Abstract
The principles of tissue engineering (TE) are widely used for bone regeneration concepts. Three-dimensional (3D) cultivation of autologous human mesenchymal stromal cells (MSCs) on porous scaffolds is the basic prerequisite to generate newly formed bone tissue. Quantitative reverse transcription polymerase chain reaction (qRT-PCR) is a specific and sensitive analytical tool for the measurement of mRNA-levels in cells or tissues. For an accurate quantification of gene expression levels, stably expressed reference genes (RGs) are essential to obtain reliable results. Since the 3D environment can affect a cell's morphology, proliferation, and gene expression profile compared with two-dimensional (2D) cultivation, there is a need to identify robust RGs for the quantification of gene expression. So far, this issue has not been adequately investigated. The aim of this study was to identify the most stably expressed RGs for gene expression analysis of 3D-cultivated human bone marrow-derived MSCs (BM-MSCs). For this, we analyzed the gene expression levels of n=31 RGs in 3D-cultivated human BM-MSCs from six different donors compared with conventional 2D cultivation using qRT-PCR. MSCs isolated from bone marrow aspirates were cultivated on human cancellous bone cube scaffolds for 14 days. Osteogenic differentiation was assessed by cell-specific alkaline phosphatase (ALP) activity and expression of osteogenic marker genes. Expression levels of potential reference and target genes were quantified using commercially available TaqMan® assays. mRNA expression stability of RGs was determined by calculating the coefficient of variation (CV) and using the algorithms of geNorm and NormFinder. Using both algorithms, we identified TATA box binding protein (TBP), transferrin receptor (p90, CD71) (TFRC), and hypoxanthine phosphoribosyltransferase 1 (HPRT1) as the most stably expressed RGs in 3D-cultivated BM-MSCs. Notably, genes that are routinely used as RGs, for example, beta actin (ACTB) and ribosomal protein L37a (RPL37A), were among the least stable genes. We recommend the combined use of TBP, TFRC, and HPRT1 for the accurate and robust normalization of qRT-PCR data of 3D-cultivated human BM-MSCs.
Introduction
The discipline of bone tissue engineering (TE) involves the combined use of three-dimensional (3D) scaffold materials, growth factors, and osteoprogenitor cells.1 As a key cellular component, mesenchymal stromal cells (MSCs) are often used for the implementation of 3D cell-based concepts in the field of bone regenerative medicine.2,3 MSCs represent a proliferating and undifferentiated cell source with the potential to differentiate toward diverse mesenchymal lineages, including osteoblasts, chondrocytes, adipocytes, and myocytes.4 This cell type is characterized by plastic adherence when maintained in standard culture conditions, specific cell surface marker patterns, and the differentiation potential in vitro mentioned earlier.5 MSCs, in addition to the bone marrow, can be isolated from different fetal and postnatal tissue sources and organs, including bone marrow, adipose tissue, skeletal muscle, brain, liver, kidney, pancreas, lung, umbilical cord blood, placenta, dental pulp, and synovial membrane.6–9 Therefore, it has been suggested that the distribution of MSCs is related to their existence in a perivascular niche.7
In an in vivo 3D environment, surrounding cells typically maintain their ellipsoidal structure and organization. However, in an in vitro two-dimensional (2D) environment, they appear in monolayers with a flattened morphology. Gene expression-mediated changes influence cell shape, functionality, and intercellular communication. On comparing monolayer cultured MSCs with 3D multicellular spheroids, it was shown that the 3D environment altered cell size, expression levels of cell surface antigens, and the osteogenic differentiation potential,10 and enhanced the suppression of inflammatory responses.11 Cells grown in 3D cultures showed reduced proliferation rate, altered their adhesion structures, and changed the transcription levels of distinct adhesion molecules.12
In addition, a 3D environment can affect in vitro cell mechanobiology and extracellular matrix proteins, for example, collagens and fibrins.13 Furthermore, as opposed to 2D cell-loaded substrates, 3D-matrix interactions display enhanced cell biological activities and differ in cytoskeletal composition.14 Three-dimensional environment of MSCs appears to promote their stem cell potential and therapeutic benefit in applications ranging from regenerative medicine to anti-inflammatory treatments and cancer therapy.15 In order to translate scientific results into clinical applications, 3D cell-based models and even 3D-organotypic models are needed.16
Gene expression analysis is a common method to investigate molecular processes and regulatory events in cells, tissues, and organs. Within this analytical method, the application of quantitative reverse transcription polymerase chain reaction (qRT-PCR) is well established.17,18 Since this technique is rather specific and sensitive, the experiments need to be performed carefully with regard to critical steps, for example, sample collection and storage, RNA isolation, starting material quantity and quality, primer concentration, the RT-qPCR assay used (one or two step), and overall transcriptional activity differences between cells.19 To correct for these parameters, different normalization methods have been introduced; for example, standardizing the cell number, a known amount of RNA, or the use of reference genes (RGs). Most researchers refer to the latter method for target gene normalization.
Selecting suitable RGs as internal controls to ensure an accurate gene expression analysis is a crucial step in gene expression analysis. In this context, using single, nonvalidated RGs for qPCR analysis can lead to a systemic source of error, causing incorrect results and misinterpretation of gene expression data.20 Ideally, RGs exhibit a stable gene expression under the experimental conditions given, and should be validated for each new experimental setup. Currently, various statistical algorithms exists to determine the most stable RG for qRT-PCR normalization, such as NormFinder,21 Global Pattern Recognition,22 Bestkeeper,©23 and geNorm.24 However, there is no standardized method for the selection of RGs. Most researchers use a combination of the methods mentioned earlier to determine RG expression stability. Some scientists use microarray data in order to find appropriate RG.25,26
Commonly used RGs, for example, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and beta actin (ACTB), were observed to be less stable or regulated under certain culture conditions in MSCs, chondrocytes, and fibroblasts.27–30 We recently showed that the combined use of DNA-damage-inducible alpha (GADD45A), pumilio homolog 1 (PUM1), and large ribosomal protein P0 (RPLP0) significantly improved gene expression accuracy compared with classically used RGs, ACTB, and GAPDH in 2D osteogenically stimulated bone marrow-derived MSCs (BM-MSCs).31
Numerous putative RGs have been reported for a wide variety of human cell lines and primary cells under different experimental conditions, almost exclusively under standardized 2D cultivation. However, for 3D culture of human BM-MSCs, commonly used in regenerative medicine and TE approaches, sufficient data for the analysis of RGs are still lacking. Thus, we investigated the gene expression stability of 31 potential RGs in an effort to identify the most stable RGs in 3D cultivated human BM-MSCs.
Materials and Methods
Isolation and cultivation of MSCs
Human BM-MSCs were isolated from n=6 healthy Caucasian donors (three men, three women, mean age 31.4 years) after obtaining their informed consent (approval no. EK263122004). BM-MSCs were isolated from the mononuclear cell fraction and expanded as previously described.32 Cells were cultured in Dulbecco's modified Eagle's medium (DMEM, low glucose; Life Technologies GmbH, Darmstadt, Germany) supplemented with 10% fetal calf serum (Sigma-Aldrich, Hamburg, Germany) and 1% penicillin/streptomycin (Life Technologies GmbH) with or without osteogenic supplements (i.e., 100 nM dexamethasone, 10 mM beta-glycerophosphate, and 50 μM ascorbic acid-2-phosphate; Sigma-Aldrich) and maintained in a humidified atmosphere at 37°C and 5% CO2. No osteogenic supplements were used for histology, morphology, and all gene expression experiments. Only for biochemical analysis of ALP activity, osteogenic supplements were used to compare the additional effects of supplements and 3D culturing. The study was performed with second-passage BM-MSCs of all donors. After reaching 80–90% confluence, cells were seeded for 2D cultivation at a density of 4.8×104 cells per well in tissue culture plastic six-well plates (Greiner, Frickenhausen, Germany). Peracetic acid-treated human cancellous bone (HCB) cubes with an approximate edge length of 5 mm (Fig. 1B; German Institute for Cell and Tissue Replacement, Berlin, Germany) with a conferred admission for medicinal drugs were used as 3D matrices.33 Fifty microliters of cell suspensions with 1×106 BM-MSCs from each donor were seeded per HCB in a drop-wise manner. After a 2 h adherence period in the incubator, the cubes were covered carefully with 6 mL of cell culture medium. Cells were maintained at 37°C in a saturated humidity atmosphere with 5% CO2. The next day, seeded HCBs were transferred into new six-well plates. Triplicates were used for each 2D and 3D sample. The cells were cultured for 14 days, changing the medium twice a week. Cell morphology and adherence were monitored by light microscopy.
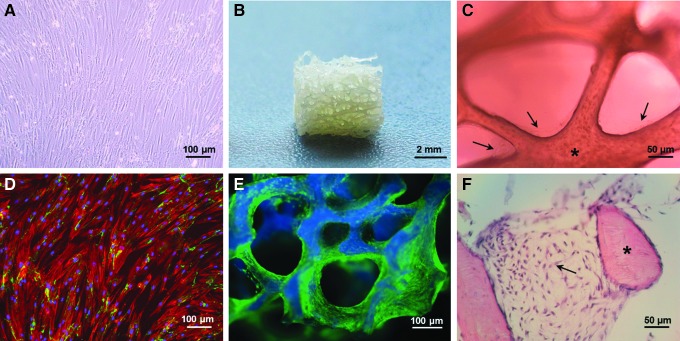
FIG. 1.
Two-dimensionally (2D) cultured bone marrow-derived mesenchymal stromal cells (BM-MSCs) of one donor after 14 days in culture (A). Macrograph of a human cancellous bone (HCB) cube with an approximate edge length of 5 mm (B). Light microscopic image of 3D cultivated BM-MSCs on HCB, 100× (C). Fluorescence microscopic picture of 2D cultivated BM-MSCs depicting nuclei (Hoechst 33342, blue), cytoskeleton (Alexa 555 phalloidin, red), and the extracellular matrix protein fibronectin (anti-mouse fibronectin-AK and Alexa Fluor® 488, green), 100× (D). Fluorescence stereoscopic image of HCB (blue) and vital BM-MSCs stained with calcein (green) (E). Histological section of HCB with adherent BM-MSCs after Giemsa stain, 200× (F). Asterisks mark HCB, and arrows highlight BM-MSCs. Color images available online at www.liebertpub.com/tec
RNA isolation and cDNA synthesis
Total RNA was isolated with the RNeasy Mini Kit (Qiagen, Hilden, Germany) after 14 days of cultivation. After rinsing with phosphate-buffered saline (PBS), 2D cultured BM-MSCs were harvested from cell culture plates by using a cell scraper. Cells were centrifuged for 5 min at 2000 g, and the PBS was removed. Three-dimensional samples were rinsed with PBS, and excess PBS was aspirated with a filter paper. All samples were frozen immediately at −80°C until later use. For the isolation of total RNA, BM-MSCs from 3D samples were lysed during a centrifugation step at 16,100 g for 1 min at room temperature (RT) in lysis buffer. After centrifugation, the lysate was transferred into new reaction tubes and further steps were performed according to the manufacturer's instructions. During the isolation, an on-column DNase digestion with RNase-Free DNase Set (Qiagen) was performed. RNA concentration and quality was determined at 260 nm and 260/280 nm using a NanoDrop 2000c spectrophotometer (PEQLAB Biotechnologie GmbH, Erlangen, Germany). The 260/280 nm ratio values were between 1.9 and 2.1. cDNA synthesis from 200 ng RNA for each sample was performed using Superscript II Reverse Transcriptase and with oligo-dT (Invitrogen, Karlsruhe, Germany) according to the manufacturer's instructions. An equal volume of cDNA was pooled from each BM-MSC donor and used as templates in qPCR.
Quantitative real-time polymerase chain reaction
Human Endogenous Control TaqMan® assays with 32 FAM labeled probes (Table 1) spotted in triplicate in a 96-well format were used for quantitative gene expression analysis of RGs (Life Technologies GmbH; Applied Biosystems, Darmstadt, Germany). No osteogenic supplements were used for all gene expression experiments. qPCRs were performed with TaqMan Fast Universal PCR Master Mix (2×) according to the manufacturer's instructions using a 7500 Real-Time PCR Fast System (Applied Biosystems). A volume of 0.96 μL template cDNA corresponding to 19.2 ng of RNA was used for each well of the qPCR array. All reactions were carried out in a total volume of 10 μL with a thermal profile of an initial cycle for 20 s at 95°C for enzyme activation followed by 40 cycles with 3 s at 95°C denaturation, 30 s at 60°C for annealing and elongation, and terminated by a holding stage at 4°C. Technical triplicates were run for all samples. For the no template control (NTC), the three wells with the probe of 18 s RNA was chosen as this assay does not provide extra wells for the NTC.
Table 1.
Reference and Target Genes Investigated in this Study, Providing Gene Symbol, Name, Function, Gene ID, and Assay Number
| Gene symbol | Name of reference gene | Function | Entrez gene ID | Assay ID |
|---|---|---|---|---|
| ABL1 | c-abl oncogene 1, nonreceptor tyrosine kinase | Protein tyrosine kinase activity | 25 | Hs00245445_m1 |
| ACTB | Actin, beta | Structural constituent of cytoskeleton | 60 | Hs99999903_m1 |
| B2M | Beta-2-microglobulin | Protein binding | 567 | Hs99999907_m1 |
| CASC3 | Cancer susceptibility candidate 3 | RNA binding, exon junction complex | 22794 | Hs00201226_m1 |
| CDKN1A | Cyclin-dependent kinase inhibitor 1A (p21, Cip1) | Regulator of cell cycle | 1026 | Hs00355782_m1 |
| CDKN1B | Cyclin-dependent kinase inhibitor 1B (p27, Kip1) | Regulator of cell cycle | 1027 | Hs00153277_m1 |
| EIF2B1 | Eukaryotic translation initiation factor 2B, subunit 1 alpha, 26kDa | Translational initiation | 1967 | Hs00426752_m1 |
| ELF1 | E74-like factor 1 (ets domain transcription factor) | Positive regulation of transcription | 1997 | Hs00152844_m1 |
| GADD45A | Growth arrest and DNA-damage-inducible, alpha | Signal transduction in response to DNA damage | 1647 | Hs00169255_m1 |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | Glycolysis, regulation of translation | 2597 | Hs99999905_m1 |
| GUSB | Glucuronidase, beta | Carbohydrate metabolic process | 2990 | Hs99999908_m1 |
| HMBS | Hydroxymethylbilane synthase | Heme biosynthetic process | 3145 | Hs00609297_m1 |
| HPRT1 | Hypoxanthine phosphoribosyltransferase 1 | Generation of purine nucleotides | 3251 | Hs99999909_m1 |
| IPO8 | Importin 8 | Intracellular protein transport | 10526 | Hs00183533_m1 |
| MRPL19 | Mitochondrial ribosomal protein L19 | Translation | 9801 | Hs00608519_m1 |
| MT-ATP6 | Mitochondrially encoded ATP synthase 6 | ATP synthase complex | 4508 | Hs02596862_g1 |
| PES1 | Pescadillo ribosomal biogenesis factor 1 | Cell proliferation, rRNA processing | 23481 | Hs00362795_g1 |
| PGK1 | Phosphoglycerate kinase 1 | Glycolysis, gluconeogenesis | 5230 | Hs99999906_m1 |
| POLR2A | Polymerase (RNA) II (DNA directed) polypeptide A, 220 kDa | Transcription initiation | 5430 | Hs00172187_m1 |
| POP4 | Processing of precursor 4, ribonuclease P/MRP subunit (S. cerevisiae) | tRNA processing, mRNA cleavage | 10775 | Hs00198357_m1 |
| PPIA | Peptidylprolyl isomerase A (cyclophilin A) | Protein folding | 5478 | Hs99999904_m1 |
| PSMC4 | Proteasome (prosome, macropain) 26S subunit, ATPase, 4 | ATP binding, proteolysis | 5704 | Hs00197826_m1 |
| PUM1 | Pumilio homolog 1 (Drosophila) | Regulation of translation | 9698 | Hs00206469_m1 |
| RPL30 | Ribosomal protein L30 | Translation | 6156 | Hs00265497_m1 |
| RPL37A | Ribosomal protein L37a | Translation | 6168 | Hs01102345_m1 |
| RPLP0 | Ribosomal protein, large, P0 | Translation | 6175 | Hs99999902_m1 |
| RPS17 | Ribosomal protein S17 | Structural constituent of ribosome | 6218 | Hs00734303_g1 |
| TBP | TATA box binding protein | Transcription factor binding | 6908 | Hs99999910_m1 |
| TFRC | Transferrin receptor (p90, CD71) | Transferrin transport | 7037 | Hs99999911_m1 |
| UBC | Ubiquitin C | Protease binding, endosomal transport | 7316 | Hs00824723_m1 |
| YWHAZ | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | Signal transduction | 7534 | Hs00237047_m1 |
| Gene symbol | Name of target gene | Function | Entrez gene ID | Assay ID |
|---|---|---|---|---|
| ALPL | Alkaline phosphatase, liver/bone/kidney | Skeletal system development | 249 | Hs01029144_m1 |
| BGLAP | Bone gamma-carboxyglutamate (gla) protein | Structural constituent of bone | 632 | Hs00609452_g1 |
| IBSP | Integrin-binding sialoprotein | Extracellular matrix organization | 3381 | Hs00173720_m1 |
| RUNX-2 | Runt-related transcription factor 2 | Osteoblast differentiation | 860 | Hs00231692_m1 |
| SPP1 | Secreted phosphoprotein 1 | Extracellular matrix binding | 6696 | Hs00959010_m1 |
Expression of the osteogenic marker genes alkaline phosphatase (ALP), liver/bone/kidney (ALPL, Hs01029144_m1), bone gamma-carboxyglutamate (gla) protein (BGLAP, Hs00609452_g1), integrin-binding sialoprotein (IBSP, Hs00173720_m1), runt-related transcription factor 2 (RUNX-2, Hs00231692_m1), and secreted phosphoprotein 1 (SPP1, Hs00959010_m1) were analyzed as target genes using the same cDNA pool, protocols, and instrument as mentioned earlier with TaqMan Gene Expression Assays (Life Technologies GmbH; Applied Biosystems). Data analysis was performed using the 7500 software v2.0.5 (Applied Biosystems). Gene expression of a total of n=31 RGs was evaluated on the basis of cycle threshold (Ct) values from BM-MSCs cultivated for 14 days under 2D versus 3D culture conditions. Results were exported to Microsoft Excel for analysis and presented as mean±SD. SD of ΔCt values were calculated using an equation with SD1 and SD2 being the SD, and n1 and n2 being the sample sizes of 3D (1) and 2D (2), respectively.
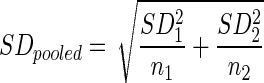 |
Gene expression of osteogenic target genes was calculated as a relative fold difference using the delta-delta Ct method, where the 3D cultivated BM-MSCs represent the sample and the 2D cultivated BM-MSCs represent the control.34
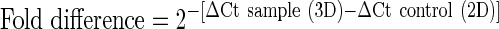 |
The 3D and 2D gene expression levels were paired by original wells. For each experimental unit, three wells (triplicate) were generated. The three ratios were paired for the calculation from those three wells (delta Ct 3D_1 and delta Ct 2D_1; delta Ct3D_2 and delta Ct 2D_2; and delta Ct 3D_3 and delta Ct 2D_3) and then, the average was obtained from the three ratios. A fold difference of one equates to no regulation. Downregulation of gene expression corresponds to values below one. Values above one mean an upregulation of gene expression.
Determination of cell-specific ALP activity
Osteogenic differentiation was validated by measurement of cell-specific ALP activity. The activity of the enzyme was measured in cell extracts of 2D and 3D samples using p-nitrophenylphosphate (pNpp) and quantified by colorimetric measurement (SpectraFluorPlus, Tecan, Switzerland). Cells were washed with PBS and lysed in 500 μL lysis buffer (1.5 M Tris–HCl, pH 10, containing 1 mM ZnCl2, 1 mM MgCl2, and 1% Triton X-100) at 4°C for 20 min. The 3D samples were additionally sonicated for 1 min for better lysis results. After incubation, the lysates of 2D samples were transferred from six-well plates into new reaction tubes and centrifuged at 20,000 g and 4°C for 30 min. The lysates of 3D samples were transferred after centrifugation into new reaction tubes to separate them from HCB cubes. A fifty-microliter lysate was incubated with 100 μL 3.6 mM pNpp in 100 mM diethanolamine, pH 9.8, containing 0.1% Triton X-100 at 37°C for 30 min. The reaction was stopped with 100 μL 100 mM NaOH, and the amount of p-nitrophenol (pNp) was determined spectrophotometrically at 405 nm (SpectraFluorPlus). The final cell-specific ALP activity was calculated as μmol pNp/30 min per 1×106 cells. Triplicates were used for each experimental unit. Total DNA quantification was assessed by Pico Green Assay (Invitrogen). The same lysate as for the determination of cell-specific ALP level was used for DNA quantification. One hundred fifty microliters of Pico Green solution were incubated with 50 μL of cell lysate in a black microtiter plate. After 5 min of incubation in the dark, the fluorescence was measured at an excitation of 490 nm and an emission of 520 nm with a plate reader (SpectraFluorPlus). Relative fluorescence units were correlated with the cell number using a calibration line. Triplicates were used for each experimental unit.
Immunofluorescence staining
To represent cell morphology, 2D cultured BM-MSCs were characterized by immunofluorescence staining of cytoskeleton by Alexa Fluor® 555 Phalloidin selectively labels F-actin (Life Technologies GmbH, Molecular Probes; Darmstadt, Germany) and extracellular matrix protein fibronectin with anti-mouse fibronectin-AK (Sigma-Aldrich, Munich, Germany) and Alexa Fluor 488. After 7 days of cultivation, BM-MSCs were washed with PBS and fixed with 4% paraformaldehyde for 10 min at RT. Thereafter, cells were permeabilized for 15 min with 0.4% Triton at RT and then incubated in 3% goat serum in PBS for 1 h to block unspecific binding sites. Then, the incubation was carried out with the antibody anti-mouse fibronectin (dilution 1:1000) overnight at 4°C. The next day, the cells were washed with PBS and incubated with Alexa Fluor 488 for 60 min at RT on an orbital shaker in the dark. Staining of nuclei was performed with Hoechst 33342 (7.5 mg/mL final concentration) for 5 min in the dark (Life Technologies GmbH, Molecular Probes). Images were taken with a computer-driven digital camera on an inverted microscope Leica DMI4000B (Leica Microsystems, Wetzlar, Germany).
Determination of cell viability
For determination of cell viability, the LIVE/DEAD® Viability/Cytotoxicity assay was used (Invitrogen). Three-dimensional samples were washed with PBS, and BM-MSCs on HCB were stained with 4 μM calcein-AM and 8 μM ethidium homodimer-1 for 45 min at RT in the dark. The cell-permeable calcein leads to green fluorescence due to the presence of intracellular esterase activity in living cells. Ethidium homodimer-1 enters cells with damaged membranes and binds to nucleic acids, resulting in a 40-fold enhancement of red fluorescence on binding to nucleic acids of dead cells. After staining, the dye was removed and the sample was carefully rinsed with PBS. Viability was visualized via fluorescence microscopy analysis using a stereoscope Zeiss Discovery V.20 (Zeiss, Jena, Germany) with Ex/Em of 494/517 nm and 528/617 nm.
Histology
To demonstrate cell adherence of BM-MSCs on the HCB cubes, samples were analyzed histologically. Fourteen days after seeding, specimens were washed with PBS and fixed in 70% ethanol at RT. After dehydration in increasing ethanol concentrations (2 h in 70%, 80%, 96%, and 100%) and xylol (1 h) at RT in the Leica tissue processor (Leica Microsystems), the samples were embedded in Technovit 9100N (Heraeus-Kulzer, Friedrichsdorf, Germany). A low-temperature (–5°C) polymerization was performed in plastic vessels (Heraeus-Kulzer, Hanau, Germany). Slices of 10 μm thickness were obtained from the cured specimen blocks using an RM2155 microtome (Leica, Nussloch, Germany). The mounted sections were deacrylated twice for 20 min with 2-methoxyethylacetate (Merck Biomaterials, Darmstadt, Germany) followed by incubation for 20 min in xylol twice, 5 min in acetone, 10 min in 80% ethanol, and rehydration with distilled water.
Giemsa staining was used to show cell morphology and adherence. Nuclei appear dark purple and cytoplasm appear light pink. Slices were stained at 60°C for 30 min. The stain was differentiated with acetic acid for 30 s, followed by a short incubation in 96% and 6 min in 100% ethanol. Dehydration was finished with xylol, and slices were mounted in DePex mounting medium.
Measurement of RG stability
The RG stability was calculated using two well-established algorithms, geNorm and NormFinder software packages. geNorm enables a ranking of suitable RGs according to their stability measured by the stability value M.24 The algorithm used is the “average pairwise variation” of a control gene with all other control genes in the study. The lowest M-value corresponds to the gene with the least expression variation and represents the most stable internal control in the experiment.
NormFinder applies a robust mathematical model that attempts to find the optimal RG out of a group of candidate genes.21 The model can analyze expression data obtained through any quantitative method; for example, real-time RT-PCR and microarray-based expression analysis. The analysis resulted in variability values of the most stable RG and in an optimum pair of RGs. The resulting pair might have compensating expression, so that one gene, for example, is slightly overexpressed in one group, but the other gene is correspondingly lower expressed in the same group. So, the optimum pair may not include the optimum single gene. In contrast to geNorm, NormFinder takes information of groupings of samples into account, such as untreated and treated or sick and healthy.
Ct-values of all samples were entered in the respective program, and the stability values were calculated for each RG. The analysis was performed twice, first for Ct-values of 2D and 3D samples and second for 3D samples alone.
As a third method, the coefficient of variation (CV) was calculated for each gene. The CV equals the standard deviation divided by the mean Ct-value and is expressed as a percentage. The CV is a statistical tool for comparing the degree of variation between genes, even if the mean values drastically differ from each other.35
Statistics
Statistical data analysis was calculated using GraphPad Prism 5.0 software (La Jolla, CA). Gene expression data were analyzed as follows. Shapiro–Wilk normality test was used to test whether the values come from a Gaussian distribution. If the values were normally distributed, unpaired t-test with Welch's correction was used. Nonparametric Mann–Whitney test was applied when values were not normally distributed. Data are presented as mean±standard deviation (SD).
Biochemical data were analyzed with two-way ANOVA with Bonferroni post-tests. Data are presented as mean±standard error of the mean (SEM). Statistical significance of all data was considered if p<0.05.
Results
Cell morphology, adherence, and viability of 2D and 3D cultured BM-MSCs
Cell morphology, adherence, and viability of BM-MSCs were analyzed after 14 days under 2D or 3D cultivation conditions without osteogenic supplements. Two-dimensional cultured BM-MSCs of all six donors showed a typical spindle-shaped cell morphology (Fig. 1A, D, example of one donor). BM-MSCs were able to adhere and proliferate on the HCB (Fig. 1C, E, F) over a period of 14 days. After 14 days of cultivation, a dense vital cellular network was formed (Fig. 1E), which partially closed the pores of the HCBs (Fig. 1F). BM-MSCs were able to assume the organization of a 3D structure on the HCBs.
Expression levels of RGs from 2D and 3D cultivated BM-MSCs
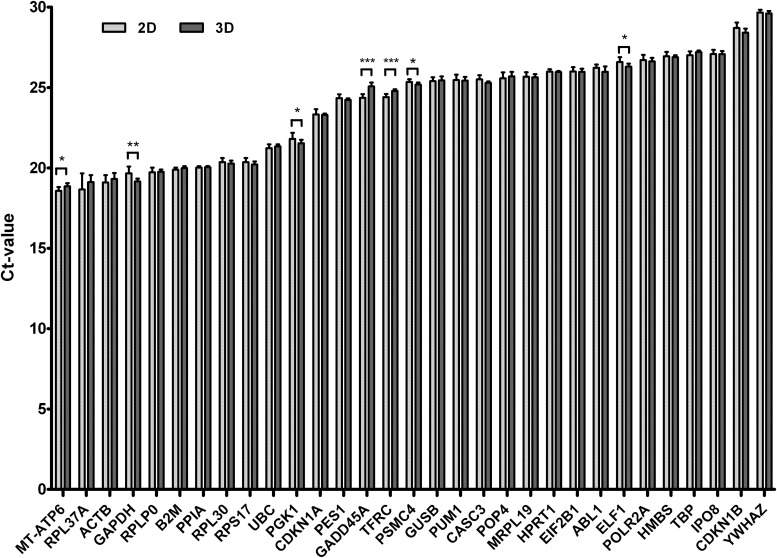
The expression levels of 31 known and potential RGs from 2D and 3D cultivated BM-MSCs without osteogenic supplements were analyzed and represented in Figure 2. Table 2 summarizes the mean Ct-values with their standard deviations of all RGs from both cultivation methods, including the ΔCt-values. The RG with the highest expression level of 2D and 3D cultivated BM-MSCs was the gene for mitochondrially encoded ATP synthase 6 (MT-ATP6) with a mean Ct-value of 18.58±0.22 and 18.85±0.18, respectively. The lowest gene expression was measured for the RG tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide and (YWHAZ) with a mean Ct-value of 29.67±0.15 and 29.61±0.15 for BM-MSCs in 2D and 3D culture, respectively. Notably, the RGs MT-ATP6, GAPDH, phosphoglycerate kinase 1 (PGK1), growth arrest and DNA-damage-inducible, alpha (GADD45A), TFRC, proteasome (prosome, macropain) 26S subunit, ATPase, 4 (PSMC4), and E74-like factor 1 (ets domain transcription factor) (ELF1) showed significant differences in gene expression of 2D and 3D cultured BM-MSCs (Fig. 2). The largest gene expression difference between both cultivation methods was detected for GADD45A with a delta Ct-value of 0.71 (2D: 24.37±0.21; 3D: 25.08±0.23) followed by GAPDH with 0.5 (2D: 19.66±0.4; 3D: 19.16±0.16). The RG candidate IPO8 showed the lowest gene expression difference between 2D and 3D cultivated BM-MSCs with a delta Ct-value of only 0.0097. Eukaryotic translation initiation factor 2B, subunit 1 alpha, 26 kDa (EIF2B1), and hypoxanthine phosphoribosyltransferase 1 (HPRT1) had the second and third lowest delta Ct-values of 0.013 and 0.022, respectively. The gene ribosomal protein L37a (RPL37A) was the RG with the highest standard deviation, and thus with the greatest variation of the mean Ct-values of 2D and 3D cultivated BM-MSCs with 18.66±0.95 and 19.12±0.41, respectively.
FIG. 2.
Cycle threshold (Ct) values of n=31 reference genes in two-dimensionally and three-dimensionally cultivated bone marrow-derived mesenchymal stromal cells (mean±standard deviation). Significance with *p<0.05, **p<0.01, and ***p<0.001.
Table 2.
Ct-Values of N =31 Putative Reference Genes in Two- and Three-Dimensionally Cultivated Human Bone Marrow-Derived Mesenchymal Stromal Cells After 14 Days (Mean±Standard Deviation)
| Symbol | Ct-values 2D [mean±SD] | Ct-values 3D [mean±SD] | ΔCt2D–3D values [mean±SDtotal] | |||
|---|---|---|---|---|---|---|
| ABL1 | 26.22 | 0.20 | 25.98 | 0.32 | 0.23 | 0.016 |
| ACTB | 19.10 | 0.43 | 19.31 | 0.35 | −0.01 | 0.034 |
| B2M | 19.89 | 0.12 | 19.99 | 0.12 | 0.24 | 0.003 |
| CASC3 | 25.51 | 0.25 | 25.29 | 0.08 | 0.24 | 0.008 |
| CDKN1A | 23.33 | 0.31 | 23.30 | 0.08 | −0.03 | 0.012 |
| CDKN1B | 28.71 | 0.32 | 28.41 | 0.23 | 0.27 | 0.018 |
| EIF2B1 | 25.99 | 0.26 | 25.98 | 0.18 | −0.29 | 0.011 |
| ELF1 | 26.59 | 0.28 | 26.31 | 0.17 | −0.13 | 0.012 |
| GADD45A | 24.36 | 0.21 | 25.07 | 0.23 | −0.74 | 0.011 |
| GAPDH | 19.66 | 0.40 | 19.16 | 0.16 | 0.49 | 0.021 |
| GUSB | 25.40 | 0.23 | 25.45 | 0.23 | −0.10 | 0.012 |
| HMBS | 26.95 | 0.26 | 26.89 | 0.11 | 0.06 | 0.009 |
| HPRT1 | 25.98 | 0.15 | 25.96 | 0.07 | 0.07 | 0.003 |
| IPO8 | 27.09 | 0.24 | 27.08 | 0.18 | −0.05 | 0.010 |
| MRPL19 | 25.67 | 0.26 | 25.64 | 0.18 | 0.08 | 0.011 |
| MT-ATP6 | 18.58 | 0.22 | 18.85 | 0.18 | −0.40 | 0.009 |
| PES1 | 24.34 | 0.23 | 24.23 | 0.10 | 0.10 | 0.007 |
| PGK1 | 21.80 | 0.36 | 21.53 | 0.21 | 0.23 | 0.019 |
| POLR2A | 26.71 | 0.30 | 26.63 | 0.21 | 0.06 | 0.015 |
| POP4 | 25.57 | 0.35 | 25.70 | 0.26 | −0.13 | 0.021 |
| PPIA | 20.02 | 0.08 | 20.05 | 0.07 | −0.03 | 0.001 |
| PSMC4 | 25.35 | 0.16 | 25.19 | 0.12 | 0.12 | 0.004 |
| PUM1 | 25.46 | 0.32 | 25.43 | 0.22 | −0.05 | 0.016 |
| RPL30 | 20.36 | 0.25 | 20.27 | 0.18 | −0.19 | 0.010 |
| RPL37A | 18.66 | 0.95 | 19.12 | 0.41 | 0.18 | 0.119 |
| RPLP0 | 19.72 | 0.28 | 19.76 | 0.13 | −0.20 | 0.011 |
| RPS17 | 20.36 | 0.24 | 20.21 | 0.18 | 0.04 | 0.010 |
| TBP | 27.01 | 0.22 | 27.20 | 0.10 | −0.16 | 0.006 |
| TFRC | 24.41 | 0.18 | 24.79 | 0.11 | −0.40 | 0.005 |
| UBC | 21.23 | 0.23 | 21.34 | 0.11 | 0.04 | 0.007 |
| YWHAZ | 29.67 | 0.16 | 29.61 | 0.15 | 0.04 | 0.005 |
Ct, cycle threshold.
Effect of 3D cultivation of BM-MSCs on the stability of RG expression
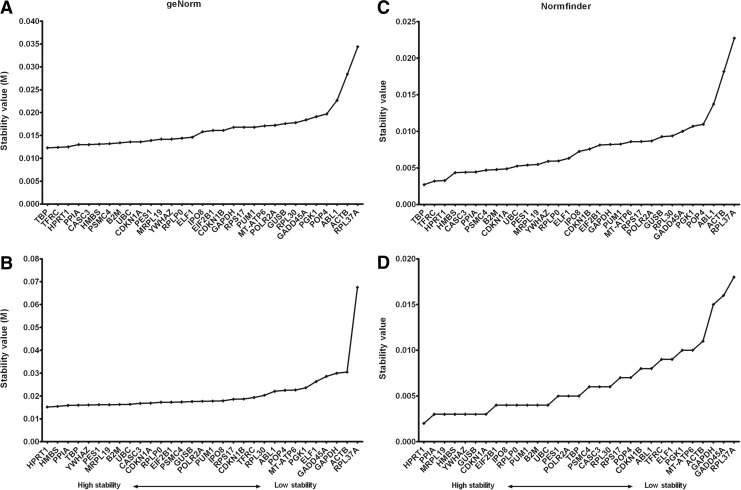
The expression stability of the n=31 RG in BM-MSCs was measured with geNorm and NormFinder after 14 days of 2D and 3D cultivation (Fig. 3). The stability values were calculated separately for the 3D cultivation data and for combined gene expression data of 2D and 3D cultivated BM-MSCs to provide all data depending on which experimental design will be chosen from the research community. The stability values of geNorm, NormFinder and the calculation of the CV after 3D cultivation are ranked and summarized in Table 2.
FIG. 3.
Gene expression stability values of putative reference genes. Calculation of reference gene stability in three-dimensionally (top) and two- and three-dimensionally (button) cultivated bone marrow-derived mesenchymal stromal cells using the geNorm (A, B) and NormFinder (C, D). Low stability values mean high gene expression stability.
Using the algorithms of geNorm and NormFinder, we identified TATA box binding protein (TBP), transferrin receptor (p90, CD71) (TFRC), and HPRT1 as the three most stable RGs in 3D-cultivated BM-MSCs. HPRT1 was the most stable RG using CV calculation method followed by CASC3 and CDNK1 with CV of 0.28%, 0.35%, and 0.37%, respectively (Table 3). Comparing the three methods of stability calculation, the three RGs with the highest stability, TBP, TFRC, and HPRT1, are within the first eight ranks. The RGs RPL37A, ACTB, and ABL1 were the least stably expressed genes in all three calculation methods. The order of stability ranks within the 31 RGs is almost identical, on comparing geNorm and NormFinder analysis (Table 3). There is little difference with a maximum shift of two positions. The calculation of the CV showed more variations in the RG order compared with geNorm and NormFinder analysis except for the last five genes.
Table 3.
Comparison of Reference Gene Expression Stability of Three-Dimensionally Cultivated Bone Marrow-Derived Mesenchymal Stromal Cells Assessed by Genorm, NormFinder, and Coefficient of Variation
| geNorm | NormFinder | Coefficient of variation | ||||
|---|---|---|---|---|---|---|
| Gene symbol | Stability value M | Rank | Stability value | Rank | % | Rank |
| TBP | 0.0123 | 1 | 0.0027 | 1 | 0.37 | 4 |
| TFRC | 0.0124 | 2 | 0.0032 | 2 | 0.46 | 8 |
| HPRT1 | 0.0125 | 3 | 0.0032 | 3 | 0.28 | 1 |
| PPIA | 0.0130 | 4 | 0.0044 | 6 | 0.38 | 5 |
| CASC3 | 0.0130 | 5 | 0.0044 | 5 | 0.35 | 2 |
| HMBS | 0.0131 | 6 | 0.0041 | 4 | 0.43 | 7 |
| PSMC4 | 0.0132 | 7 | 0.0046 | 7 | 0.50 | 9 |
| B2M | 0.0134 | 8 | 0.0047 | 8 | 0.62 | 12 |
| UBC | 0.0136 | 9 | 0.0052 | 10 | 0.57 | 11 |
| CDKN1A | 0.0136 | 10 | 0.0048 | 9 | 0.37 | 3 |
| PES1 | 0.0139 | 11 | 0.0053 | 11 | 0.43 | 6 |
| MRPL19 | 0.0142 | 12 | 0.0054 | 12 | 0.82 | 17 |
| YWHAZ | 0.0142 | 13 | 0.0059 | 13 | 0.54 | 10 |
| RPLP0 | 0.0144 | 14 | 0.0059 | 14 | 0.69 | 13 |
| ELF1 | 0.0146 | 15 | 0.0063 | 15 | 0.70 | 14 |
| IPO8 | 0.0158 | 16 | 0.0072 | 16 | 0.71 | 15 |
| EIF2B1 | 0.0161 | 17 | 0.0081 | 18 | 0.73 | 16 |
| CDKN1B | 0.0161 | 18 | 0.0075 | 17 | 0.87 | 19 |
| GAPDH | 0.0168 | 19 | 0.0082 | 19 | 0.90 | 20 |
| RPS17 | 0.0168 | 20 | 0.0086 | 22 | 0.93 | 22 |
| PUM1 | 0.0168 | 21 | 0.0082 | 20 | 0.91 | 21 |
| MT-ATP6 | 0.0171 | 22 | 0.0085 | 21 | 1.03 | 26 |
| POLR2A | 0.0172 | 23 | 0.0086 | 23 | 0.82 | 18 |
| GUSB | 0.0176 | 24 | 0.0092 | 24 | 0.96 | 25 |
| RPL30 | 0.0178 | 25 | 0.0093 | 25 | 0.94 | 23 |
| GADD45A | 0.0184 | 26 | 0.0100 | 26 | 0.95 | 24 |
| PGK1 | 0.0191 | 27 | 0.0106 | 27 | 1.04 | 27 |
| POP4 | 0.0197 | 28 | 0.0109 | 28 | 1.09 | 28 |
| ABL1 | 0.0227 | 29 | 0.0137 | 29 | 1.31 | 29 |
| ACTB | 0.0284 | 30 | 0.0181 | 30 | 1.92 | 30 |
| RPL37A | 0.0344 | 31 | 0.0227 | 31 | 2.28 | 31 |
Genes are ranked for gene expression stability. The three most stable RGs are marked in bold.
When the stability of RG expression of 2D and 3D cultured BM-MSCs was analyzed in combination, geNorm detected HPRT1, HMBS, and peptidylprolyl isomerase A (cyclophilin A) (PPIA) as the three most stably expressed RGs (Fig. 3). The NormFinder analysis revealed HPRT1, PPIA, and mitochondrial ribosomal protein L19 (MRPL19) as the three most stably expressed RGs when analyzing both Ct-values of 2D and 3D cultures. Surprisingly, the two most widely used RGs GAPDH and ACTB were among the three least stably expressed genes in the geNorm analysis. Again, the RG RPL37A had the lowest gene expression stability when analyzed with geNorm and NormFinder. GADD45A and GAPDH were the second and third least stable RGs in the NormFinder analysis.
Effect of RG expression stability on the expression of osteogenic target genes
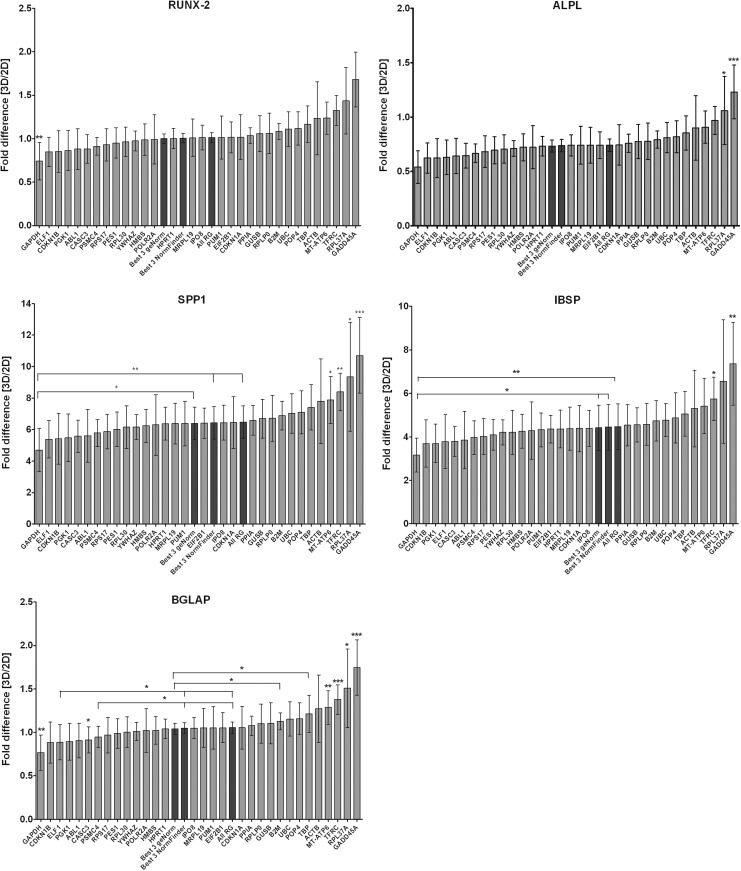
Gene expression levels of the osteogenic target genes RUNX-2, ALPL, SPP1, IBSB, and BGLAP were measured after 14 days in 2D and 3D cultures without osteogenic supplements. The gene expression levels were then calculated with each of the 31 RGs as an internal control, with a combination of the three most stable RGs after analysis with geNorm and NormFinder and with a combination of all 31 RGs (Fig. 4). The fold difference was calculated with the 2D cultured BM-MSCs as a control and the 3D cultivated BM-MSCs representing the sample.
FIG. 4.
Gene expression levels of osteogenic marker genes in three-dimensionally cultivated bone marrow-derived mesenchymal stromal cells after 14 days of cultivation. Relative fold differences of runt-related transcription factor 2 (RUNX-2), alkaline phosphatase, liver/bone/kidney (ALPL), secreted phosphoprotein 1 (SPP1), integrin-binding sialoprotein (IBSP), and bone gamma-carboxyglutamate (gla) protein (BGLAP) were calculated using the ΔΔCt method with two-dimensionally cultured MSCs as a control. Normalization was performed with 31 single reference genes (RGs), the combination of the three most stable RGs from the geNorm and NormFinder analysis, and the mean of all 31 RGs (bars are represented in dark gray). Error bars represent the standard deviation of the mean from three individual experiments in triplicate. Significance was calculated with *p<0.05, **p<0.01, and ***p<0.001.
The osteogenic target genes SPP1 and IBSP were generally upregulated under 3D conditions with whatever normalization combination was chosen. It should be noted that the upregulation of SPP1 and IBSP was due only to the 3D cultivation; no osteogenic supplements were used in the culture medium. The fold difference values for SPP1 ranged from 4.7±1.37 to 10.71±2.4. If normalization with all 31 RGs was performed, the fold difference was 6.48±1.04 and for the three most stable RGs of the geNorm and NormFinder normalization, the fold difference was 6.4±1.03 and 6.44±1.03. The results show a clear and significant underestimation of the SPP1 expression in the normalization with GAPDH (fold change of 4.7±1.37) and a strong and highly significant overestimation in the normalization with GADD45A (fold change of 10.71±2.4). The fold changes in the normalization with RPL37A, TFRC, and MT-ATP6 were also significantly higher than in the normalization with all RGs or with the three most stable RGs of the geNorm and NormFinder analysis.
Depending on which RG was normalized, the fold changes of IBSP ranged from 3.17±0.78 to 7.37±1.91 after 3D cultivation. The normalization with the three most stable RGs of the geNorm and NormFinder analysis and all RGs resulted in a fold change of 4.47±1.04, 4.42±1.04, and 4.44±1.05, respectively. Again in the normalization with GAPDH, a significant underestimation of the fold change was found with 3.17±0.78 compared with the normalization with all RGs or with the three most stable ones of geNorm and NormFinder. A significant overvaluation in gene expression was found when normalization was performed with GADD45A (7.37±1.91) and TFRC (5.75±0.99) compared with the normalization with all RGs or with the three most stable ones of geNorm and NormFinder.
The osteogenic target genes RUNX-2 and BGLAP (Fig. 4) were not regulated by 3D cultivation compared with 2D cultivated BM-MSCs. Fold differences of 1.01±0.05, 1.00±0.05, and 1.01±0.06 were measured for RUNX-2 when normalized to all RG, geNorm, or NormFinder, respectively. A significant (p<0.05) underestimation of RUNX-2 expression was detected when normalization with GAPDH (0.74±0.21), ELF1 (0.85±0.12), and PSMC4 (0.91±0.1) was chosen compared with the normalization with all RGs or with the three most stable ones of geNorm and NormFinder. A highly significant (p<0.001) overestimation of the fold difference was measured when normalized with GADD45A (1.68±0.32), TFRC (1.32±0.17), and MT-ATP6 (1.24±0.19) compared with fold differences of the normalization with all RGs or with the three most stable ones of geNorm and NormFinder. When RPL37A (1.44±0.38) was taken for normalization, a significant difference of the fold change was measured compared with the normalization with all RGs (with p<0.01) or with the three most stable ones of geNorm and NormFinder (with p<0.05). A significant difference (p<0.05) of RUNX-2 expression was measured when normalization was performed with TBP and B2M but only compared with the normalization with the three most stable RG of the geNorm analysis.
Gene expression of BGLAP was not regulated by 3D cultivation as seen by a fold difference of 1.05±0.06, 1.04±0.06, and 1.05±0.06 when normalized to all RGs, geNorm, or NormFinder, respectively (Fig. 4). A significant underestimation of the fold difference was measured for the normalization with GAPDH (0.77±0.2) and CASC3 (0.91±0.15) compared with the normalization with all RGs or with the three most stable RGs of geNorm and NormFinder. When normalized to ELF1 (0.89±0.2) and PSMC4 (0.95±0.12), a significant underestimation of the fold difference was detected compared with the normalization with the three most stable RGs of NormFinder and all RGs. A highly significant (p<0.001) overestimation of the fold difference was measured when normalized with GADD45A (1.75±0.32), TFRC (1.38±0.17), and MT-ATP6 (1.29±0.39) compared with fold differences of the normalization with all RGs or with the three most stable ones of geNorm and NormFinder. The normalization to MT-ATP6 (1.29±0.39, p<0.01) and RPL37A (1.51±0.45, p<0.05) resulted in a significant higher expression of BGLAP compared with the three other normalization methods. Compared with the normalization with geNorm, a significant overvaluation of the fold change with the normalization of TBP and B2M was detected.
The osteogenic target gene ALPL was downregulated by the 3D cultivation method compared with conventional 2D cultured BM-MSCs when normalized to all RG (0.74±0.06), geNorm (0.73±0.06), and NormFinder (0.74±0.06) (Fig. 4). An overestimation of downregulation was detected for the normalization with GAPDH with a fold change of 0.54±0.15, p<0.01 when compared with the normalization methods mentioned earlier. When ELF1 was used as an internal control, a significant difference in the fold change of ALPL (0.62±0.14) was measured compared with the normalization with all RGs and the three most stable RGs of the NormFinder analysis. When normalized to CASC3 (0.64±0.12) and PSMC4 (0.67±.09), a significant overestimation of downregulation was detected compared with the normalization with all RGs. A highly significant and significant overestimation of the fold changes for ALPL was measured when normalized to the potential RGs GADD45 (1.23±0.25, p<0.001), TFRC (0.97±0.13, p<0.001), RPL37A (1.06±0.31, p<0.05), and MT-ATP6 (0.91±0.15, p<0.01 for GeNorm and NormFinder, p<0.05 for all RG) compared with the fold differences of the normalization with all RGs or with the three most stable ones of geNorm and NormFinder. The normalization with GADD45 even resulted in a seemingly upregulation of expression of the osteogenic target gene ALPL.
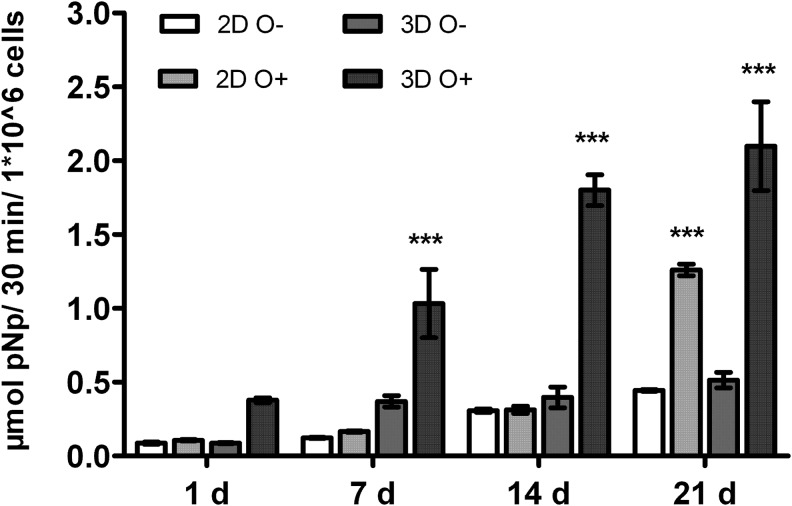
Effect of 3D cultivation on the level of cell-specific ALP activity
The level of cell-specific ALP activity was measured after 1, 7, 14, and 21 days of 3D and 2D cultivation with or without osteogenic supplements (Fig. 5). Three-dimensional cultivation itself did not increase the ALP activity level significantly (day 14 and 21: 0.4±0.12 and 0.51±0.09 μmol pNp/30 min/1×106 cells) compared with the ALP activity level in 2D cultivated BM-MSCs (day 14 and 21: 0.31±0.02 and 0.44±0.01 μmol pNp/30 min/1×106 cells). A highly significant increase of ALP activity was detected after 7 (1.03±0.4 μmol pNp/30 min/1×106 cells), 14 (1.8±0.18 μmol pNp/30 min/1×106 cells), and 21 days (2.1±0.52 μmol pNp/30 min/1×106 cells) in 3D culture with the osteogenic supplements in the medium compared with all other culture conditions. On day 21, we could measure a significant increase of the ALP activity level in 2D osteogenically stimulated BM-MSCs (1.26±0.07 μmol pNp/30 min/1×106 cells) compared with 2D BM-MSCs without osteogenic supplements (0.44±0.01 μmol pNp/30 min/1×106 cells).
FIG. 5.
Activity of alkaline phosphatase in two- and three-dimensionally cultured bone marrow-derived mesenchymal stromal cells over a cultivation period of 21 days with and without osteogenic supplements in culture medium (mean±standard deviation). Significance was calculated with 2-way ANOVA and Bonferroni post-tests with ***p<0.001.
Discussion
The principles of TE are widely used for the concepts of complex tissue regeneration.36 Aiming at musculoskeletal repair, the cultivation of autologous human MSCs on 3D matrices is a basic prerequisite.3 Fluorescence-based qPCR is the most commonly used molecular technology for the quantification of gene expression.20 To obtain consistent and biologically relevant gene expression data, the use of stable RGs is absolutely essential. Numerous candidate RGs have been reported for a wide variety of human cell lines and primary cells under different experimental conditions, but almost exclusively under standard 2D cultivation. However, analysis of RGs for the 3D culture of human BM-MSCs frequently used in the field of TE research has remained insufficient.
In this study, we compared gene expression stability of n=31 putative RGs from 3D cultivated human BM-MSCs of six different donors compared with the conventional 2D cultivation using qRT-PCR. Since no standardized method for the selection of the most stable RGs exists, we analyzed gene expression data with the two well-established algorithms geNorm and NormFinder. In addition, we calculated the CV by comparing the degree of variation between genes. TBP, TFRC, and HPRT1 were identified as the three most stable RGs for the 3D culture of human BM-MSCs. In line with our results, Foldager et al. found that TBP was the RG with the highest expression stability in 3D cultivated human chondrocytes.29 In a study on human adipose-derived stem cells during hypoxic culture, osteogenic and chondrogenic differentiation, Fink et al. identified TBP being among the most stable RGs.28 The present data are in line with an earlier study of Kulkarni et al., in which TBP and HPRT1 were determined to be the most stable RGs in human cornea (46). Recently, we identified TBP as the most stable RG in a study of 13 patients with idiopathic hip osteoarthritis compared with 15 age-matched healthy donors.37
The transferrin receptor (p90, CD71) encoded by the TFRC gene is required for iron delivery from transferrin to cells. Higher expression of TFRC has been identified as a negative prognostic marker for numerous tumor types and lymphomas.38 Majidzadeh et al. verified TFRC and ACTB as the best combination of RGs analyzing tissue of human breast cancer patients.39 In contrast, Cabiati et al. identified TFRC as the least stable RG among ten candidates in cardiac and pulmonary tissues of obese and hyperglycemic rats.40 The contrary results of TFRC gene expression stability depending on which species and tissue was analyzed show the importance of verifying the stability of internal control genes for each experimental setup. We do not recommend using TFRC as a RG for the combined analysis of 2D and 3D cultivated BM-MSCs, as the gene expression was significantly upregulated for BM-MSCs cultivated under 2D conditions. Furthermore, the stability values of TFRC were rather high using geNorm and NormFinder analyses of the combined 3D/2D dataset that correspond to low gene expression stability.
Using the CV for calculation of expression stability, we identified HPRT1, CASC3, and CDKN1A as the most stable RGs. When analyzing the combined gene expression data of 2D and 3D cultivated BM-MSCs by geNorm, we found that HPRT1, HMBS, and PPIA; and in NormFinder analysis, HPRT1, PPIA, and MRPL19 were the three most stable RGs. The present results demonstrate that regardless of the analysis method, cultivation method, or data set (3D or 2D+3D) used, HPRT1 was one of the most stable RGs (3D, combined 2D/3D). These findings are consistent with a study of Amable et al., who showed that HPRT1 was the RG with the highest stability in human bone marrow-derived MSCs and dermal fibroblasts under different expansion and differentiation conditions.41 PPIA gene encoding for the PPIA is responsible for protein folding processes and was identified as the second and third most stable RG in the combined analysis of 2D and 3D cultured BM-MSCs. In line with our findings, Rienzo et al. determined PPIA as an appropriate internal RG in endothelial and osteosarcoma cells for tumor neovascularization studies.42 Accordingly, Sorby et al. identified HPRT1 and PPIA to be suitable for normalizing gene expression data in metastatic and nonmetastatic colon cancer patients.43 The usefulness of PPIA as a RG has also been validated in other studies.44,45 In a 3D hydrogel cell culture system with alginate and porcine MSCs, PPIA and TBP were selected as the most stable RGs according to geNorm, Bestkeeper, and NormFinder analysis.46
Commonly used RGs, for example, GAPDH, ACTB, and 18sRNA, are frequently used for gene expression studies but are often less stable under certain experimental conditions.26,30,47–51 In our study, all three analyzing methods, geNorm, NormFinder, and CV, identified ABL1, ACTB, and RPL37A as the three least stable RGs in 3D cultivated BM-MSCs. In the combined analysis of 2D and 3D cultivated BM-MSCs, the well-established RGs GAPDH and ACTB and the gene encoding for the ribosomal protein RPL37A were among the three least stable RGs when analyzed by geNorm algorithm. In a subset of human ovarian cancer cell lines, the most frequently used GAPDH was among the least stably expressed RGs.52 De Jonge et al. demonstrated that none of the commonly used RGs (ACTB, GAPDH, HPRT1, and B2M) were among the top 50 most stable RGs after a meta-analysis of 13,629 human gene arrays.26 In addition, Liu et al. showed that ACTB was the least stable RG in MSCs under dynamic hydrostatic pressure and postulated that cell deformation is expected to perturb the cytoskeleton.53 Wang et al. found that YWHAZ and RPL13A, not ACTB or GAPDH, were the most stably expressed RGs in different passages of human umbilical cord mesenchymal stem cells.51 The low gene expression stability of GAPDH and ACTB is consistent with our findings, where the 3D cultivation of BM-MSCs resulted in a significantly higher gene expression of GAPDH compared with 2D and influenced the expression stability of structural gene ACTB being the second least stable RG in the analysis. Sturzenbaum and Kille postulated that the use of ACTB is principally not advisable in situations where tissues undergo extensive morphological changes.54 The ribosomal gene RPL37A was revealed as the least stable one under 3D conditions, regardless of which analysis program was used. In contrast to our results, Maess et al. identified RPL37A as the most stable RG when measuring gene expression during THP-1 monocyte differentiation.55 In our analysis, the RG ABL1, a homologue of the transforming gene of the Abelson murine leukemia virus and a proto-oncogene, was among the three least stable RGs. Interestingly, this result is consistent with the findings of Kolkova et al., where ABL1 turned out to be the least stable gene found by NormFinder analysis in human ovarian tumor tissue.56
The combined analysis of 2D and 3D cultivated BM-MSCs with NormFinder again determined not only GAPDH, but also GADD45A and RPL37A as the least stable RGs. In contrast, the DNA-damage-inducible gene GADD45A was identified as the most stable RG in a previous investigation of our group in osteogenically induced BM-MSCs (32). Since the gene expression of GADD45A was significantly downregulated in 3D culture of BM-MSCs in comparison to 2D, we do not recommend the use of GADD45A as an internal control for gene expression analysis. Our results show that there is no gold standard RG which can be widely used for qRT-PCR normalization in human BM-MSC experiments.
The choice of stable RGs is crucial for the correct interpretation of target gene expression obtained by qRT-PCR. This was shown by several studies when normalizing gene expression data by substituting different RGs, which resulted in distinct transcript variation.31,43,57–59 When evaluating the osteogenic response of 3D cultivated BM-MSCs on cancellous bone compared with 2D by measuring the gene expression of the osteogenic target genes RUNX-2, ALPL, SPP1, IBSP, and BGLAP, we detected significant over- and underestimation of the fold change values when analyzing with less stable RGs, for example, GAPDH, GADD45A, and RPL37A. As proposed by Vandesompele et al., the use of multiple RGs enables a more accurate normalization of gene expression data.24 In addition, Jacob et al. recommended the use of ideally three RGs selected by at least three stability algorithms.52 To analyze the target gene expression levels in our study, we choose a combination of the three most stable RGs according to NormFinder and geNorm analyses or a combination of all 31 RGs. The fold changes of the osteogenic target gene SPP1, also known as osteopontin, being responsible for extracellular matrix binding, ranged from 4.7 when normalized to GAPDH to 10.7 when GADD45A was used as an internal control. The normalization with the three most stable RG from the geNorm and NormFinder analyses resulted in a fold change of 6.4 for SPP1. This clearly shows the huge impact of choosing an accurate RG and the risk of over- or underestimation of target gene expression differences when using less stable RGs. The target genes RUNX-2, ALPL, SPP1, and BGLAP showed significant differences in fold change values when analyzed using GAPDH, GADD45A, TFRC, RPL37A, and MT-ATP6 as RGs compared with the analysis with the three most stable RGs from the geNorm and NormFinder analysis or with all 31 RGs. While the gene expression level of GAPDH was significantly increased by the 3D cultivation of BM-MSCs, the levels of GADD45A, TFRC, and MT-ATP6 were significantly decreased compared with 2D BM-MSCs.
The induction of the differentiation toward the osteogenic lineage shown by an increase of SPP1 and IBSP gene expression levels by six and four-fold differences, respectively, was only induced by the 3D cultivation of BM-MSCs. No osteogenic supplements were used for cultivation. The osteogenic target genes RUNX-2 and BGLAP were not regulated, which may be due to the time point of analyses (14 days), as RUNX-2 is an early marker and BGLAP is a late marker of osteoblastic differentiation.60 The upregulation of SPP1 gene expression was shown earlier by our group, where BM-MSCs were cultivated on the same matrices and gene expression was measured by microarray and qRT-PCR technology.61 The results are in line with a recent study of Kabiri et al., who observed upregulated BMP-2 expression levels in BM-MSCs cultured as 3D-microaggregates when compared with 2D-monolayer enhancing osteogenic differentiation.62 Neither the 3D environment influenced the gene expression level of the osteogenic target gene ALPL, nor did we detect an increase of the corresponding protein activity. ALP activity levels were only significantly enhanced by the use of osteogenic supplements under 2D conditions on day 21 but already elevated after 7 days in combination with 3D culture increasing over the course of time (14, 21 days).
In a genome-wide gene expression analysis of cancer cells, Zschenker and co-authors observed significant changes in gene expression profiles of 3D as compared with 2D cell culture conditions in extracellular matrix genes.63 Notably, next to the 3D cultivation method itself, RG expression can be additionally dependent on the scaffold biomaterial. This was shown by Chooi et al., who investigated the stability of RGs in rabbit chondrocytes cultured on alginate versus agarose matrices.46 Interestingly, they identified the same RGs, that is, TBP and HPRT1, as the most stable ones using a 3D agarose matrix as was shown in this study using HCB as a scaffold. On alginate scaffolds TBP, RPL5, and RPL18 were selected as the most stable RGs. In addition to the 3D environment, the stiffness of the substrate can have an impact on RG expression. In a study investigating the effect of subendothelial matrix stiffening on endothelial cell function, Chen et al. found that B2M, HPRT1, and YWHAZ are a set of stably expressed RGs. The authors do not recommend the use of GAPDH and ACTB due to their low expression stabilities.64
Taken together, testing the stability of RG candidates is crucial for each experimental setup and should be performed before the analysis of gene expression data. The choice of unfavorable, less stable RGs will create biased results, based on misleading fold differences and can lead to misinterpretation of research results.
Conclusions
In conclusion, 3D cultivation has a strong influence on the gene expression of several candidate RGs in human BM-MSCs. Our results suggest the combined use of TBP, TFRC, and HPRT1 for the normalization of gene expression data from 3D cultivated human BM-MSCs. For a combined analysis of 2D and 3D cultivated BM-MSCs, HPRT1, HMBS, and PPIA were the most stable RGs. In contrast, the commonly used RGs ACTB, GAPDH as well as ABL1 and RPL37A proved disadvantageous in our setting and would lead to a misinterpretation of gene expression data. Furthermore, we found that the osteogenic target genes SPP1 and IBSP were upregulated by a 3D environment compared with 2D cultivation of BM-MSCs without the use of osteogenic medium supplements. In summary, it is essential to test RG expression stability to control for RG-based errors in gene expression analysis depending on the experimental setting.
Acknowledgments
The authors are grateful for the technical assistance rendered by Cornelia Liebers as well as by Prof. Martin Bornhäuser, MD and to Katrin Müller of the medical clinic I, University Hospital Carl Gustav Carus at Technische Universität Dresden for providing the BM-MSCs. Gratitude is expressed to Dr. Denis Corbeil of the Biotechnology Center at Technische Univeristät Dresden for providing laboratory infrastructure. This project was supported by the German Academic Exchange Service/Federal Ministry of Education and Research (D/09/04774) and the DFG-Center for Regenerative Therapies Dresden at the Technische Universität Dresden (Seed Grant, 043_261578).
Disclosure Statement
No competing financial interests exist.
References
- 1.Amini A.R., Laurencin C.T., and Nukavarapu S.P.Bone tissue engineering: recent advances and challenges. Crit Rev Biomed Eng 40,363, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caplan A.I.Review: mesenchymal stem cells: cell-based reconstructive therapy in orthopedics. Tissue Eng 7/8,1198, 2005 [DOI] [PubMed] [Google Scholar]
- 3.Noth U., Rackwitz L., Steinert A.F., and Tuan R.S.Cell delivery therapeutics for musculoskeletal regeneration. Adv Drug Deliv Rev 62,765, 2010 [DOI] [PubMed] [Google Scholar]
- 4.Pittenger M.F., Mackay A.M., Beck S.C., Jaiswal R.K., Douglas R., Mosca J.D., Moorman M.A., Simonetti D.W., Craig S., and Marshak D.R.Multilineage potential of adult human mesenchymal stem cells. Science 284,143, 1999 [DOI] [PubMed] [Google Scholar]
- 5.Dominici M., Le B.K., Mueller I., Slaper-Cortenbach I., Marini F., Krause D., Deans R., Keating A., Prockop D., and Horwitz E.Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 8,315, 2006 [DOI] [PubMed] [Google Scholar]
- 6.Campagnoli C., Roberts I.A., Kumar S., Bennett P.R., Bellantuono I., and Fisk N.M.Identification of mesenchymal stem/progenitor cells in human first-trimester fetal blood, liver, and bone marrow. Blood 98,2396, 2001 [DOI] [PubMed] [Google Scholar]
- 7.da Silva M.L., Chagastelles P.C., and Nardi N.B.Mesenchymal stem cells reside in virtually all post-natal organs and tissues. J Cell Sci 119,2204, 2006 [DOI] [PubMed] [Google Scholar]
- 8.Kern S., Eichler H., Stoeve J., Kluter H., and Bieback K.Comparative analysis of mesenchymal stem cells from bone marrow, umbilical cord blood, or adipose tissue. Stem Cells 24,1294, 2006 [DOI] [PubMed] [Google Scholar]
- 9.Sakaguchi Y., Sekiya I., Yagishita K., and Muneta T.Comparison of human stem cells derived from various mesenchymal tissues: superiority of synovium as a cell source. Arthritis Rheum 52,2521, 2005 [DOI] [PubMed] [Google Scholar]
- 10.Frith J.E., Thomson B., and Genever P.G.Dynamic three-dimensional culture methods enhance mesenchymal stem cell properties and increase therapeutic potential. Tissue Eng Part C Methods 16,735, 2010 [DOI] [PubMed] [Google Scholar]
- 11.Bartosh T.J., Ylostalo J.H., Mohammadipoor A., Bazhanov N., Coble K., Claypool K., Lee R.H., Choi H., and Prockop D.J.Aggregation of human mesenchymal stromal cells (MSCs) into 3D spheroids enhances their antiinflammatory properties. Proc Natl Acad Sci U S A 107,13724, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Birgersdotter A., Sandberg R., and Ernberg I.Gene expression perturbation in vitro—a growing case for three-dimensional (3D) culture systems. Semin Cancer Biol 15,405, 2005 [DOI] [PubMed] [Google Scholar]
- 13.Pedersen J.A., and Swartz M.A.Mechanobiology in the third dimension. Ann Biomed Eng 33,1469, 2005 [DOI] [PubMed] [Google Scholar]
- 14.Cukierman E., Pankov R., Stevens D.R., and Yamada K.M.Taking cell-matrix adhesions to the third dimension. Science 294,1708, 2001 [DOI] [PubMed] [Google Scholar]
- 15.Saleh F.A., and Genever P.G.Turning round: multipotent stromal cells, a three-dimensional revolution? Cytotherapy 13,903, 2011 [DOI] [PubMed] [Google Scholar]
- 16.Schmeichel K.L., and Bissell M.J.Modeling tissue-specific signaling and organ function in three dimensions. J Cell Sci 116,2377, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bustin S.A., Benes V., Nolan T., and Pfaffl M.W.Quantitative real-time RT-PCR-a perspective. J Mol Endocrinol. 34,597, 2005 [DOI] [PubMed] [Google Scholar]
- 18.Bustin S.A.Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J Mol Endocrinol 29,23, 2002 [DOI] [PubMed] [Google Scholar]
- 19.Nolan T., Hands R.E., and Bustin S.A.Quantification of mRNA using real-time RT-PCR. Nat Protoc 1,1559, 2006 [DOI] [PubMed] [Google Scholar]
- 20.Bustin S.A., Benes V., Garson J., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G., Wittwer C.T., Schjerling P., Day P.J., Abreu M., Aguado B., Beaulieu J.F., Beckers A., Bogaert S., Browne J.A., Carrasco-Ramiro F., Ceelen L., Ciborowski K., Cornillie P., Coulon S., Cuypers A., De B.S., De C.L., De C.J., De N.H., De S.W., Deckers K., Dheedene A., Durinck K., Ferreira-Teixeira M., Fieuw A., Gallup J.M., Gonzalo-Flores S., Goossens K., Heindryckx F., Herring E., Hoenicka H., Icardi L., Jaggi R., Javad F., Karampelias M., Kibenge F., Kibenge M., Kumps C., Lambertz I., Lammens T., Markey A., Messiaen P., Mets E., Morais S., Mudarra-Rubio A., Nakiwala J., Nelis H., Olsvik P.A., Perez-Novo C., Plusquin M., Remans T., Rihani A., Rodrigues-Santos P., Rondou P., Sanders R., Schmidt-Bleek K., Skovgaard K., Smeets K., Tabera L., Toegel S., Van A.T., Van den B.W., Van der M.J., Van G.M., Van P.G., Van P.M., Van R.N., Vergult S., Wauman J., Tshuikina-Wiklander M., Willems E., Zaccara S., Zeka F., and Vandesompele J.The need for transparency and good practices in the qPCR literature. Nat Methods 10,1063, 2013 [DOI] [PubMed] [Google Scholar]
- 21.Andersen C.L., Jensen J.L., and Orntoft T.F.Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64,5245, 2004 [DOI] [PubMed] [Google Scholar]
- 22.Akilesh S., Shaffer D.J., and Roopenian D.Customized molecular phenotyping by quantitative gene expression and pattern recognition analysis. Genome Res 13,1719, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pfaffl M.W., Tichopad A., Prgomet C., and Neuvians T.P.Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol Lett 26,509, 2004 [DOI] [PubMed] [Google Scholar]
- 24.Vandesompele J., De P.K., Pattyn F., Poppe B., Van R.N., De P.A., and Speleman F.Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3,34.1–34.12, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monaco E., Bionaz M., de Lima A.S., Hurley W.L., Loor J.J., and Wheeler M.B.Selection and reliability of internal reference genes for quantitative PCR verification of transcriptomics during the differentiation process of porcine adult mesenchymal stem cells. Stem Cell Res Ther 1,1, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Jonge H.J., Fehrmann R.S., de Bont E.S., Hofstra R.M., Gerbens F., Kamps W.A., de Vries E.G., van der Zee A.G., te Meerman G.J., and ter E.A.Evidence based selection of housekeeping genes. PLoS One 2,e898, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Curtis K.M., Gomez L.A., Rios C., Garbayo E., Raval A.P., Perez-Pinzon M.A., and Schiller P.C.EF1alpha and RPL13a represent normalization genes suitable for RT-qPCR analysis of bone marrow derived mesenchymal stem cells. BMC Mol Biol 11,61, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fink T., Lund P., Pilgaard L., Rasmussen J.G., Duroux M., and Zachar V.Instability of standard PCR reference genes in adipose-derived stem cells during propagation, differentiation and hypoxic exposure. BMC Mol Biol 9,2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Foldager C.B., Munir S., Ulrik-Vinther M., Soballe K., Bunger C., and Lind M.Validation of suitable house keeping genes for hypoxia-cultured human chondrocytes. BMC Mol Biol 10,2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmittgen T.D., and Zakrajsek B.A.Effect of experimental treatment on housekeeping gene expression: validation by real-time, quantitative RT-PCR. J Biochem Biophys Methods 46,69, 2000 [DOI] [PubMed] [Google Scholar]
- 31.Jacobi A., Rauh J., Bernstein P., Liebers C., Zou X., and Stiehler M.Comparative analysis of reference gene stability in human mesenchymal stromal cells during osteogenic differentiation. Biotechnol Prog 29,1034, 2013 [DOI] [PubMed] [Google Scholar]
- 32.Oswald J., Boxberger S., Jorgensen B., Feldmann S., Ehninger G., Bornhauser M., and Werner C.Mesenchymal stem cells can be differentiated into endothelial cells in vitro. Stem Cells 22,377, 2004 [DOI] [PubMed] [Google Scholar]
- 33.Pruss A., Baumann B., Seibold M., Kao M., Tintelnot K., von Versen R., Radtke H., Dorner T., Pauli G., and Gobel U.B.Validation of the sterilization procedure of allogeneic avital bone transplants using peracetic acid-ethanol. Biologicals 29,59, 2001 [DOI] [PubMed] [Google Scholar]
- 34.Pfaffl M.W.A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29,e45, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Novak J.P., Sladek R., and Hudson T.J.Characterization of variability in large-scale gene expression data: implications for study design. Genomics 79,104, 2002 [DOI] [PubMed] [Google Scholar]
- 36.Atala A., Kasper F.K., and Mikos A.G.Engineering complex tissues. Sci Transl. Med 4,160rv12, 2012 [DOI] [PubMed] [Google Scholar]
- 37.Schildberg T., Rauh J., Bretschneider H., and Stiehler M.Identification of suitable reference genes in bone marrow stromal cells from osteoarthritic donors. Stem Cell Res 11,1288, 2013 [DOI] [PubMed] [Google Scholar]
- 38.Ploszynska A., Ruckemann-Dziurdzinska K., Jozwik A., Mikosik A., Lisowska K., Balcerska A., and Witkowski J.M.Cytometric evaluation of transferrin receptor 1 (CD71) in childhood acute lymphoblastic leukemia. Folia Histochem. Cytobiol 50,304, 2012 [DOI] [PubMed] [Google Scholar]
- 39.Majidzadeh A., Esmaeili R., and Abdoli N.TFRC and ACTB as the best reference genes to quantify Urokinase Plasminogen Activator in breast cancer. BMC Res Notes 4,215, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cabiati M., Raucci S., Caselli C., Guzzardi M.A., D'Amico A., Prescimone T., Giannessi D., and Del R. S.Tissue-specific selection of stable reference genes for real-time PCR normalization in an obese rat model. J Mol Endocrinol 48,251, 2012 [DOI] [PubMed] [Google Scholar]
- 41.Amable P.R., Teixeira M.V., Carias R.B., Granjeiro J.M., and Borojevic R.Identification of appropriate reference genes for human mesenchymal cells during expansion and differentiation. PLoS One 8,e73792, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rienzo M., Schiano C., Casamassimi A., Grimaldi V., Infante T., and Napoli C.Identification of valid reference housekeeping genes for gene expression analysis in tumor neovascularization studies. Clin Transl Oncol 15,211, 2013 [DOI] [PubMed] [Google Scholar]
- 43.Sorby L.A., Andersen S.N., Bukholm I.R., and Jacobsen M.B.Evaluation of suitable reference genes for normalization of real-time reverse transcription PCR analysis in colon cancer. J Exp Clin Cancer Res 29,144, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tatsumi K., Ohashi K., Taminishi S., Okano T., Yoshioka A., and Shima M.Reference gene selection for real-time RT-PCR in regenerating mouse livers. Biochem Biophys Res Commun 374,106, 2008 [DOI] [PubMed] [Google Scholar]
- 45.Nishimura M., Nikawa T., Kawano Y., Nakayama M., and Ikeda M.Effects of dimethyl sulfoxide and dexamethasone on mRNA expression of housekeeping genes in cultures of C2C12 myotubes. Biochem Biophys Res Commun 367,603, 2008 [DOI] [PubMed] [Google Scholar]
- 46.Chooi W.H., Zhou R., Yeo S.S., Zhang F., and Wang D.A.Determination and validation of reference gene stability for qPCR analysis in polysaccharide hydrogel-based 3D chondrocytes and mesenchymal stem cell cultural models. Mol Biotechnol 54,623, 2013 [DOI] [PubMed] [Google Scholar]
- 47.Bas A., Forsberg G., Hammarstrom S., and Hammarstrom M.L.Utility of the housekeeping genes 18S rRNA, beta-actin and glyceraldehyde-3-phosphate-dehydrogenase for normalization in real-time quantitative reverse transcriptase-polymerase chain reaction analysis of gene expression in human T lymphocytes. Scand J Immunol 59,566, 2004 [DOI] [PubMed] [Google Scholar]
- 48.Suzuki T., Higgins P.J., and Crawford D.R.Control selection for RNA quantitation. Biotechniques 29,332, 2000 [DOI] [PubMed] [Google Scholar]
- 49.Kulkarni B., Mohammed I., Hopkinson A., and Dua H.S.Validation of endogenous control genes for gene expression studies on human ocular surface epithelium. PLoS One 6,e22301, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pfister C., Tatabiga M.S., and Roser F.Selection of suitable reference genes for quantitative real-time polymerase chain reaction in human meningiomas and arachnoidea. BMC Res Notes 4,275, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Y., Han Z., Yan S., Mao A., Wang B., Ren H., Chi Y., and Han Z.Evaluation of suitable reference gene for real-time PCR in human umbilical cord mesenchymal stem cells with long-term in vitro expansion. In Vitro Cell Dev Biol Anim 46,595, 2010 [DOI] [PubMed] [Google Scholar]
- 52.Jacob F., Guertler R., Naim S., Nixdorf S., Fedier A., Hacker N.F., and Heinzelmann-Schwarz V.Careful selection of reference genes is required for reliable performance of RT-qPCR in human normal and cancer cell lines. PLoS One 8,e59180, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu J., Zou L., Wang J., and Zhao Z.Validation of beta-actin used as endogenous control for gene expression analysis in mechanobiology studies. Stem Cells 27,2371, 2009 [DOI] [PubMed] [Google Scholar]
- 54.Sturzenbaum S.R., and Kille P.Control genes in quantitative molecular biological techniques: the variability of invariance. Comp Biochem Physiol B Biochem Mol Biol 130,281, 2001 [DOI] [PubMed] [Google Scholar]
- 55.Maess M.B., Sendelbach S., and Lorkowski S.Selection of reliable reference genes during THP-1 monocyte differentiation into macrophages. BMC Mol Biol 11,90, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kolkova Z., Arakelyan A., Casslen B., Hansson S., and Kriegova E.Normalizing to GADPH jeopardises correct quantification of gene expression in ovarian tumours—IPO8 and RPL4 are reliable reference genes. J Ovarian Res 6,60, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Caradec J., Sirab N., Keumeugni C., Moutereau S., Chimingqi M., Matar C., Revaud D., Bah M., Manivet P., Conti M., and Loric S.‘Desperate house genes’: the dramatic example of hypoxia. Br J Cancer 102,1037, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Piehler A.P., Grimholt R.M., Ovstebo R., and Berg J.P.Gene expression results in lipopolysaccharide-stimulated monocytes depend significantly on the choice of reference genes. BMC Immunol 11,21, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rho H.W., Lee B.C., Choi E.S., Choi I.J., Lee Y.S., and Goh S.H.Identification of valid reference genes for gene expression studies of human stomach cancer by reverse transcription-qPCR. BMC Cancer 10,240, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Galli C., Passeri G., and Macaluso G.M.Osteocytes and WNT: the mechanical control of bone formation. J Dent Res 89,331, 2010 [DOI] [PubMed] [Google Scholar]
- 61.Stiehler M., Seib F.P., Rauh J., Goedecke A., Werner C., Bornhauser M., Gunther K.P., and Bernstein P.Cancellous bone allograft seeded with human mesenchymal stromal cells: a potential good manufacturing practice-grade tool for the regeneration of bone defects. Cytotherapy 12,658, 2010 [DOI] [PubMed] [Google Scholar]
- 62.Kabiri M., Kul B., Lott W.B., Futrega K., Ghanavi P., Upton Z., and Doran M.R.3D mesenchymal stem/stromal cell osteogenesis and autocrine signalling. Biochem Biophys Res Commun 419,142, 2012 [DOI] [PubMed] [Google Scholar]
- 63.Zschenker O., Streichert T., Hehlgans S., and Cordes N.Genome-wide gene expression analysis in cancer cells reveals 3D growth to affect ECM and processes associated with cell adhesion but not DNA repair. PLoS One 7,e34279, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen G., Zhao L., Feng J., You G., Sun Q., Li P., Han D., and Zhou H.Validation of reliable reference genes for real-time PCR in human umbilical vein endothelial cells on substrates with different stiffness. PLoS One 8,e67360, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]