Summary
MicroRNAs (miRNAs) control expression of thousands of genes in plants and animals. miRNAs function by guiding Argonaute proteins to complementary sites in messenger RNAs (mRNAs) targeted for repression. We determined crystal structures of human Argonaute-2 (Ago2) bound to a defined guide RNA with and without target RNAs representing miRNA recognition sites. These structures suggest a stepwise mechanism, in which Ago2 primarily exposes guide nucleotides 2–5 for initial target pairing. Pairing to nt 2–5 promotes conformational changes that expose nt 2–8 and 13–16 for further target recognition. Interactions with the guide-target minor groove allow Ago2 to interrogate target RNAs in a sequence-independent manner, while an adenosine binding-pocket opposite guide nt 1 further facilitates target recognition. Spurious slicing of miRNA targets is avoided through an inhibitory coordination of one catalytic magnesium ion. These results explain the conserved nucleotide pairing patterns in animal miRNA target sites first observed over two decades ago.
Introduction
miRNAs are small (~22 nt) RNAs with regulatory roles in plants and animals. miRNAs function within RNA-Induced Silencing Complexes (RISCs), which contain a member of the Argonaute protein family (1). Argonaute uses the miRNA as a guide for identifying complementary target mRNAs, which then leads to silencing of the targeted messages via translational repression and degradation (2). Over 1,000 miRNAs are encoded in the human genome and more than 30% of mammalian protein-coding genes contain a conserved miRNA target site (3). Consequently, miRNAs contribute to diverse physiological processes in mammals, including epithelial regeneration (4), cardiac function (5), ovulation (6), and the progression of cancer (7).
Perfect complementarity between miRNAs and their targets is not necessary for silencing and some miRNA nucleotides are more important than others (8, 9). Specifically, pairing to the miRNA “seed region” (nt 2–7 or 2–8, from the 5′ end) is the most evolutionarily conserved feature of miRNA targets in animals (10-14). Crystal structures of human Argonaute proteins show nt 2–6 of the guide RNA bound a in prearranged A-form conformation, which was proposed to minimize the entropic cost associated with forming a stable duplex with target RNAs (15-17). However, nucleotides outside of the seed region in these structures were mostly disordered, and no structure of any eukaryotic Argonaute bound to a target RNA has been reported.
Guide RNAs are threaded through the N-PAZ channel
Structural insights into eukaryotic Argonaute proteins have been thwarted by the challenge of separating Argonaute from co-purifying cellular RNAs (15, 18). Although a protocol for purifying RNA-free Ago2 has been reported (19), the method generates relatively small amounts of material and is difficult to reproduce. As an alternative, we adapted a biochemical method for purifying RISC loaded with a specified guide from cell lysates (20) to produce milligram quantities of recombinant Ago2 bound to a defined 21 nt guide RNA. The ratio of Ago2 protein to guide RNA in the purified samples is 1:1.2±0.2 and pre-steady state kinetics showed that 99.0±8.6% of the Ago2 molecules are catalytically active against the specified target RNA (fig. S1).
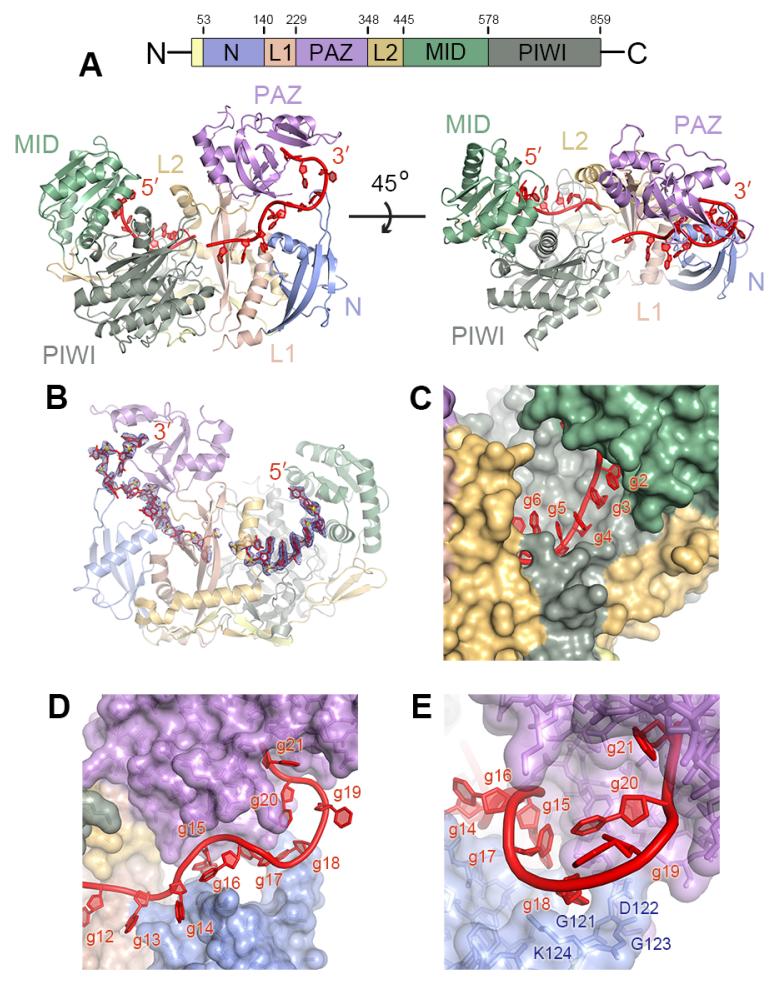
Guide-selected Ago2 samples crystallized in a condition similar to that used to determine structures of human Ago1 and Ago2 (15, 19). However, in contrast to previous structures, we observed unambiguous electron density for most of the guide RNA, with only four guide (g) nucleotides (nt g8–g11) disordered (Fig. 1A, B). The guide 5′ end is anchored in the Ago2 MID domain and nucleotides g2–g7 are splayed in a helical conformation (Fig. 1C). Nucleotides g12–g20 extend from the center of the protein to the 3′ nucleotide-binding site in the PAZ domain (21). Nucleotides g14–g18 are threaded through a narrow channel formed between the PAZ and N domains of Ago2 (Fig. 1D). The channel ends at g18, where the RNA turns and extends into the 3′ binding pocket. Consistent with the ability of Ago2 to bind guide RNAs of any nucleotide sequence, the majority of contacts are made through hydrogen bonds and salt linkages to the RNA sugar-phosphate backbone (fig. S2). Base stacking between g14–g18 is completely disrupted by interactions with the N-PAZ channel, and the Watson-Crick edges of g15, g17 and g18 face the interior of the complex (Fig. 1D). Therefore, in the observed conformation, interactions with Ago2 prevent the 3′ half of the guide RNA from base pairing with complementary targets.
Fig. 1.
Structure of the Ago2-guide complex. (A) Schematic of the Ago2 primary sequence. Front and top views of human Ago2 bound to a defined guide RNA (red). Ago2 contains a large central cleft between two lobes (N-PAZ and MID-PIWI) connected by two linker domains (L1 and L2). (B) Guide RNA omit map contoured at 2 σ (blue mesh). (C) Nucleotides g2-g5 are fully exposed, while Ago2 occludes nucleotides g6 and g7. (D) The 3′ half of the guide is treaded through the N-PAZ channel. (E) View down the N-PAZ channel.
The ordered guide RNA was accompanied by a reduction in the relative temperature factors in the Ago2 N domain compared to previous structures (fig. S3). Improvements in the electron density map allowed us to observe amino acid residues 119–125, which fold into a hairpin loop that forms the end of the N-PAZ channel and directs the guide 3′ end into the PAZ domain through contacts to g18 (Fig. 1E). An expansion of positively charged residues in the analogous loop in Arabidopsis Ago4, Ago6, and Ago9 has been proposed as an explanation for how these Agos preferentially retain longer (~24 nt) guide RNAs (22). The improved map also revealed an error surrounding residues 110-118 in our original Ago2 structures (15) that was propagated to subsequent structures of human Ago1 and Ago2 (16, 19) (figs. S4 and S5).
Structural basis for miRNA target recognition
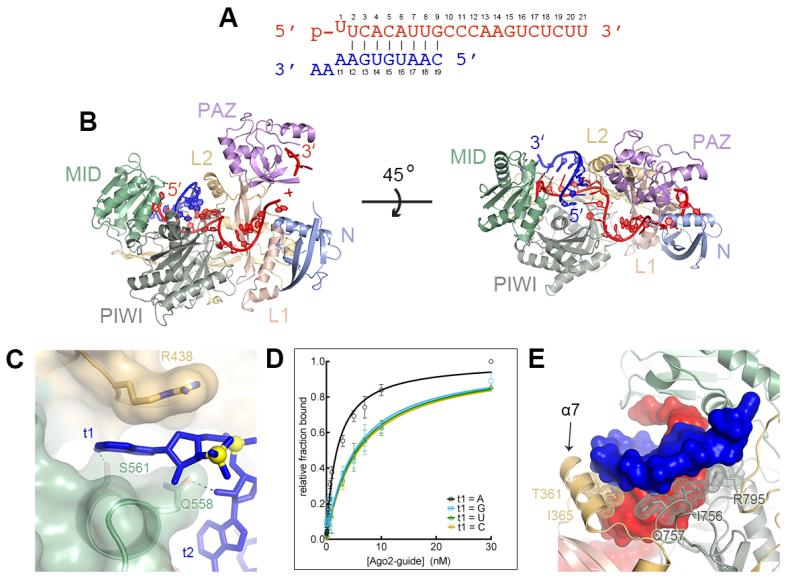
We determined crystal structures of guide-selected Ago2 bound to short target RNAs (11 nt) with complementarity to nucleotides g2–g7, g2–g8, or g2–g9. The target RNAs included an adenosine nucleotide in the t1 position (the target nucleotide opposite g1) and two nucleotides 3′ of t1 (Fig. 2A). Target RNAs bound the complex through Watson-Crick pairing with the seed, forming an A-form duplex in the front half of the Ago2 central cleft (Fig. 2B and S6). The t1-adenine inserts into narrow pocket between the MID and L2 domains of Ago2 where Ser-561 hydrogen bonds to the adenine N6 amine (Fig. 2C). The pocket appears to specifically recognize adenosine nucleotides because a target RNA with a t1-A bound Ago2 with almost 3-fold higher affinity than equivalent targets with U, G or C t1 nucleotides (Fig. 2D). This may explain why t1 adenosines enhance miRNA-mediated repression and are conserved in many mammalian miRNA target sites (3, 11). No electron density was apparent for the two nucleotides 3′ of t1, indicating Ago2 does not make defined contacts to target RNAs downstream of miRNA binding sites.
Fig. 2.
Structure of Ago2 bound to seed-matched target RNAs. (A) Sequences of guide (red) and target RNAs (blue). (B) Front and top views of Ago2 bound to guide and target RNAs. (C) Binding pocket for t1 adenine between L2 and MID domains. (D) Equilibrium binding data for target RNAs bearing different t1 nucleotides. Mean values from ≥ three independent replicates ± standard error shown. (E) Ago2 interrogates the guide-target minor groove. Protein is shown as a ribbon, RNA in surface representation, and interacting side-chains as sticks with dots. Helix-7 (α7) indicated.
Ago2 makes several hydrogen bonds/salt linkages to seed paired target RNAs (fig. S7), but the major mode of recognition by the protein appears to be shape complementarity to the minor groove of the guide-target duplex. Aliphatic segments of residues R795, I756, Q757 within the PIWI domain, and I365 and T361 on helix-7 of the L2 domain line the minor groove, making extensive hydrophobic and van der Waals interactions with positions 2–7 of the guide-target duplex (Fig. 2D). These minor groove contacts may explain why G:U wobble base pairs, in which the guanine unpaired exocyclic amino alters minor groove shape and electrostatic potential (23), reduce Argonaute target affinity and are not commonly observed in miRNA target sites (13, 24, 25).
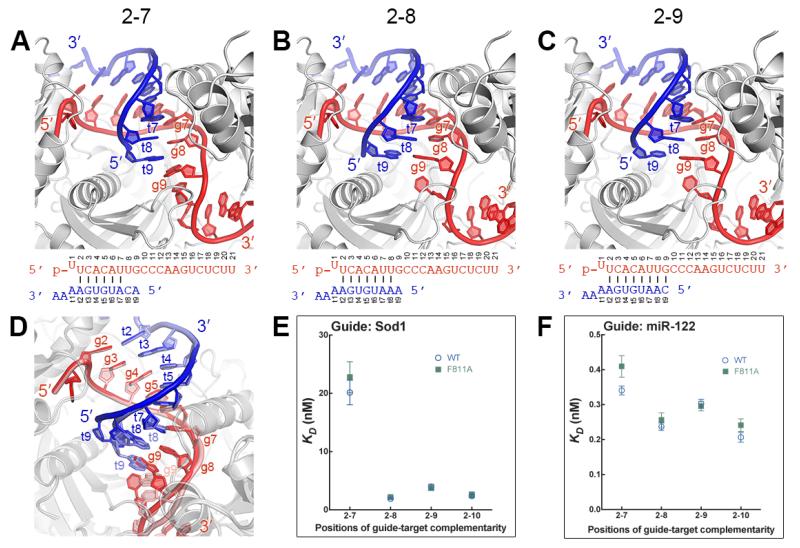
In contrast to positions 2–7, Ago2 does not contact the minor groove at positions 8 and 9, suggesting the protein is more tolerant of mismatches in this region (Fig. 3 A-C). Indeed, the guide-target duplex with g8–g9 mismatches has distortions away from A-form due to staggering of the mismatched bases (Fig. 3A). The distortions are limited to positions 8 and 9, indicating that pairing status at g8 and g9 does not perturb guide-target duplex structure in positions 2–7 (Fig. 3D). However, binding experiments show that pairing to g8 can substantially contribute to the affinity of Ago2 for target RNAs (Fig. 3E and S8). An unrelated guide RNA displayed a smaller difference between the affinities for g2–g7 and g2–g8 complementary target RNAs, revealing that the degree to which pairing to g8 influences target affinity is dependent on the seed sequence (Fig. 3F).
Fig. 3.
Structural analysis of seed-pairing. (A-C) Ago2-guide-target complexes with pairing to g2–g7 (A), g2–g8 (B), or g2–g9 (C). (D) Alignment of g2–g9 structure (guide, red; target, blue) with g2–g7 structure (guide pink, target light blue). (E and F) Dissociation constants of wild type (WT) and F811A Ago2 proteins binding target RNAs with various degrees of guide complementarity. Ago2 was loaded guide RNAs derived from either Sod1 (E) or miR122 (F). Mean of independent triplicates, ±SEM.
Narrowing of the central cleft restricts pairing past g8
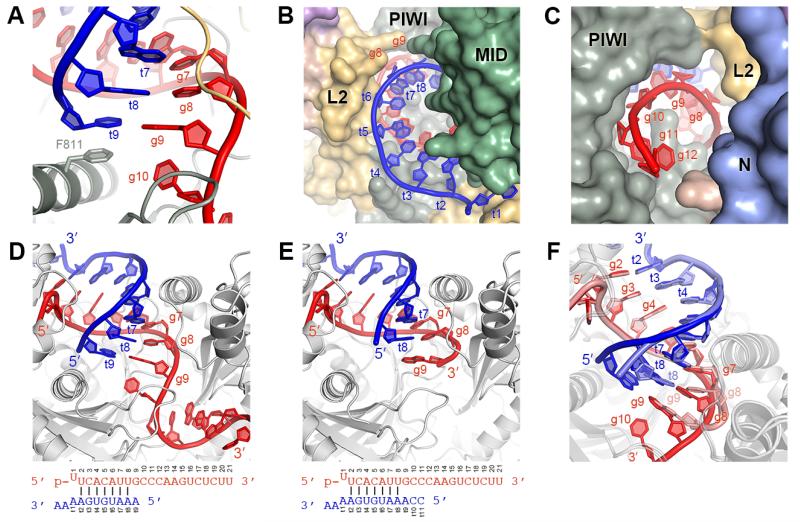
While complementarity to g8 increased the affinity of Ago2 for target RNAs, extending complementarity to g9 and g10 did not enhance affinity further (Fig. 3E–F). In fact, both sequences displayed a modest decrease in affinity for targets complementary to g2–g9 compared to g2–g8, indicating that pairing to g9 is actually detrimental to stability of the Ago2-guide-target complex. t9 stacks against Phe-811 in the 2-9 paired structure (Fig. 4A). However, an F811A mutation did not markedly alter the affinity for full-length target RNAs (Fig. 3E–F). Moreover, the complex lacks sufficient space necessary to accommodate target nucleotides beyond t9 (Fig. 4B-C), and the t9 nucleotide was disordered in crystals containing a longer target RNA, which included mismatches to g11 (Fig. 4D-E). We conclude that the observed conformation of Ago2 can only accommodate pairing to g9 when using short targets that end at t9 (like those used to facilitate crystallization), and that pairing to g9 on longer targets (like those in the 3′ UTR of an mRNA) requires further opening of the Ago2 central cleft. We suggest that opening the cleft involves conformational changes responsible for the decrease in target affinity associated with pairing to g9. This model may explain why complementarity beyond g8 is not a well-conserved feature of vertebrate miRNA targets sites (11).
Fig. 4.
Ago2 restricts guide-target pairing beyond the seed. (A) Phe-811 stacks against t9 of short target RNAs. (B and C) The central cleft viewed from the MID (B) and N domain (C), showing narrowing of the cleft after g8 (g13–g16 omitted for clarity). (D–E) Ago2-guide complex bound to target RNAs paired to g2–g8 that end at t9 (D) or t11 (E). (F) Superposition of structures in (D) and (E).
A structural model for miRNA targeting
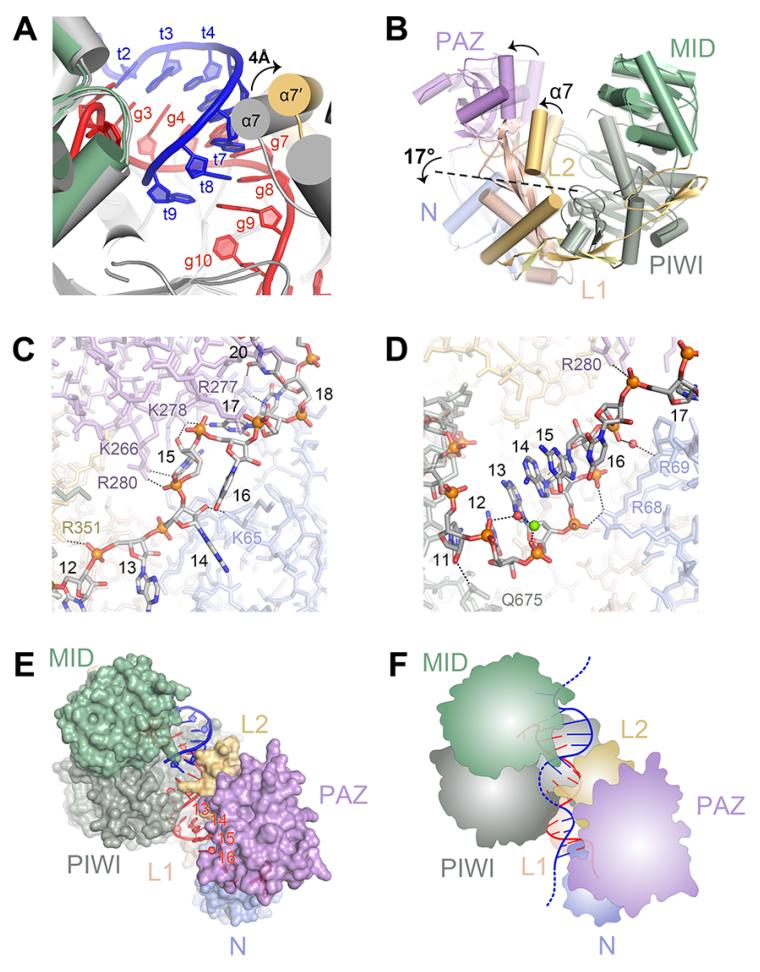
Structures of human Ago1 and Ago2 and yeast Ago1 indicate that, in the absence of target RNA, eukaryotic Argonaute proteins kink their guide RNAs at the end of the seed region (15-18). The kink appears to be stabilized by helix-7, which inserts a hydrophobic residue (I365 in Ago2) between g6 and g7. In moving from the guide-only to the target-bound conformation, helix-7 shifts ~4 Å to interact with the minor groove of the guide-target duplex (Fig. 5A). The movement of helix-7 is required to avoid steric clashes with target nucleotides t6 and t7 and is therefore necessary for target pairing beyond g5 (fig. S9). Movement of helix-7 also releases the constraints on the guide RNA, relaxing the kink and allowing g6 and g7 to adopt an A-form conformation for target pairing. Thus, the guide and protein act synergistically to recognize target RNAs— movement of helix-7 to accommodate targets beyond g5 also releases g6 and g7, facilitating the additional base pairing. Formation of these base pairs generates a guide-target duplex with a minor groove that provides a new binding surface for helix-7, thereby stabilizing the opened conformation. Conversely, mismatches or G:U wobble pairs would present a distorted minor groove to helix-7, which would then be more likely to shift back towards the guide RNA and displace the mispaired target.
Fig. 5.
Comparison of Ago2-guide and Ago2-guide-target structures. (A) Helix-7 (α7) shifts to accommodate pairing to target RNAs. The Ago2-guide structure (gray) aligned to the Ago2-guide-target structure (colored). (B) The PAZ domain and helix-7 move as a rigid body. Superposition of protein components from Ago2-guide (semi-transparent) and Ago2-guide-target (opaque) structures. Arrows indicate movement from guide-only to guide-target structures. Dashed line marks hinge in the L1/L2 domains. (C-D) Contacts to the guide RNA supplemental region in the guide-only (C) and target-bound (D) structures. (E) The supplemental region (g13-g16) adopts an exposed helical conformation in the Ago2-guide-target structure. (F) Cartoon model for seed plus supplemental pairing.
Seed pairing opens the N-PAZ channel for supplemental pairing
Superimposing guide-only and guide-target structures of Ago2 indicates that helix-7 and the PAZ domain move as a discrete rigid body relative to the MID, PIWI and N domains (Fig. 5B). The hinge for this conformational change resides in the base of helix-7 and extends across the beta-sheet in the L1 domain. We suggest that the movements in helix-7, induced by seed pairing, are propagated to positional shifts in the PAZ domain, leading to a widening of the N-PAZ channel. Mutation of Phe-181, which resides in the hinge, inhibits small RNA duplex unwinding during RISC-loading (26), suggesting that related conformational changes are involved in passenger strand removal.
Widening of the N-PAZ channel is accompanied by repositioning of the 3′ half of the guide RNA, with g11–g16 shifting to adopt a near A-form conformation (Fig. 5C and D). Helical stacking is disrupted after g16 by Pro-67 of the N domain and electron density for g17–g19 is weak, indicative of conformational heterogeneity. However, density for g20–g21 is visible with the 3′ end of the guide bound to the PAZ domain. The Watson-Crick faces of g13–g16, which can supplement repression of target sites with weak seed pairing (27), are splayed out towards the solvent in a manner reminiscent of the seed region in guide-only structures (Fig. 5D). We suggest that seed pairing is coupled to a rearrangement in the 3′ half of the guide that facilitates target interactions in the supplemental region. Nucleotides g12–g13 are partially occluded by a PIWI domain loop (residues 602–608), inidicating that target pairing to the supplemental region may nucleate at g14–g16 and extend back into the central cleft. We suggest that target RNAs paired to the seed may exit the central cleft and extend over the middle of the PIWI domain to pair with the supplemental region on the other side (Fig. 5E and F). This would the complex to maximize guide-target interactions while avoiding the topological issues and entropic costs associated with wrapping the target and guide RNAs around each other within the central cleft (3). Consistent with this model, complementarity to g9–g12 is not correlated with miRNA-mediated repression (27), and the affinity of mouse Ago2 for a fully complementary target RNA is less than that of a similar target that pairs only to the seed and supplemental regions (24).
An inactivated magnesium ion in the slicer active site
In addition to miRNA-mediated repression, Ago2 can silence targets by catalyzing an endonucleolytic “slicing” reaction (28, 29). Slicing is more specific than miRNA-mediated repression because target cleavage requires extensive complementarity between guide and target RNAs (25, 30, 31). Slicing specificity in T. thermophilus Argonaute (TtAgo) is achieved through a conformational switch involving insertion of a catalytic “glutamate finger” into the active site, triggered by extensive guide-target pairing (32). While it is possible that human Ago2 undergoes similar conformational transitions, the equivalent glutamate residue (Glu-637) is inserted into the active site in all available Ago2 structures, irrespective of target pairing status (fig. S10). Therefore, the mechanisms underlying slicing specificity in humans are less clear.
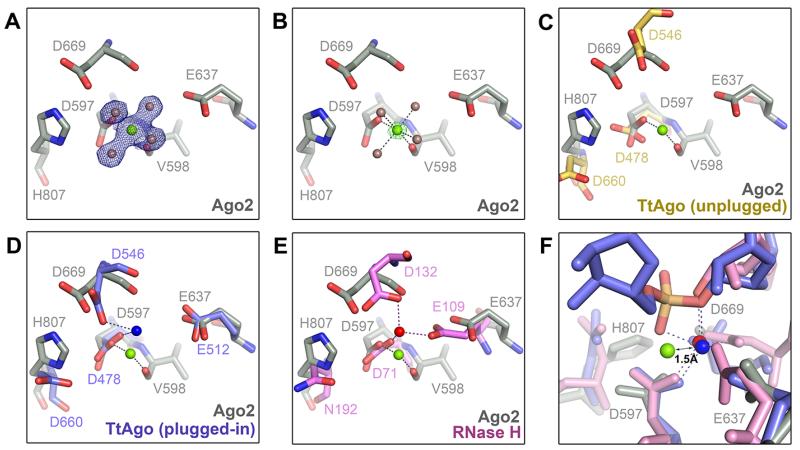
We observed electron density for a metal ion in the active site of guide-selected Ago2 (Fig. 6A, B). The metal is coordinated to four water molecules, the carboxylate side chain of Asp-597, and the main chain carbonyl of Val-598, with a coordination geometry (octahedral) and inter-atomic distances (2.1 Å) characteristic of a magnesium ion (33). Comparison with TtAgo suggests the magnesium is the functional equivalent of catalytic metal “B” in TtAgo guide-target structures (32) (Fig. 6C, D). However, the coordination of metal B in TtAgo differs from that in Ago2—in TtAgo the metal is chelated by the carboxylate groups of Asp-478 and Asp-546 (equivalent to Asp-597 and Asp-669 in Ago2, respectively), with no interactions to the main chain (32). Comparison to structures of bacterial RNase H indicates that metal B in TtAgo is in a conserved, cleavage-compatible position (Fig. 6D, E). We therefore suggest that the magnesium observed in Ago2 is in an inactivated position, stabilized by coordination to the main chain carbonyl of Val-598.
Fig. 6.
Inactive magnesium ion in the Ago2 active site. (A and B). Magnesium ion (green) is bound to the Asp-597 carboxylate side chain, the Val-598 main chain carbonyl, and four water molecules (brown spheres). 2Fo-Fc map (blue mesh) contoured at 1.5 σ (A), and Fo-Fc magnesium omit map (green mesh) at 15 σ (B). (C) Active site of Ago2 (gray) aligned with unplugged active site of TtAgo (yellow) (PDB ID 3DLH). (D) Ago2 active site aligned with plugged-in TtAgo (blue) (PDB ID 3HVR). Metals ions shown as spheres. (E) Ago2 active site aligned with B. halodurans RNase H (pink with red magnesium ion; PDB ID 2G8H). (F) Alignment of Ago2 (gray with green magnesium), TtAgo (blue) and B. halodurans RNase H (pink) shows the Ago2 magnesium shifted 1.5 Å from the active position.
We propose that chelating to the scissile phosphate in complementary target RNAs induces a ~1.5 Å shift in the position of the magnesium into an active coordination similar to that observed in TtAgo and RNase H (Fig. 6F). The shift likely involves an exchange of the main chain carbonyl bond for a bond to catalytic residue Asp-669 (28), mediated by Asp-659. We suggest that precise positioning of the scissile phosphate is required to move the metal ion into the activated position, which is consistent with the observation that guide-target mismatches near the cleavage site can dramatically reduce catalytic activity (24, 30, 34-36). This model also explains how structural elements in the PIWI and N domains, which likely influence positioning of the guide-target duplex within the central cleft, contribute significantly to slicing activity (16, 17, 37).
Discussion
The structures presented here show Ago2 in two distinct conformational states— one in which the majority of the guide RNA is sequestered from target pairing, and a second with the full seed and supplemental regions exposed. Although the RNAs in previous structures of human Ago1 and Ago2 were mostly disordered, the proteins crystallized in conformations similar to the guide-sequestered structure reported here, indicating this is a well-populated conformation (15-17, 19). We propose that the guide-sequestered state limits spurious interactions with cellular RNAs by confining pairing potential to guide nucleotides g2–g5 (Fig 1C). In this model, Ago2 primarily uses g2–g5 to identify candidate target sites (fig. S11), which are then further interrogated by g6–g7 and helix-7. Target interactions can be stabilized further still by pairing to g8 and/or g13–g16 and through interactions with t1 adenine nucleotides, which may also facilitate the initial identification of target sites (11). This stepwise targeting model is similar to the mechanism of nucleic acids annealing in free solution, where extended duplex structures are nucleated by small segments (3–4 base pairs) of complementarity (38).
The observations that Ago2 is loaded with small RNA duplexes (39-41) and cleaves target RNAs with extensive complementarity (28, 29) indicate the protein adopts at least one additional conformational state with a widened central cleft to accommodate guide-target pairing beyond g8. Extended target pairing probably requires release of the guide 3′ end from the PAZ domain, as in TtAgo (fig. S12) (32, 42). Furthermore, the identification of miRNA target sites with central or 3′ complementarity and limited seed pairing suggests the protein may adopt additional stabile conformational states (43-45). However, because 80% of miRNA-target interactions appear to function through seed pairing (44) it is likely that the structures presented here represent the majority of miRNA targeting events in human cells.
Supplementary Material
TABLE 1.
| Ago2 crystal | 4W5N | 4W5O | 4W5Q | 4W5R | 4W5T |
|---|---|---|---|---|---|
| Target RNA | guide only | 2-9 paired | 2-8 paired | 2-8 paired (long) | 2-7 paired |
| Beamline | SSRL 11-1 | SSRL 12-2 | SSRL 11-1 | APS 24-ID-E | SSRL 12-2 |
| Space Group | P1211 | P1211 | P1211 | P1211 | P1211 |
| Unit Cell Dimensions | |||||
| A (Å) | 63.12 | 55.81 | 55.50 | 55.38 | 55.64 |
| B (Å) | 108.87 | 116.99 | 116.65 | 116.18 | 116.47 |
| C (Å) | 68.07 | 69.77 | 70.07 | 69.68 | 69.82 |
| α (°) | 90.00 | 90.00 | 90.00 | 90.00 | 90.00 |
| β (°) | 106.93 | 92.45 | 92.26 | 92.17 | 92.34 |
| γ (°) | 90.00 | 90.00 | 90.00 | 90.00 | 90.00 |
| Ago2 Molecules per ASU | 1 | 1 | 1 | 1 | 1 |
| Data Collection | |||||
| Wavelength (Å) | 0.97945 | 0.97950 | 0.97944 | 0.97918 | 0.97950 |
| Resolution (Å) | 38.98-2.90 (3.08-2.90) |
39.00-1.80 (1.84-1.80) |
38.88-3.10 (3.31-3.10) |
116.16-2.50 (2.64-2.50) |
38.82-2.50 (2.64-2.50) |
| No Reflections | |||||
| Total | 67638 | 276820 | 55568 | 113374 | 104468 |
| Unique | 19381 | 80397 | 16052 | 30428 | 29434 |
| Completeness (%) | 98.8 (98.8) | 97.7 (92.0) | 98.9 (98.9) | 99.8 (99.9) | 95.8 (97.6) |
| Redundancy | 3.5 (3.6) | 3.4 (3.4) | 3.5 (3.4) | 3.7 (3.7) | 3.5 (3.6) |
| I/σI | 15.6 (2.6) | 11.5 (2.2) | 10.6 (2.5) | 9.0 (1.9) | 12.5 (3.3) |
| Rmerge | 7.0 (44.7) | 5.5 (53.5) | 9.4 (39.6) | 14.1 (75.8) | 7.4 (39.8) |
| Rpim | 6.8 (43.1) | 5.1 (47.9) | 9.0 (7.5) | 8.5 (45.7) | 4.6 (24.3) |
| Refinement | |||||
| Resolution (Å) | 38.98-2.90 | 39.00-1.80 | 35.29-3.10 | 69.63-2.50 | 35.29-2.50 |
| R-free/R-factor | 25.33/21.73 | 19.65/16.72 | 23.25/19.03 | 23.4/19.46 | 21.51/17.28 |
| R.M.S. Deviation | |||||
| Bond Distances (Å) | 0.010 | 0.007 | 0.011 | 0.006 | 0.009 |
| Bond Angles (°) | 1.035 | 1.142 | 1.218 | 0.7820 | 1.194 |
| Number of Atoms | |||||
| Non-hydrogen, protein | 6461 | 6419 | 6439 | 6442 | 6471 |
| Non-hydrogen, RNA | 355 | 595 | 572 | 406 | 527 |
| Phenol | 21 | 28 | 28 | 21 | 28 |
| Isopropanol | 0 | 8 | 0 | 8 | 0 |
| Phosphate | 0 | 5 | 0 | 0 | 0 |
| Mg | 1 | 3 | 3 | 2 | 2 |
| Water | 34 | 433 | 0 | 126 | 156 |
| Ramachandran Plot | |||||
| Most Favored Regions | 93.83% | 96.20% | 93.95% | 95.70% | 96.11% |
| Additionally Allowed | 5.67% | 3.68% | 5.55% | 4.05% | 3.64% |
| Generously Allowed | 0.50% | 0.13% | 0.5% | 0.25% | 0.25% |
Acknowledgments
We are grateful to P.D. Zamore and J.R. Williamson for generous advice and scientific insights. Diffraction data were collected at beam lines 11-1 and 12-2 at the Stanford Synchrotron Radiation Lightsource and 24-ID-E at the Advanced Photon Source, supported by NIGMS grant P41 GM103403 and DOE contract number DE-AC02-06CH11357. NTS is a predoctoral fellow of the American Heart Association and an Achievement Rewards for College Scientists (ARCS) scholar. The work was supported by NIH grant R01 GM104475 to IJM.
References
- 1.Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chivukula RR, Shi G, Acharya A, Mills EW, Zeitels LR, Anandam JL, Abdelnaby AA, Balch GC, Mansour JC, Yopp AC, Maitra A, Mendell JT. An Essential Mesenchymal Function for miR-143/145 in Intestinal Epithelial Regeneration. Cell. 2014;157:1104–1116. doi: 10.1016/j.cell.2014.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xin M, Olson EN, Bassel-Duby R. Mending broken hearts: cardiac development as a basis for adult heart regeneration and repair. Nature reviews. Molecular cell biology. 2013;14:529–541. doi: 10.1038/nrm3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hasuwa H, Ueda J, Ikawa M, Okabe M. miR-200b and miR-429 function in mouse ovulation and are essential for female fertility. Science. 2013;341:71–73. doi: 10.1126/science.1237999. [DOI] [PubMed] [Google Scholar]
- 7.Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482:347–355. doi: 10.1038/nature10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 9.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 10.Lai EC. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002;30:363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- 11.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 12.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 13.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 15.Schirle NT, MacRae IJ. The crystal structure of human Argonaute2. Science. 2012;336:1037–1040. doi: 10.1126/science.1221551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faehnle CR, Elkayam E, Haase AD, Hannon GJ, Joshua-Tor L. The making of a slicer: activation of human Argonaute-1. Cell Rep. 2013;3:1901–1909. doi: 10.1016/j.celrep.2013.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakanishi K, Ascano M, Gogakos T, Ishibe-Murakami S, Serganov AA, Briskin D, Morozov P, Tuschl T, Patel DJ. Eukaryote-specific insertion elements control human ARGONAUTE slicer activity. Cell Rep. 2013;3:1893–1900. doi: 10.1016/j.celrep.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakanishi K, Weinberg DE, Bartel DP, Patel DJ. Structure of yeast Argonaute with guide RNA. Nature. 2012;486:368–374. doi: 10.1038/nature11211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elkayam E, Kuhn CD, Tocilj A, Haase AD, Greene EM, Hannon GJ, Joshua-Tor L. The structure of human argonaute-2 in complex with miR-20a. Cell. 2012;150:100–110. doi: 10.1016/j.cell.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flores-Jasso CF, Salomon WE, Zamore PD. Rapid and specific purification of Argonaute-small RNA complexes from crude cell lysates. RNA. 2013;19:271–279. doi: 10.1261/rna.036921.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma JB, Ye K, Patel DJ. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature. 2004;429:318–322. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poulsen C, Vaucheret H, Brodersen P. Lessons on RNA silencing mechanisms in plants from eukaryotic argonaute structures. Plant Cell. 2013;25:22–37. doi: 10.1105/tpc.112.105643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Varani G, McClain WH. The G x U wobble base pair. A fundamental building block of RNA structure crucial to RNA function in diverse biological systems. EMBO reports. 2000;1:18–23. doi: 10.1093/embo-reports/kvd001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wee LM, Flores-Jasso CF, Salomon WE, Zamore PD. Argonaute divides its RNA guide into domains with distinct functions and RNA-binding properties. Cell. 2012;151:1055–1067. doi: 10.1016/j.cell.2012.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes & development. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwak PB, Tomari Y. The N domain of Argonaute drives duplex unwinding during RISC assembly. Nature structural & molecular biology. 2012;19:145–151. doi: 10.1038/nsmb.2232. [DOI] [PubMed] [Google Scholar]
- 27.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Molecular cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 29.Rivas FV, Tolia NH, Song JJ, Aragon JP, Liu J, Hannon GJ, Joshua-Tor L. Purified Argonaute2 and an siRNA form recombinant human RISC. Nature structural & molecular biology. 2005;12:340–349. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- 30.Elbashir SM, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. The EMBO journal. 2001;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doench JG, Petersen CP, Sharp PA. siRNAs can function as miRNAs. Genes & development. 2003;17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sheng G, Zhao H, Wang J, Rao Y, Tian W, Swarts DC, van der Oost J, Patel DJ, Wang Y. Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:652–657. doi: 10.1073/pnas.1321032111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng H, Chordia MD, Cooper DR, Chruszcz M, Muller P, Sheldrick GM, Minor W. Validation of metal-binding sites in macromolecular structures with the CheckMyMetal web server. Nat Protoc. 2014;9:156–170. doi: 10.1038/nprot.2013.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holen T, Amarzguioui M, Wiiger MT, Babaie E, Prydz H. Positional effects of short interfering RNAs targeting the human coagulation trigger Tissue Factor. Nucleic acids research. 2002;30:1757–1766. doi: 10.1093/nar/30.8.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amarzguioui M, Holen T, Babaie E, Prydz H. Tolerance for mutations and chemical modifications in a siRNA. Nucleic acids research. 2003;31:589–595. doi: 10.1093/nar/gkg147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haley B, Zamore PD. Kinetic analysis of the RNAi enzyme complex. Nature structural & molecular biology. 2004;11:599–606. doi: 10.1038/nsmb780. [DOI] [PubMed] [Google Scholar]
- 37.Hauptmann J, Dueck A, Harlander S, Pfaff J, Merkl R, Meister G. Turning catalytically inactive human Argonaute proteins into active slicer enzymes. Nature structural & molecular biology. 2013;20:814–817. doi: 10.1038/nsmb.2577. [DOI] [PubMed] [Google Scholar]
- 38.Ross PD, Sturtevant JM. The Kinetics of Double Helix Formation from Polyriboadenylic Acid and Polyribouridylic Acid. Proceedings of the National Academy of Sciences of the United States of America. 1960;46:1360–1365. doi: 10.1073/pnas.46.10.1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 40.Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 41.Leuschner PJ, Ameres SL, Kueng S, Martinez J. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO reports. 2006;7:314–320. doi: 10.1038/sj.embor.7400637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y, Juranek S, Li H, Sheng G, Wardle GS, Tuschl T, Patel DJ. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. 2009;461:754–761. doi: 10.1038/nature08434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shin C, Nam JW, Farh KK, Chiang HR, Shkumatava A, Bartel DP. Expanding the microRNA targeting code: functional sites with centered pairing. Molecular cell. 2010;38:789–802. doi: 10.1016/j.molcel.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grosswendt S, Filipchyk A, Manzano M, Klironomos F, Schilling M, Herzog M, Gottwein E, Rajewsky N. Unambiguous Identification of miRNA:Target Site Interactions by Different Types of Ligation Reactions. Molecular cell. 2014;54:1042–1054. doi: 10.1016/j.molcel.2014.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Helwak A, Kudla G, Dudnakova T, Tollervey D. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell. 2013;153:654–665. doi: 10.1016/j.cell.2013.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.