC1. A complete view of nutrient sensing mechanisms includes addressing potential cross regulation between different nutrient sensing pathways, but also incorporating the regulation by other signaling events. For example, we know some consequences of chronic inflammation in deregulating nutrient sensing mechanisms and the signaling cascades downstream, as occurring in the obese state, but how exercise modulates nutrient inputs, or how aging effects nutrient sensing abilities, remain to be determined. From the experimental point of view, advances in genomics will likely contribute insight on clinical conditions secondary to deregulated nutrient sensing, such as the identification of novel mutations and polymorphisms in humans. Finally, nutrient abundance not only affects the onset of diabetes, but also influences cancer development and the process of aging. Nutrient sensing and metabolism in cancer cells has received a new wave of attention, in part thanks to the advances in next generation sequencing and metabolomics. Cancer cells are exposed to limited nutrients due to poor vasculature, and deregulated proliferation poses energetic and nutrient demands and liabilities, which act in concert with aberrant activation of growth signals. On the other hand, one of the most successful interventions against the onset of aging is limitation in nutrient intake, or caloric restriction112. Hence, understanding normal nutrient sensing mechanisms is a prerequisite for designing better interventions against human disease beyond diabetes.
ACKNOWLEDGEMENTS
Supported by grants from the National Institutes of Health (R01 CA129105, CA103866 and AI047389; R21 AG042876) and awards from the American Federation for Aging, Starr Foundation, Koch Institute Frontier Research Program, and the Ellison Medical Foundation to D.M.S., and fellowships from the Charles King’s Trust Foundation / Simeon J. Fortin Fellowship to A.E. W.C.C is supported by American Cancer Society - Ellison Foundation Postdoctoral Fellowship (PF-13-356-01-TBE). D.M.S. is an investigator of Howard Hughes Medical Institute.
REFERENCES
- 1.Wu G, Morris SM. Arginine metabolism: nitric oxide and beyond. Biochem J. 1998;336(Pt 1):1–17. doi: 10.1042/bj3360001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reeds PJ. Dispensable and indispensable amino acids for humans. J. Nutr. 2000;130:1835S–40S. doi: 10.1093/jn/130.7.1835S. [DOI] [PubMed] [Google Scholar]
- 3.Richieri GV, Kleinfeld AM. Unbound free fatty acid levels in human serum. The Journal of Lipid Research. 1995;36:229–240. [PubMed] [Google Scholar]
- 4.Itoh Y, et al. Free fatty acids regulate insulin secretion from pancreatic beta cells through GPR40. Nature. 2003;422:173–176. doi: 10.1038/nature01478. [DOI] [PubMed] [Google Scholar]
- 5.Hirasawa A, et al. Free fatty acids regulate gut incretin glucagon-like peptide-1 secretion through GPR120. Nat Med. 2005;11:90–94. doi: 10.1038/nm1168. [DOI] [PubMed] [Google Scholar]
- 6.Oh DY, et al. GPR120 is an omega-3 fatty acid receptor mediating potent anti-inflammatory and insulin-sensitizing effects. Cell. 2010;142:687–698. doi: 10.1016/j.cell.2010.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ichimura A, et al. Dysfunction of lipid sensor GPR120 leads to obesity in both mouse and human. Nat Med. 2012;483:350–354. doi: 10.1038/nature10798. [DOI] [PubMed] [Google Scholar]
- 8.Oh DY, et al. A Gpr120-selective agonist improves insulin resistance and chronic inflammation in obese mice. Nat Med. 2014;20:942–947. doi: 10.1038/nm.3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pepino MY, Kuda O, Samovski D, Abumrad NA. Structure-function of CD36 and importance of fatty acid signal transduction in fat metabolism. Annu. Rev. Nutr. 2014;34:281–303. doi: 10.1146/annurev-nutr-071812-161220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laugerette F, et al. CD36 involvement in orosensory detection of dietary lipids, spontaneous fat preference, and digestive secretions. J Clin Invest. 2005;115:3177–3184. doi: 10.1172/JCI25299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cartoni C, et al. Taste preference for fatty acids is mediated by GPR40 and GPR120. Journal of Neuroscience. 2010;30:8376–8382. doi: 10.1523/JNEUROSCI.0496-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin C, et al. The lipid-sensor candidates CD36 and GPR120 are differentially regulated by dietary lipids in mouse taste buds: impact on spontaneous fat preference. PLoS ONE. 2011;6:e24014. doi: 10.1371/journal.pone.0024014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pepino MY, Love-Gregory L, Klein S, Abumrad NA. The fatty acid translocase gene CD36 and lingual lipase influence oral sensitivity to fat in obese subjects. The Journal of Lipid Research. 2012;53:561–566. doi: 10.1194/jlr.M021873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown MS, Goldstein JL. A receptor-mediated pathway for cholesterol homeostasis. Science. 1986;232:34–47. doi: 10.1126/science.3513311. [DOI] [PubMed] [Google Scholar]
- 15.Brown AJ, Sun L, Feramisco JD, Brown MS, Goldstein JL. Cholesterol addition to ER membranes alters conformation of SCAP, the SREBP escort protein that regulates cholesterol metabolism. Mol Cell. 2002;10:237–245. doi: 10.1016/s1097-2765(02)00591-9. This paper demonstrates the functional regulation of the SCAP protein conformation by cholesterol levels within ER membrane, providing strong support to its cholesterol sensing ability.
- 16.Radhakrishnan A, Sun L-P, Kwon HJ, Brown MS, Goldstein JL. Direct binding of cholesterol to the purified membrane region of SCAP: mechanism for a sterol-sensing domain. Mol Cell. 2004;15:259–268. doi: 10.1016/j.molcel.2004.06.019. [DOI] [PubMed] [Google Scholar]
- 17.Feramisco JD, et al. Intramembrane aspartic acid in SCAP protein governs cholesterol-induced conformational change. Proc Natl Acad Sci USA. 2005;102:3242–3247. doi: 10.1073/pnas.0500206102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang T, et al. Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell. 2002;110:489–500. doi: 10.1016/s0092-8674(02)00872-3. [DOI] [PubMed] [Google Scholar]
- 19.Radhakrishnan A, Goldstein JL, McDonald JG, Brown MS. Switch-like control of SREBP-2 transport triggered by small changes in ER cholesterol: a delicate balance. Cell Metab. 2008;8:512–521. doi: 10.1016/j.cmet.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Motamed M, et al. Identification of luminal Loop 1 of Scap protein as the sterol sensor that maintains cholesterol homeostasis. J Biol Chem. 2011;286:18002–18012. doi: 10.1074/jbc.M111.238311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, Motamed M, Seemann J, Brown MS, Goldstein JL. Point mutation in luminal loop 7 of Scap protein blocks interaction with loop 1 and abolishes movement to Golgi. J Biol Chem. 2013;288:14059–14067. doi: 10.1074/jbc.M113.469528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeon T-I, Osborne TF. SREBPs: metabolic integrators in physiology and metabolism. Trends Endocrinol. Metab. 2012;23:65–72. doi: 10.1016/j.tem.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sever N, Yang T, Brown MS, Goldstein JL, DeBose-Boyd RA. Accelerated degradation of HMG CoA reductase mediated by binding of insig-1 to its sterol-sensing domain. Mol Cell. 2003;11:25–33. doi: 10.1016/s1097-2765(02)00822-5. [DOI] [PubMed] [Google Scholar]
- 24.Song B-L, Sever N, DeBose-Boyd RA. Gp78, a membrane-anchored ubiquitin ligase, associates with Insig-1 and couples sterol-regulated ubiquitination to degradation of HMG CoA reductase. Mol Cell. 2005;19:829–840. doi: 10.1016/j.molcel.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 25.Birsoy K, et al. Cellular program controlling the recovery of adipose tissue mass: An in vivo imaging approach. Proc Natl Acad Sci USA. 2008;105:12985–12990. doi: 10.1073/pnas.0805621105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wrann CD, et al. FOSL2 promotes leptin gene expression in human and mouse adipocytes. J Clin Invest. 2012;122:1010–1021. doi: 10.1172/JCI58431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clément K, et al. A mutation in the human leptin receptor gene causes obesity and pituitary dysfunction. Nature. 1998;392:398–401. doi: 10.1038/32911. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y, et al. Positional cloning of the mouse obese gene and its human homologue. Nature. 1994;372:425–432. doi: 10.1038/372425a0. In this seminal paper, mouse Ob gene and its human homolog Leptin are identified.
- 29.Lee GH, et al. Abnormal splicing of the leptin receptor in diabetic mice. Nature. 1996;379:632–635. doi: 10.1038/379632a0. [DOI] [PubMed] [Google Scholar]
- 30.Scherer PE, Williams S, Fogliano M, Baldini G, Lodish HF. A novel serum protein similar to C1q, produced exclusively in adipocytes. J Biol Chem. 1995;270:26746–26749. doi: 10.1074/jbc.270.45.26746. [DOI] [PubMed] [Google Scholar]
- 31.Hu E, Liang P, Spiegelman BM. AdipoQ is a novel adipose-specific gene dysregulated in obesity. J Biol Chem. 1996;271:10697–10703. doi: 10.1074/jbc.271.18.10697. [DOI] [PubMed] [Google Scholar]
- 32.Shehzad A, Iqbal W, Shehzad O, Lee YS. Adiponectin: regulation of its production and its role in human diseases. Hormones (Athens) 2012;11:8–20. doi: 10.1007/BF03401534. [DOI] [PubMed] [Google Scholar]
- 33.Maeda N, et al. Diet-induced insulin resistance in mice lacking adiponectin/ACRP30. Nat Med. 2002;8:731–737. doi: 10.1038/nm724. [DOI] [PubMed] [Google Scholar]
- 34.Kadowaki T, et al. Adiponectin and adiponectin receptors in insulin resistance, diabetes, and the metabolic syndrome. J Clin Invest. 2006;116:1784–1792. doi: 10.1172/JCI29126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Waki H, Tontonoz P. Endocrine functions of adipose tissue. Annu Rev Pathol. 2007;2:31–56. doi: 10.1146/annurev.pathol.2.010506.091859. [DOI] [PubMed] [Google Scholar]
- 36.Takahashi M, et al. Genomic structure and mutations in adipose-specific gene, adiponectin. Int J Obes Relat Metab Disord. 2000;24:861–868. doi: 10.1038/sj.ijo.0801244. [DOI] [PubMed] [Google Scholar]
- 37.Hara K, et al. Genetic variation in the gene encoding adiponectin is associated with an increased risk of type 2 diabetes in the Japanese population. Diabetes. 2002;51:536–540. doi: 10.2337/diabetes.51.2.536. [DOI] [PubMed] [Google Scholar]
- 38.Kondo H, et al. Association of adiponectin mutation with type 2 diabetes: a candidate gene for the insulin resistance syndrome. Diabetes. 2002;51:2325–2328. doi: 10.2337/diabetes.51.7.2325. [DOI] [PubMed] [Google Scholar]
- 39.Ibba M, Soll D. Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 40.Dong J, Qiu H, Garcia-Barrio M, Anderson J, Hinnebusch AG. Uncharged tRNA activates GCN2 by displacing the protein kinase moiety from a bipartite tRNA-binding domain. Mol Cell. 2000;6:269–279. doi: 10.1016/s1097-2765(00)00028-9. [DOI] [PubMed] [Google Scholar]
- 41.Berlanga JJ, Santoyo J, De Haro C. Characterization of a mammalian homolog of the GCN2 eukaryotic initiation factor 2alpha kinase. Eur J Biochem. 1999;265:754–762. doi: 10.1046/j.1432-1327.1999.00780.x. [DOI] [PubMed] [Google Scholar]
- 42.Scheuner D, et al. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol Cell. 2001;7:1165–1176. doi: 10.1016/s1097-2765(01)00265-9. [DOI] [PubMed] [Google Scholar]
- 43.Zhang P, et al. The GCN2 eIF2alpha kinase is required for adaptation to amino acid deprivation in mice. Mol Cell Biol. 2002;22:6681–6688. doi: 10.1128/MCB.22.19.6681-6688.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maurin A-C, et al. The GCN2 kinase biases feeding behavior to maintain amino acid homeostasis in omnivores. Cell Metab. 2005;1:273–277. doi: 10.1016/j.cmet.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 45.Hao S, et al. Uncharged tRNA and sensing of amino acid deficiency in mammalian piriform cortex. Science. 2005;307:1776–1778. doi: 10.1126/science.1104882. [DOI] [PubMed] [Google Scholar]
- 46.Guo F, Cavener DR. The GCN2 eIF2alpha kinase regulates fatty-acid homeostasis in the liver during deprivation of an essential amino acid. Cell Metab. 2007;5:103–114. doi: 10.1016/j.cmet.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 47.Thoreen CC, et al. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485:109–113. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garami A, et al. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol Cell. 2003;11:1457–1466. doi: 10.1016/s1097-2765(03)00220-x. [DOI] [PubMed] [Google Scholar]
- 50.Inoki K, Li Y, Xu T, Guan K-L. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 2003;17:1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tee AR, Manning BD, Roux PP, Cantley LC, Blenis J. Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr Biol. 2003;13:1259–1268. doi: 10.1016/s0960-9822(03)00506-2. [DOI] [PubMed] [Google Scholar]
- 52.Zhang Y, et al. Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat Cell Biol. 2003;5:578–581. doi: 10.1038/ncb999. [DOI] [PubMed] [Google Scholar]
- 53.Hara K, et al. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J Biol Chem. 1998;273:14484–14494. doi: 10.1074/jbc.273.23.14484. Explores the amino acid essentiality for mTORC1 activation and specific amino acid requirements, independent of the growth factor mediated regulation of activity.
- 54.Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan K-L. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10:935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sancak Y, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. 50 and 51 report the Identification of the Rag GTPases as the direct link of amino acids levels and mTORC1, regulating its subcellular localization.
- 56.Sancak Y, et al. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bar-Peled L, Schweitzer LD, Zoncu R, Sabatini DM. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell. 2012;150:1196–1208. doi: 10.1016/j.cell.2012.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bar-Peled L, et al. A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science. 2013;340:1100–1106. doi: 10.1126/science.1232044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Panchaud N, Péli-Gulli M-P, De Virgilio C. Amino acid deprivation inhibits TORC1 through a GTPase-activating protein complex for the Rag family GTPase Gtr1. Sci Signal. 2013;6:ra42. doi: 10.1126/scisignal.2004112. [DOI] [PubMed] [Google Scholar]
- 60.Tsun Z-Y, et al. The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol Cell. 2013;52:495–505. doi: 10.1016/j.molcel.2013.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Petit CS, Roczniak-Ferguson A, Ferguson SM. Recruitment of folliculin to lysosomes supports the amino acid-dependent activation of Rag GTPases. J Cell Biol. 2013;202:1107–1122. doi: 10.1083/jcb.201307084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chantranupong L, et al. The Sestrins Interact with GATOR2 to Negatively Regulate the Amino-Acid-Sensing Pathway Upstream of mTORC1. Cell Rep. 2014 doi: 10.1016/j.celrep.2014.09.014. doi:10.1016/j.celrep.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Peng M, Yin N, Li MO. Sestrins Function as Guanine Nucleotide Dissociation Inhibitors for Rag GTPases to Control mTORC1 Signaling. Cell. 2014;159:122–133. doi: 10.1016/j.cell.2014.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kitamoto K, Yoshizawa K, Ohsumi Y, Anraku Y. Dynamic aspects of vacuolar and cytosolic amino acid pools of Saccharomyces cerevisiae. J. Bacteriol. 1988;170:2683–2686. doi: 10.1128/jb.170.6.2683-2686.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Binda M, et al. The Vam6 GEF controls TORC1 by activating the EGO complex. Mol Cell. 2009;35:563–573. doi: 10.1016/j.molcel.2009.06.033. [DOI] [PubMed] [Google Scholar]
- 66.Harms E, Gochman N, Schneider JA. Lysosomal pool of free-amino acids. Biochemical and Biophysical Research Communications. 1981;99:830–836. doi: 10.1016/0006-291x(81)91239-0. [DOI] [PubMed] [Google Scholar]
- 67.Neuhaus EM, Almers W, Soldati T. Morphology and dynamics of the endocytic pathway in Dictyostelium discoideum. Molecular Biology of the Cell. 2002;13:1390–1407. doi: 10.1091/mbc.01-08-0392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee JH, et al. De novo somatic mutations in components of the PI3K-AKT3-mTOR pathway cause hemimegalencephaly. Nat Genet. 2012;44:941–945. doi: 10.1038/ng.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bohn G, et al. A novel human primary immunodeficiency syndrome caused by deficiency of the endosomal adaptor protein p14. Nat Med. 2007;13:38–45. doi: 10.1038/nm1528. [DOI] [PubMed] [Google Scholar]
- 70.Efeyan A, Zoncu R, Sabatini DM. Amino acids and mTORC1: from lysosomes to disease. Trends Mol Med. 2012;18:524–533. doi: 10.1016/j.molmed.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bachmanov AA, Beauchamp GK. Taste receptor genes. Annu. Rev. Nutr. 2007;27:389–414. doi: 10.1146/annurev.nutr.26.061505.111329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Damak S, et al. Detection of sweet and umami taste in the absence of taste receptor T1r3. Science. 2003;301:850–853. doi: 10.1126/science.1087155. [DOI] [PubMed] [Google Scholar]
- 73.Nelson G, et al. An amino-acid taste receptor. Nature. 2002;416:199–202. doi: 10.1038/nature726. [DOI] [PubMed] [Google Scholar]
- 74.Chaudhari N, Roper SD. The cell biology of taste. J Cell Biol. 2010;190:285–296. doi: 10.1083/jcb.201003144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wu SV, et al. Expression of bitter taste receptors of the T2R family in the gastrointestinal tract and enteroendocrine STC-1 cells. Proc Natl Acad Sci USA. 2002;99:2392–2397. doi: 10.1073/pnas.042617699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wauson EM, et al. The G Protein-Coupled Taste Receptor T1R1/T1R3 Regulates mTORC1 and Autophagy. Mol Cell. 2012 doi: 10.1016/j.molcel.2012.08.001. doi:10.1016/j.molcel.2012.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Printz RL, Magnuson MA, Granner DK. Mammalian glucokinase. Annu. Rev. Nutr. 1993;13:463–496. doi: 10.1146/annurev.nu.13.070193.002335. [DOI] [PubMed] [Google Scholar]
- 78.Nordlie RC, Foster JD, Lange AJ. Regulation of glucose production by the liver. Annu. Rev. Nutr. 1999;19:379–406. doi: 10.1146/annurev.nutr.19.1.379. [DOI] [PubMed] [Google Scholar]
- 79.Ogunnowo-Bada EO, Heeley N, Brochard L, Evans ML. Brain glucose sensing, glucokinase and neural control of metabolism and islet function. Diabetes Obes Metab. 2014;16(Suppl 1):26–32. doi: 10.1111/dom.12334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gloyn AL. Glucokinase (GCK) mutations in hyper- and hypoglycemia: maturity-onset diabetes of the young, permanent neonatal diabetes, and hyperinsulinemia of infancy. Hum. Mutat. 2003;22:353–362. doi: 10.1002/humu.10277. [DOI] [PubMed] [Google Scholar]
- 81.Postic C, et al. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. J Biol Chem. 1999;274:305–315. doi: 10.1074/jbc.274.1.305. [DOI] [PubMed] [Google Scholar]
- 82.Thorens B, Mueckler M. Glucose transporters in the 21st Century. AJP: Endocrinology and Metabolism. 2010;298:E141–5. doi: 10.1152/ajpendo.00712.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Santer R, et al. Mutations in GLUT2, the gene for the liver-type glucose transporter, in patients with Fanconi-Bickel syndrome. Nat Genet. 1997;17:324–326. doi: 10.1038/ng1197-324. [DOI] [PubMed] [Google Scholar]
- 84.De Vos A, et al. Human and rat beta cells differ in glucose transporter but not in glucokinase gene expression. J Clin Invest. 1995;96:2489–2495. doi: 10.1172/JCI118308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chang-Chen KJ, Mullur R, Bernal-Mizrachi E. Beta-cell failure as a complication of diabetes. Rev Endocr Metab Disord. 2008;9:329–343. doi: 10.1007/s11154-008-9101-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Leto D, Saltiel AR. Regulation of glucose transport by insulin: traffic control of GLUT4. Nat Rev Mol Cell Biol. 2012;13:383–396. doi: 10.1038/nrm3351. [DOI] [PubMed] [Google Scholar]
- 87.Hardie DG. AMP-activated protein kinase--an energy sensor that regulates all aspects of cell function. Genes Dev. 2011;25:1895–1908. doi: 10.1101/gad.17420111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol. 2012;13:251–262. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Efeyan A, et al. Regulation of mTORC1 by the Rag GTPases is necessary for neonatal autophagy and survival. Nature. 2013;493:679–683. doi: 10.1038/nature11745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zoncu R, et al. mTORC1 Senses Lysosomal Amino Acids Through an Inside-Out Mechanism That Requires the Vacuolar H+-ATPase. Science. 2011;334:678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang F, et al. Molecular mechanism for the umami taste synergism. Proc Natl Acad Sci USA. 2008;105:20930–20934. doi: 10.1073/pnas.0810174106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nelson G, et al. Mammalian sweet taste receptors. Cell. 2001;106:381–390. doi: 10.1016/s0092-8674(01)00451-2. Identification of the T1R2 + T1R3 as the sweet taste receptor by means of mouse transgenesis and heterologous expression in cultured cells.
- 93.Mace OJ, Affleck J, Patel N, Kellett GL. Sweet taste receptors in rat small intestine stimulate glucose absorption through apical GLUT2. The Journal of Physiology. 2007;582:379–392. doi: 10.1113/jphysiol.2007.130906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dyer J, Salmon KSH, Zibrik L, Shirazi-Beechey SP. Expression of sweet taste receptors of the T1R family in the intestinal tract and enteroendocrine cells. Biochem Soc Trans. 2005;33:302–305. doi: 10.1042/BST0330302. [DOI] [PubMed] [Google Scholar]
- 95.Nettleton JA, et al. Diet soda intake and risk of incident metabolic syndrome and type 2 diabetes in the Multi-Ethnic Study of Atherosclerosis (MESA) Diabetes Care. 2009;32:688–694. doi: 10.2337/dc08-1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Klionsky DJ. Autophagy as a Regulated Pathway of Cellular Degradation. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Egan DF, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kim J, Kundu M, Viollet B, Guan K-L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mammucari C, et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 100.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Thoreen CC, et al. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem. 2009;284:8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kuma A, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. Shows the essentiality of autophagy as a critical mechanism to mobilize internal energy stores and adapt to the interruption of trans-placental nutrient supply in neonates.
- 103.Komatsu M, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yu L, et al. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature. 2010;465:942–946. doi: 10.1038/nature09076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Naito T, Kuma A, Mizushima N. Differential contribution of insulin and amino acids to the mTORC1-autophagy pathway in the liver and muscle. Journal of Biological Chemistry. 2013 doi: 10.1074/jbc.M113.456228. doi:10.1074/jbc.M113.456228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kroemer G, Mariño G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tsukamoto S, et al. Autophagy is essential for preimplantation development of mouse embryos. Science. 2008;321:117–120. doi: 10.1126/science.1154822. [DOI] [PubMed] [Google Scholar]
- 108.Komatsu M, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 109.Karsli-Uzunbas G, et al. Autophagy is required for glucose homeostasis and lung tumor maintenance. Cancer Discovery. 2014;4:914–927. doi: 10.1158/2159-8290.CD-14-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Singh R, et al. Autophagy regulates lipid metabolism. Nature. 2009 doi: 10.1038/nature07976. doi:10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 112.de Cabo R, Carmona-Gutierrez D, Bernier M, Hall MN, Madeo F. The search for antiaging interventions: from elixirs to fasting regimens. Cell. 2014;157:1515–1526. doi: 10.1016/j.cell.2014.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]