Summary
HAP2, a male gamete-specific protein conserved across vast evolutionary distances has garnered considerable attention as a potential membrane fusogen required for fertilization in taxa ranging from protozoa and green algae to flowering plants and invertebrate animals [1–6]. However, its presence in Tetrahymena thermophilaa ciliated protozoan with seven sexes or mating types that bypasses the production of male gametes raises interesting questions regarding the evolutionary origins of gamete-specific functions in sexually dimorphic species. Here we show that HAP2 is expressed in all seven mating types of T. thermophila and that fertility is only blocked when the gene is deleted from both cells of a mating pair. HAP2 deletion strains of complementary mating types can recognize one another and form pairs, however pair stability is compromised and membrane pore formation at the nuclear exchange junction is blocked. The absence of pore formation is consistent with previous studies suggesting a role for HAP2 in gamete fusion in other systems. We propose a model in which each of the several hundred membrane pores established at the conjugation junction of mating Tetrahymena represents the equivalent of a male/female interface, and that pore formation is driven on both sides of the junction by the presence of HAP2. Such a model supports the idea that many of the disparate functions of sperm and egg were shared by the “isogametes” of early eukaryotes, and became partitioned to either male or female sex cells later in evolution.
Results
Genetic evidence for HAP2 function in multiple mating types
Tetrahymena thermophila has seven sexes or mating types, any of which can mate with the other six, but not with itself [7, 8]. Cells become competent to mate following nutritional starvation. When mixed together, starved cells of complementary mating types undergo a period of morphological transformation, producing a region of smooth, deciliated membrane at their anterior ends where they subsequently pair [9–11]. Approximately 1hr after mixing (30°C), cells begin to form loose associations that are easily disrupted. By 2hrs, these loose associations give way to tight pairing, and a series of meiotic events ensues in which two haploid pronuclei, one stationary, and one migratory, are generated in each cell of the mating pair (diagrammed in Figure S1). The transformation of a loose adhesion zone into a mature, mechanically robust mating junction coincides with the appearance of 0.1–0.2 µm diameter intercellular pores that form as a result of several hundred independent membrane fusion events. Over time these pores expand up to 10X their initial diameter allowing the exchange of migratory pronuclei [11,12]. Following exchange, migratory and stationary pronuclei fuse, membrane integrity is restored, and cells disengage to complete the program of conjugal development.
The macronuclear genome sequence of T. thermophila predicts a single homolog encoding HAP2, a male gamete-specific protein thought to play a role in sperm/egg fusion in a wide range of species [1–6]. To confirm this, we obtained a full-length cDNA and deduced amino acid sequence for the protein (accession number: KJ629172). Importantly, analysis of the Tetrahymena Functional Genomics database indicated that the HAP2 gene is highly up-regulated when cells of complementary mating types are mixed [13]. With the idea that HAP2 is required for membrane pore formation in conjugating Tetrahymenawe initiated experiments to examine the effect of HAP2 deletion on mating success as defined by the completion of all the normal events of conjugation including cross fertilization and new macronucleus development. Initially, we found that deletion of the HAP2 gene from the vegetative macronucleus of T. thermophila mating type VII (strain ΔHAP2-428) had no effect on progeny development when crossed with wild type cells of mating type II (SB1969; data not shown). One interpretation of this result was that HAP2 is dispensable for mating in Tetrahymena. Alternatively, HAP2 expression could be mating-type specific (and confined to mating types other than VII), or ubiquitous in all mating types but required on only one side of a pair to allow membrane fusion and pronuclear exchange to occur. To explore this further, we performed additional crosses using HAP2 deletion strains (ΔHAP2) carrying different drug selectable markers in their germline micronuclei. Reciprocal matings were conducted between {wild type X ΔHAP2} cells, {ΔHAP2 X ΔHAP2} cells, and control {wild type X wild type} cells of different mating types, with the parents of each cross containing either cycloheximide (Cy) or 6-methyl purine (6-MP) resistance markers in their micronuclei. In such crosses, fertilization success was scored as the percentage of mating pairs that gave rise to progeny resistant to both drugs (see Figure S2). The results are shown in Table 1. Whereas reciprocal crosses between knockout cell lines and cells carrying the wild type HAP2 gene were clearly fertile (giving rise to double drug resistant progeny), deletion of HAP2 from both mating types completely blocked the appearance of cross-fertilized progeny. To test the penetrance of the infertility phenotype, we performed mass matings followed by sequential drug challenge. From mating cultures of {ΔHAP2×ΔHAP2} cell lines involving over 106 mating pairs, no double drug resistant progeny were produced. Insertion of either the wild type gene, or an HA-tagged version of the full-length HAP2 cDNA into the endogenous HAP2 locus of both cells of a {ΔHAP2 X ΔHAP2} cross restored fertility, demonstrating the necessity of HAP2 in cross-fertilization and mating success (Table 1).
Table 1.
Effects of HAP2 deletion on mating success.
| Mating | % Survival | % “Cross- fertilizers” R/R |
% “Self- fertilizers” R/S or S/R |
% “Back- outs” S/S |
|---|---|---|---|---|
| CU427.4 X CU428.2 | 96 404/423 |
90 364/404 |
2 9/404 |
8 31/404 |
| ΔHAP2a X CU427.4 | 91 514/564 |
17 86/514 |
12 63/514 |
71 365/514 |
| ΔHAP2b X CU428.2 | 91 774/846 |
28 217/774 |
7 55/774 |
65 502/774 |
| ΔHAP2a X ΔHAP2b | 96 226/235 |
0 0/226 |
4 9/226 |
96 217/226 |
| Genomic Rescue Cross | 86 284/329 |
27 77/284 |
NA | NA |
| HAP2-HA cDNA Rescue Cross | 66 217/329 |
25 54/217 |
NA | NA |
Reciprocal crosses (X) between wild type and HAP2 knockout (∆HAP2) strains were carried out in various combinations. ΔHAP2a and ΔHAP2b are, respectively, CU428.2 and CU427.4 cell lines in which the HAP2 gene has been completely replaced by a neomycin resistance cassette. For each cross, pairs were individually isolated into hanging drops and allowed to establish "synclones" (mixed clonal descendants from both exconjugants), which were characterized phenotypically as explained in detail in Figure S2. Three types of progeny synclones were phenotypically distinguished: those that successfully completed all steps of conjugation and developed a new MAC (Cross-fertilizers; R/R); those that were blocked in gamete pronucleus exchange but proceeded normally otherwise (Self-fertilizers; R/S or S/R), and those that failed to complete development of new MACs and retained their parental MAC (Back-outs; S/S). % Survival is the percent of isolated pairs that generated living cultures. The percentages of the three classes of progeny are shown in the remaining columns. The data presented for each cross are the pooled results from two-to-four independent pair isolation experiments. The actual number of pairs representing a particular outcome out of the total number of surviving pairs is shown below each percentage. Among progeny that developed a new MAC, the fraction of true progeny (i.e. cross-fertilizers) was directly correlated with the number of wild type HAP2 parents in the cross (2, 1, or 0), and the differences were highly significant statistically (c2 (6, N = 1918) = 781.7, p <0.0001). Rescue strains are cell lines in which HAP2 gene knockouts were replaced in the MAC either with the wild type copy or the cDNA version of the gene, in both parents of the cross (see Supplemental Experimental Procedures; Fig. S5). Because of the drug phenotype of the rescue strains, “Self-fertilizers” and “Back-outs” were not distinguished from one another (shown as NA, or “not applicable“).
Expression of HAP2 in all seven mating types of T. thermophila
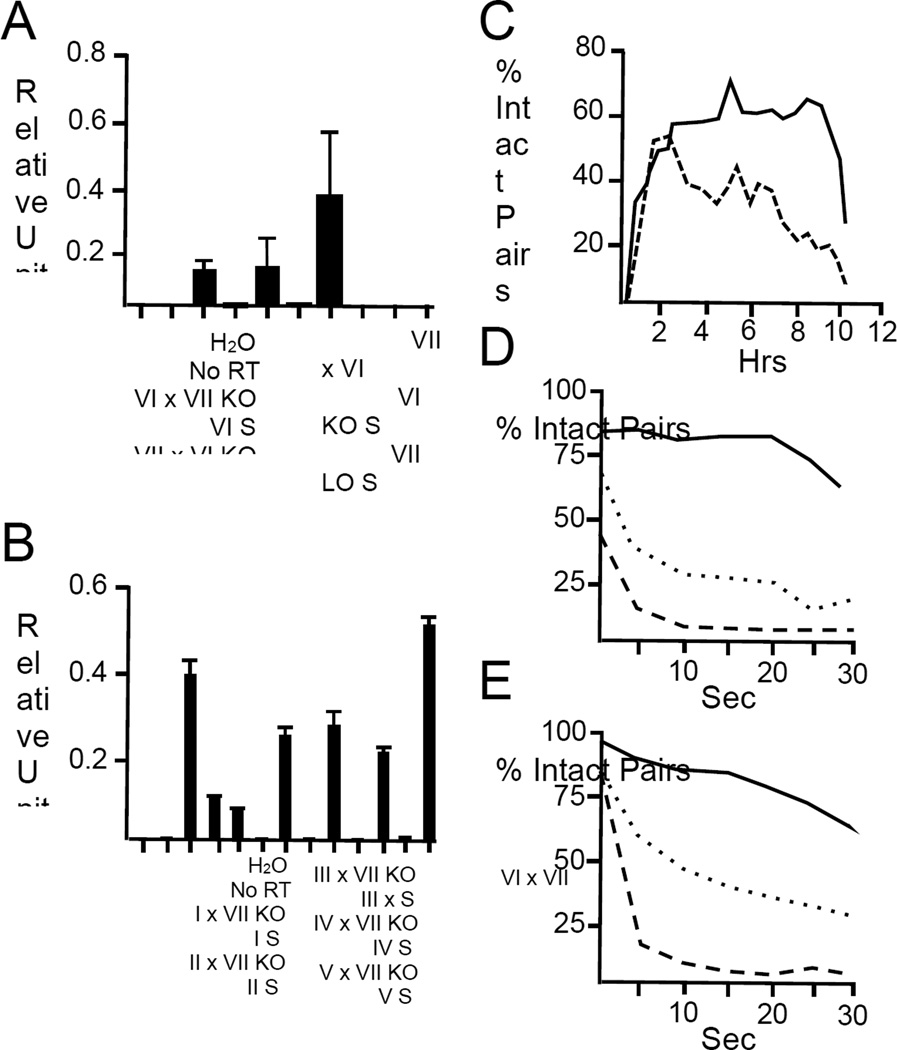
Taken together, the results described above indicate that HAP2 is functional in both members of a mating pair regardless of mating type. To test this further we conducted quantitative real-time PCR experiments to determine HAP2 mRNA levels and found that the gene was indeed expressed in reciprocal {wild type×ΔHAP2} crosses of mating types VI and VII, regardless of which mating type carried the wild type allele. In such crosses, HAP2 transcript levels were roughly half that in control {wild type×wild type} crosses (Figure 1A). Furthermore, in crosses between the ΔHAP2 knockout in mating type VII and wild type cells from all other mating types (i.e., I-V), HAP2 mRNA was always detected verifying that expression is not confined to any specific mating type. All seven sexes of T. thermophila express the HAP2 gene during early conjugal development (Figure 1B).
Figure 1. HAP2 transcript levels and pair stability in wild type and ΔHAP2 crosses.
Panel (A) shows HAP2 mRNA transcript levels in control and reciprocal {wild type X ΔHAP2} and {ΔHAP2 X ΔHAP2} crosses as determined by quantitative real-time PCR. {Wild type X ΔHAP2} crosses showed roughly half the level of expression seen in wild type control matings, while {ΔHAP2 X ΔHAP2} crosses showed no detectable HAP2 expression. Knockout (KO) strains used in this study were ΔHAP2-428 (mating type VII; Supplemental Experimental Procedures) and ΔHAP2-427 (mating type VI; Supplemental Experimental Procedures). Bars indicate the average expression levels +/− SEM (n=3). Panel (B) shows relative HAP2 transcript levels in crosses between mating type VII knockout strain, ΔHAP2-428, and wild type strains of mating types I-V (strains SB3539; B2086.2; SB281; CU438.1; C3 368.1, respectively; Supplemental Experimental Procedures). HAP2 mRNA expression in a wild type cross between mating types VII and VI (CU428.2 and CU427.4) is shown at the extreme right. Bars represent mean and standard errors for technical triplicates of cDNA samples from either mating (X) or starvation (S) cultures. Panels (C-E) show the relative stability of mating pairs in crosses between {wild type X wild type} (—); {wild type X ΔHAP2} (……); and, {ΔHAP2 X ΔHAP2} (-----) cells. In Panel C, wild type cells (CU428.2 and CU427.4), or ΔHAP2 KO strains (ΔHAP2-428 and ΔHAP2-427) were starved, mixed together at time zero, and measured for pair frequency at 30 min intervals thereafter. In Panels D-E, cells were mixed and then physically disrupted using a Vortex Genie at 2–2.5 hrs (Panel D), or 4–4.5 hrs (Panel E) after mixing. Cells were carefully withdrawn from culture vessels and gently placed on microscope slides before visualization under bright field. For each time point, 100 subjects were counted 3 separate times and the percent of intact pairs averaged for each time point. Each experiment took 30 minutes to conduct. A diagram depicting the cellular events associated with conjugation of wild type cells at different time points is shown in Figure S1.
Requirement for HAP2 in pair stability
At the light microscope level, deletion of HAP2 had little or no effect on the ability of complementary mating types to recognize one another and form pairs. Nevertheless, when left undisturbed, {ΔHAP2 X ΔHAP2} pairs came apart more rapidly than {wild type X wild type} pairs (Figure 1C). To quantify pair stability, we exposed mating cells to varying periods of mechanical agitation and counted the ratio of intact pairs to single cells in the culture. Figure 1D shows that wild type pairs were relatively insensitive to vortexing, while {ΔHAP2 X ΔHAP2} partners were highly sensitive, with over 90% of pairs separating after 5 seconds of agitation. We tested pairs from both early (2 hrs) and mid-stage matings (4 hrs), and obtained similar results, indicating that the loss of stability of {ΔHAP2 X ΔHAP2} mating pairs at 2 hrs was not simply the result of delayed development. Interestingly, {wild type X ΔHAP2} partners showed an intermediate level of sensitivity to vortexing (Figure 1E), which correlates with HAP2 expression at the transcript level.
HAP2 is required for membrane pore formation
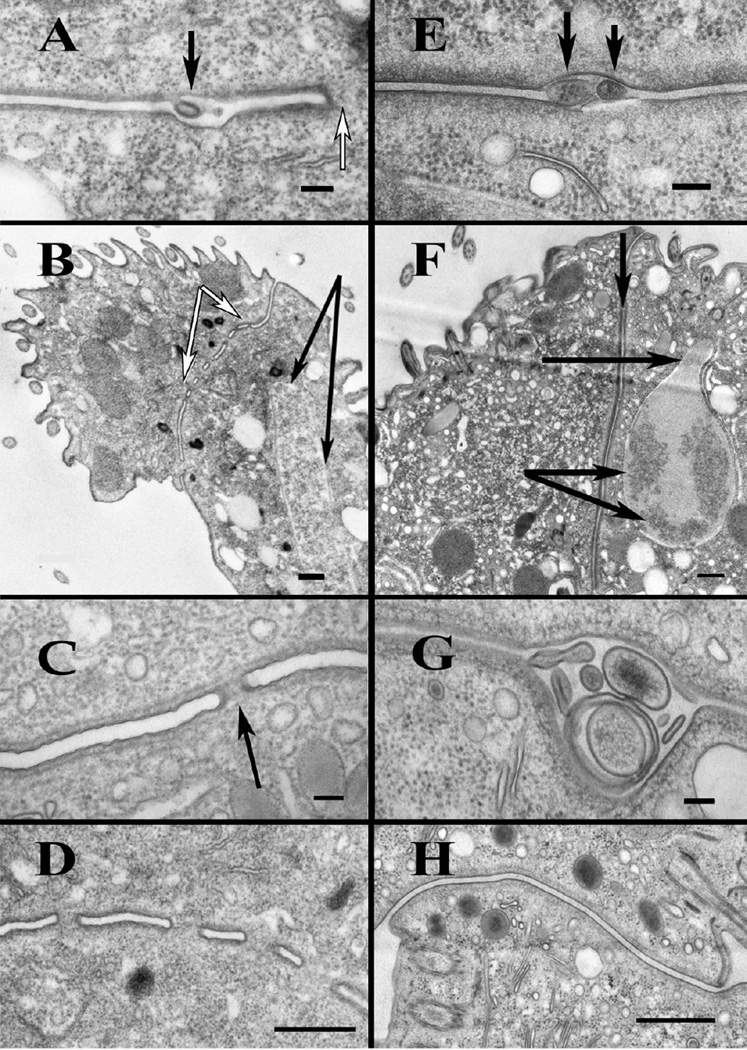
Bilateral exchange of gametic pronuclei in T. thermophila has been linked to the formation of intercellular pores at the nuclear exchange junction [12, 14]. Using transmission electron microscopy, we found that HAP2 was necessary for the formation of intercellular pores at this site, and consequently exchange of gametic pronuclei between mating cells. As shown in Figure 2A and E, respectively, {wild type×wild type} and {ΔHAP2 X ΔHAP2} mating pairs exhibited smooth adhesion zones between closely apposed plasma membranes, along with membrane outpocketings extending from both partners into the intercellular cleft between cells beginning with the loose-pairing interval at 1–1.5 hrs post-mixing. In the wild type pairs, these membrane protrusions have been shown to fuse with the plasma membrane of partnering cells (Cole, unpublished), and approximately 2 hrs after cell mixing, pores could be observed at numerous loci within the adhesion zone (Figure 2A-D). By contrast, pores were never seen in {ΔHAP2 X ΔHAP2} pairs examined at 2hrs (Figure 2E-H), or at later time points even when meiotic nuclear configurations were seen at the mating junction (Figure 2F). The presence of large, seemingly hypertrophied membrane vesicles within the junctional lumens of {ΔHAP2 X ΔHAP2} pairs were likely a consequence of the inability of membrane protrusions to fuse and form pores (Figure 2G). A proposed model for the events at the mating junction of {ΔHAP2 X ΔHAP2} and {wild type×wild type} crosses is shown in Figure S3.
Figure 2. Ultrastructure of the nuclear exchange junction.
A-D) TEM images of wild type nuclear exchange junctions. E-H) TEM images of {ΔHAP2 X ΔHAP2} mating junctions. A) Wild type junction showing a membrane tubule (black arrow) protruding into the extracellular space 2 hrs into mating. White arrow indicates adjacent pore. Scale bar = 100 nm. B) A low magnification view of a wild-type junction 4 hrs into mating, showing multiple, complete junction pores (region bracketed by white arrows). Black arrows indicate meiotic nucleus in the extended "crescent" (or prophase I) configuration. This illustrates that, in matings of wild type cells, pores are complete by the onset of Meiosis I. Scale bar = 500 nm. C) Wild type junction (2 hrs into mating), showing a typical junction pore (arrow). Scale bar = 100 nm. D) Image of wild-type (4 hr) junction showing multiple complete fusion pores. (Scale bar = 500 nm). E) {ΔHAP2 X ΔHAP2} junction at 2hrs showing protrusions of plasma membrane into extracellular space (arrows). Scale bar = 100 nm. F) Low magnification image of a {ΔHAP2 X ΔHAP2} pair during late Meiosis I (~4hrs) showing the complete junction devoid of pores. Lower arrows indicate condensed nuclear chromatin. The upper horizontal arrow indicates the "neck" of the intra-nuclear meiotic spindle full of microtubules. This pair was at late anaphase or early telophase of Meiosis I. Vertical arrow at the top, indicates the exchange junction, devoid of pores. G) {ΔHAP2 X ΔHAP2} pair at 2 hrs showing a region of the junction with swollen extracellular vesicles collecting in the junction cleft. Also note absence of pores. H) Complete {ΔHAP2 X ΔHAP2} exchange junction at 2 hrs showing complete absence of fusion pores. Scale bar = 500 nm. A diagram of membrane events at the junctions of wild type and ΔHAP2 crosses is shown in Figure S3.
HAP2 localizes to the nuclear exchange junction
Consistent with a role for HAP2 in membrane pore formation, a GFP-tagged version of the HAP2 protein localized predominantly to the interface between conjugating cells decorating the entire mating junction, excluding established pores, (Figure 3A-C; Movie S1). Immunoelectron microscopy of freeze-substituted material showed some cytoplasmic labeling of the GFP-tagged protein (consistent with a site of synthesis within the endomembrane system), as well as localization to the extracellular space between fusion pores at the conjugation junction (Figure S4). Because the chimeric GFP-tagged fusion protein was overexpressed in wild type cells in the presence of the native HAP2 protein, we conducted additional localization studies using ΔHAP2 deletion strains complemented with a C-terminal, HA-tagged version of the full-length HAP2 cDNA in both strains crossed. As shown in Figure 3E-F, the epitope-tagged version of HAP2 localized to the conjugation junction in a pattern almost identical to that of the HAP2::GFP fusion protein.
Figure 3. Localization of HAP2 in mating cells.
Panels A-D. A chimeric HAP2::GFP fusion construct under cadmium-inducible promoter control was introduced into the macronucleus of T. thermophila mating type IV cells (strain CU522). Transformants were starved, induced with CdCl2and mixed with mating type II cells under starvation conditions. At varying times thereafter, live mating cells were visualized using fluorescence optics. Panel (A) shows a mating pair 3 hrs after mixing with strong GFP-fluorescence over the mating junction. A 3-D perspective of the GFP-labeling pattern in conjugating cells can be seen in the accompanying video (see Movie S1). Scale bar = 20 µm. Panel (B) is a higher magnification image of the mating junction from a later pair (3.5 hrs) shown on edge. Lines bracket a region perforated by pores. Scale bar = 5 µm. Panel (C) shows the mating junction at 4 hrs with punctate staining (semicircular and circular profiles) on either side of the margins of the junction (white lines). This punctate staining pattern resembles autophagosomes in the same region of the mating junction shown in panel (D). Scale bar = 10 µm. Panel (D) is a transmission electron micrograph through a region near the mating junction (1.5 hrs into mating) with autophagic vacuoles (arrows) shown in cross-section. Scale bar = 500 nm. Immunogold labeling of GFP-tagged HAP2 at the ultrastructural level is shown in Figure S4. Panels E-F. A C-terminal HA-tagged version of the full length HAP2 cDNA was introduced into the endogenous HAP2 locus of ΔHAP2 knockout strains (mating types VI and VII) by homologous recombination. Cells were mated, fixed and labeled with mouse anti-HA antibodies and secondary rhodamine red-tagged goat anti-mouse IgG as described in the text. Two different mating pairs are shown in panels (E) and (F). Scale bars = 10 µm.
Discussion
The experiments described here show that HAP2 is expressed in all mating types of T. thermophila and functions in both cells of a mating pair to allow fertilization to occur. Interestingly, cells of complementary mating types can pair in the complete absence of HAP2 indicating that the protein acts downstream of mating type recognition and cell adhesion, which are likely controlled by multiple factors beginning with cell-to-cell contact [15, 16] and interaction of products of the mating type alleles [8]. Nevertheless, the fact that HAP2 both localizes to the conjugation junction, and is required for membrane pore formation, provides strong support for previous arguments that HAP2 acts as a gamete fusogen either by itself, or in conjunction with other proteins [1–6]. Finally, we observed that the physical interaction of mating cells was stabilized by HAP2. Rather than a programmed event, premature dissociation of {ΔHAP2×ΔHAP2} knockout pairs was likely the result of an inability to form pores, and highlights a novel role for membrane fusion in pair stabilization that may have functional consequences in species other than Tetrahymena with highly motile gametes.
It is worth noting that the frequencies of progeny development in {wild type X ΔHAP2} crosses, and {ΔHAP2×ΔHAP2} genomic rescue crosses (in which the HAP2 knockout construct was partially replaced in both parental strains with the native HAP2 gene), were less than that in control {wild type X wild type} matings (Table 1). In each case, this may have been due to a dosage effect since in the {wild type X ΔHAP2} crosses, HAP2 was expressed in only one cell of a mating pair, and in the genomic rescue crosses we were unable to restore the gene to its full, 45N, copy-number in the macronuclei of the rescued ΔHAP2 knockout cell lines for technical reasons. While other possibilities exist, a reduction in the number of membrane pores, or a decrease in pair stability, or both, could account for the reduction in progeny development seen in these matings.
Previous studies have shown that HAP2 function is restricted to male gametes in sexually dimorphic, anisogamous species (i.e. organisms whose gametes are dissimilar in form and function) [1–5]. Nevertheless, isogamous organisms almost certainly arose before anisogamous species [17, 18], and phenotypic traits that are now fixed in either sperm or egg (such as HAP2 activity), may very well have been shared by the gametes of ancestral, isogamous life forms. In this regard, Tetrahymena, while producing neither male nor female gametes, may offer a glimpse into how the problem of gamete fusion was initially solved in the earliest eukaryotes, namely, through the formation of one or more pores (fusion events) initiated independently by both cells of a mating pair, each exhibiting either “male” or “female” character at given points of membrane contact at their interface. In this regard, HAP2 expression patterns have been examined in three isogamous species other than Tetrahymena, the slime mold, Physarum polycephalum [2], and two algal species, Chlamydomonas reinhardii [2] and Gonium pectorale [19]. Consistent with what we describe here, HAP2 was reported to be expressed in the two mating types of Physarum, and in both plus and minus mating types of Chlamydomonas and Gonium [2, 19]. While these observations suggest that HAP2 expression is not restricted to particular mating types in isogamous species, deletion of the HAP2 gene from the minus (“male”) but not the plus mating type of C. reinhardii blocks fertilization [4], and while HAP2 is made, the protein is rapidly degraded in plus (“female”) gametes of G. pectorale following gamete activation [19]. On the one hand, the apparent vestigial nature of HAP2 expression in the plus mating types of these species (along with the molecular and ultrastructural differences in their gametes [19–22], may indicate they are in transition from isogamous to anisogamous forms [23.24]. At the same time, Tetrahymena is itself anomalous, in that it bypasses the production of “male” and “female” gametes altogether, and instead produces stationary (“female”), and migratory (“male”) pronuclei that are exchanged between complementary mating types. Thus the requirement for a male gamete-specific fusogen, HAP2, in all mating types of T. thermophila might be an adaptation to its nuclear exchange behavior rather than a characteristic of isoagmous life forms in general.
While it remains to be determined whether T. thermophila represents the exception rather than the rule in terms of HAP2 expression in isogamous species, an analysis of membrane dynamics in mating cells indicates that membrane fusion events, which are typically initiated by male gametes, are driven on both sides of the conjugation junction in mating Tetrahymena. This begins with the formation of membrane protrusions, or tubules of about the size of the singular mating structure in minus gametes of C. reinhardii (i.e. ~ 50 nm diameter), and which extend from the plasma membranes of each partner into the junction cleft. As shown here, in the absence of HAP2 these membrane events appeared to initiate normally with protrusions extending from both cells into the junction cleft. However, no pores formed, and cytoplasmic continuity was never established in the critical developmental interval just following cell adhesion and the formation of membrane protrusions. This, and the fact that HAP2 appeared targeted to a region of specialized membrane (the conjugation junction) where pore formation occurs, argues strongly for a role for HAP2 in membrane fusion.
From a practical standpoint, the ability to induce synchronous mass mating in cells that can be cultured to high density on a large scale, along with methods for isolating the conjugation junction itself [25], make Tetrahymena a potentially powerful system for examining HAP2 function at a biochemical level. It should also be noted that a recent screen of proteins up-regulated during conjugation in T. thermophila has led to the identification of a gene encoding a predicted zinc-finger domain containing protein (ZFR1; TTHERM_01285910), which localizes to the conjugation junction of mating cells and, as with HAP2, appears necessary for normal fertility and pair-stability [26]. The possibility these and other proteins act in concert to drive membrane fusion, and perhaps pore expansion, clearly bears further investigation. Finally, while these studies provide strong evidence of a role for HAP2 in membrane fusion, Tetrahymena and other ciliate species appear unique in their ability to limit cell-to-cell fusion and reverse the establishment of cytoplasmic continuity between cells during the mating process.
Supplementary Material
Acknowledgements
We thank Dr. Timothy Springer, Harvard University, and Matthew Lefebvre, St. Olaf’s College for alerting us to HAP2 as candidate gene of interest, and Ms. Catherine Devine for her excellent technical assistance during the course of this project. Research reported in this publication was supported by a National Science Foundation grant (MCB-1233315) entitled RUI: Membrane Dynamics During Cortical Development in Tetrahymena thermophila to E.S. Cole and D. Beussman; and an NSF Graduate Research Fellowship to J. Fricke Pinello under award number DGE-1144153. Any opinions, findings, and conclusions expressed in this manuscript are those of the authors and do not necessarily reflect the views of the National Science Foundation. This research was also supported by the Office Of The Director, National Institutes Of Health under award number 2 P40 OD010964-10 to T.G.C. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. T.G.C. and D.C.-H. are founding members of Tetragenetics, Inc., Cambridge, MA, and T.G.C. is a member of its scientific advisory board.
References
- 1.Johnson MA, von Besser K, Zhou Q, Smith E, Aux G, Patton D, Levin JZ, Preuss D. Arabidopsis hapless mutations define essential gametophytic functions. Genetics. 2004;2:971–982. doi: 10.1534/genetics.104.029447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mori T, Kuroiwa H, Higashiyama T, Kuroiwa T. GENERATIVE CELL SPECIFIC 1 is essential for angiosperm fertilization. Nat. Cell Biol. 2006;1:64–71. doi: 10.1038/ncb1345. [DOI] [PubMed] [Google Scholar]
- 3.von Besser K, Frank AC, Johnson MA, Preuss D. Arabidopsis HAP2 (GCS1) is a sperm-specific gene required for pollen tube guidance and fertilization. Development. 2006;23:4761–4769. doi: 10.1242/dev.02683. [DOI] [PubMed] [Google Scholar]
- 4.Liu Y, Tewari R, Ning J, Blagborough AM, Garbom S, Pei J, Grishin NV, Steele RE, Sinden RE, Snell WJ, et al. The conserved plant sterility gene HAP2 functions after attachment of fusogenic membranes in Chlamydomonas and Plasmodium gametes. Genes Dev. 2008;8:1051–1068. doi: 10.1101/gad.1656508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steele RE, Dana CE. Evolutionary history of the HAP2/GCS1 gene and sexual reproduction in metazoans. PLoS One. 2009;11:e7680. doi: 10.1371/journal.pone.0007680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong JL, Johnson MA. Is HAP2-GCS1 an ancestral gamete fusogen? Trends Cell Biol. 2010;3:134–141. doi: 10.1016/j.tcb.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 7.Nanney DL. Nucleo-Cytoplasmic Interaction during Conjugation in Tetrahymena. Biol. Bull. 1953;1:133–148. [Google Scholar]
- 8.Cervantes MD, Hamilton EP, Xiong J, Lawson MJ, Yuan D, Hadjithomas M, Miao W, Orias E. Selecting One of Several Mating Types through Gene Segment Joining and Deletion in Tetrahymena thermophila. PLoS Biol. 2013;3:e1001518. doi: 10.1371/journal.pbio.1001518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suganuma Y, Shimode C, Yamamoto H. Conjugation in Tetrahymena: formation of a special junction area for conjugation during the co-stimulation period. J. Electron Microsc. 1984;1:10–18. [PubMed] [Google Scholar]
- 10.Wolfe J. Cytoskeletal reorganization and plasma membrane fusion in conjugating Tetrahymena. J. Cell. Sci. 1985:69–85. doi: 10.1242/jcs.73.1.69. [DOI] [PubMed] [Google Scholar]
- 11.Orias E. Ciliate conjugation. In: Gall JG, editor. The Molecular Biology of Ciliated Protozoa. New York, NY: Academic Press; 1986. pp. 45–94. [Google Scholar]
- 12.Cole ES. The Tetrahymena conjugation junction. In: Baluska F, Volkmann D, Barlow P, editors. Cell-Cell Channels. New York: Springer; 2006. pp. 39–62. [Google Scholar]
- 13.Xiong J, Lu Y, Feng J, Yuan D, Tian M, Chang Y, Fu C, Wang G, Zeng H, Miao W. Database (Oxford) 0, bat008. 2013. Tetrahymena Functional Genomics Database (TetraFGD): an integrated resource for Tetrahymena functional genomics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wolfe J. The conjugation junction of Tetrahymena: Its structure and development. J. Morphol. 1982;172:159–178,. doi: 10.1002/jmor.1051720204. [DOI] [PubMed] [Google Scholar]
- 15.Bruns PJ, Palestine RF. Co-stimulation in Tetrahymena pyriformis: a developmental interaciton between specially prepared cells. Dev. Biol. 1975;42:75–83. doi: 10.1016/0012-1606(75)90315-2. [DOI] [PubMed] [Google Scholar]
- 16.Finley MJ, Bruns PJ. A nonspecific response to heterotypic cell--cell interactions. Dev. Biol. 1980;79:81–94. doi: 10.1016/0012-1606(80)90074-3. [DOI] [PubMed] [Google Scholar]
- 17.Parker GA, Baker RR, Smith VG. The origin and evolution of gamete dimorphism and the male-female phenomenon. J. Theor. Biol. 1972;3:529–553. doi: 10.1016/0022-5193(72)90007-0. [DOI] [PubMed] [Google Scholar]
- 18.Maynard-Smith J. The evolution of sex. Cambridge Univ Press; 1978. [Google Scholar]
- 19.Kawai J, Mori T, Hamaji T, Suzuki M, Olson BJ, Uemura T, Ueda T, Nakano A, Toyoda A, Fujiyama A, Nozaki H. Sex-specific posttranslational regulation of the gamete fusogen GCS1 in the isogamous volvocine alga Gonium pectorale. Eukaryot. Cell. 2014;13:648–656. doi: 10.1128/EC.00330-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goodenough UW, Weiss RL. Gametic differentiation in Chlamydomonas reinhardtii. III. Cell wall lysis and microfilament-associated mating structure activation in wild-type and mutant strains. J. Cell Biol. 1975;3:623–637. doi: 10.1083/jcb.67.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin H, Goodenough UW. Gametogenesis in the Chlamydomonas reinhardtii minus mating type is controlled by two genes, MID and MTD1. Genetics. 2007;2:913–925. doi: 10.1534/genetics.106.066167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ning J, Otto TD, Pfander C, Schwach F, Brochet M, Bushell E, Goulding D, Sanders M, Lefebvre PA, Pei J, Grishin NV, Vanderlaan G, Billker O, Snell WJ. Comparative genomics in Chlamydomonas and Plasmodium identifes an ancient nuclear envelope protein family essential for sexual reproduction in protists, fungi, plants, and vertebrates. Genes Dev. 2013;27:1198–1215. doi: 10.1101/gad.212746.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Umen JG. Evolution of sex and mating loci: an expanded view from Volvocine algae. Curr. Opin. Microbiol. 2011;6:634–641. doi: 10.1016/j.mib.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hiraide R, Kawai-Toyooka H, Hamaji T, Matsuzaki R, Kawafune K, Abe J, Sekimoto H, Umen J, Nozaki H. The evolution of male-female sexual dimorphism predates the gender-based divergence of the mating locus gene MAT3/RB. Mol. Biol. Evol. 2013;30:1038–1040. doi: 10.1093/molbev/mst018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cole ES, Anderson PC, Fulton RB, Majerus ME, Rooney MG, Savage JM, Chalker D, Honts J, Welch ME, Wentland AL, et al. A proteomics approach to cloning fenestrin from the nuclear exchange junction of tetrahymena. J. Eukaryot. Microbiol. 2008;4:245–256. doi: 10.1111/j.1550-7408.2008.00337.x. [DOI] [PubMed] [Google Scholar]
- 26.Xu J, Tian H, Wang W, Liang A. The zinc finger protein Zfr1p is localized specifically to conjugation junction and required for sexual development in Tetrahymena thermophila. PLoS One. 2012;12:e52799. doi: 10.1371/journal.pone.0052799. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.