Summary
Quorum sensing is a cell-cell communication process that bacteria use to transition between individual and social lifestyles. In vibrios, homologous small RNAs called the Qrr sRNAs function at the center of quorum-sensing pathways. The Qrr sRNAs regulate multiple mRNA targets including those encoding the quorum-sensing regulatory components luxR, luxO, luxM, and aphA. We show that a representative Qrr, Qrr3, uses four distinct mechanisms to control its particular targets: Qrr3 sRNA represses luxR through catalytic degradation, represses luxM through coupled degradation, represses luxO through sequestration, and activates aphA by revealing the ribosome-binding site while the sRNA itself is degraded. Qrr3 forms different base-pairing interactions with each mRNA target, and the particular pairing strategy determines which regulatory mechanism occurs. Combined mathematical modeling and experiments show that the specific Qrr regulatory mechanism employed governs the potency, dynamics and competition of target mRNA regulation, which in turn, defines the overall quorum-sensing response.
Introduction
Small regulatory RNAs (sRNAs) act as core regulators in many bacterial signal transduction cascades (Waters and Storz, 2009). Bacterial sRNAs function by several mechanisms. Here, we focus on trans-encoded Hfq-binding sRNAs. This class of sRNA can act positively or negatively, and non-contiguous base-pairing with mRNA targets is employed. In the case of negative regulation, trans-encoded sRNAs base-pair near the ribosome binding site of the mRNA target, leading to ribosome occlusion (Altuvia et al., 1998; Kawamoto et al., 2006; Møller et al., 2002; Udekwu et al., 2005). In most cases, occlusion is associated with RNase E recruitment and degradation of the mRNA (Massé et al., 2003; Prévost et al., 2011). In the case of positive regulation, trans-encoded sRNAs perform anti anti-sense base-pairing with the mRNA target (Fröhlich and Vogel, 2009; Majdalani et al., 1998). Binding reveals the ribosome binding site and promotes stabilization of the mRNA target, and, in turn, gene expression (McCullen et al., 2010). The RNA chaperone Hfq mediates the interactions between trans-encoded sRNAs and their mRNA targets and protects the sRNAs from RNase E-mediated degradation (Vogel and Luisi, 2011; Kawamoto et al., 2006). Hfq is thought to be limiting, leading to competition between different sRNA-mRNA pairs for its chaperone function (Fender et al., 2010; Hussein and Lim, 2011; Moon and Gottesman, 2011).
The implications of trans-encoded sRNA regulation at the systems level depend on the fate of the sRNA. First, sRNAs can undergo coupled degradation in which both the sRNA and the mRNA target are degraded following base-pairing. The RyhB sRNA exemplifies this mode of regulation (Massé et al., 2003). Second, sRNAs can act catalytically, in which the target mRNA is degraded but the sRNA is not. Thus, the sRNA is available to recycle. One such example is the MicM (ChiX) sRNA, which acts catalytically on the mRNA target ybfM (chiP) (Overgaard et al., 2009). Third, sRNAs can also act by sequestering their targets. An example of this type of regulation occurs between the sRNA Spot42 and its mRNA target galK. In this case, Spot42 specifically blocks the ribosome-binding site of galK, but no mRNA degradation occurs (Møller et al., 2002). Finally, the fates of sRNAs that act as activators have not been well characterized. In theory, activating sRNAs can be degraded, recycled, or sequestered.
The Qrr sRNAs (Quorum Regulatory RNAs) are Hfq-dependent trans-encoded sRNAs that control vibrio quorum sensing (Lenz et al., 2004). Quorum sensing is a cell-cell communication process that bacteria use to monitor changes in cell-population density and control collective behaviors such as biofilm formation and virulence factor production. Quorum sensing involves production, detection, and population-wide response to extracellular signal molecules called autoinducers (Rutherford and Bassler, 2012). In vibrio quorum-sensing circuits, several nearly identical Qrr sRNAs control multiple target mRNAs, and the Qrr sRNAs act as both positive and negative regulators. These features enable us to exploit this set of sRNAs and their particular mRNA targets to dissect pairing regimes, target preferences, and modes of regulation. The Qrr sRNAs positively control the production of the low-cell-density master regulator AphA, and they repress the production of the high-cell-density master regulator LuxR (Rutherford et al., 2011; Tu and Bassler, 2007). The Qrr sRNAs feed back to repress the genes encoding one of the quorum-sensing synthase-receptor pairs, LuxMN, and the gene encoding the transcriptional factor LuxO (Teng et al., 2011; Tu et al., 2010). The Qrr sRNAs also post-transcriptionally regulate sixteen genes outside of the quorum-sensing circuit (Shao et al., 2013).
In the present study, we show that a representative Qrr sRNA, Qrr3, uses four distinct mechanisms to regulate four different target mRNAs. The Qrr3 sRNA undergoes coupled degradation when it pairs with luxM mRNA, it uses sequestration to control luxO mRNA, it catalytically represses luxR mRNA, and it activates aphA mRNA expression while the Qrr3 sRNA itself is degraded. The mRNA targets that reduce Qrr sRNA stability (luxM and aphA) do so by remodeling the 5’-most stem-loop of the Qrr sRNA. mRNA targets that sequester the Qrr sRNA (luxO) presumably do so via tight binding to the Qrr sRNA. Indeed, we demonstrate that a particular regulatory mechanism can be converted into a different one by altering the base-pairing position or binding strength. The different sRNA-target mRNA interaction mechanisms result in distinct regulatory strength and dynamical behaviors of the mRNA targets in vivo. Furthermore, the particular regulatory mechanism used for mRNA target regulation is critical for properly timed quorum-sensing responses.
Results
The Qrr3 sRNA uses distinct mechanisms to regulate different mRNA targets
There are twenty known targets of the V. harveyi Qrr sRNAs (Shao et al., 2013), four of which, luxM, luxO, luxR, and aphA, are members of the quorum-sensing regulatory circuit and are the focus of this work (Figure 1). Little is known about how the five Qrr sRNAs choose among their mRNA targets in vivo. To investigate Qrr target preferences, we developed a competition assay in Escherichia coli. We constructed a dual reporter system on a single plasmid that encodes (1) an IPTG-inducible 5’UTR-GFP fusion to a Qrr-repressed mRNA target (the Qrr “target” in all assays), and (2) an arabinose inducible 5’UTR-mCherry fusion to a different Qrr-controlled mRNA target (the Qrr “competitor”). We transformed this dual reporter plasmid along with a second plasmid encoding anhydrotetracycline-inducible Qrr3 into E. coli (Figure S1A). The five Qrr sRNAs are similar in sequence and secondary structure and they share most of the target mRNAs (Shao et al., 2013; Tu and Bassler, 2007). We arbitrarily chose Qrr3 to use in these assays.
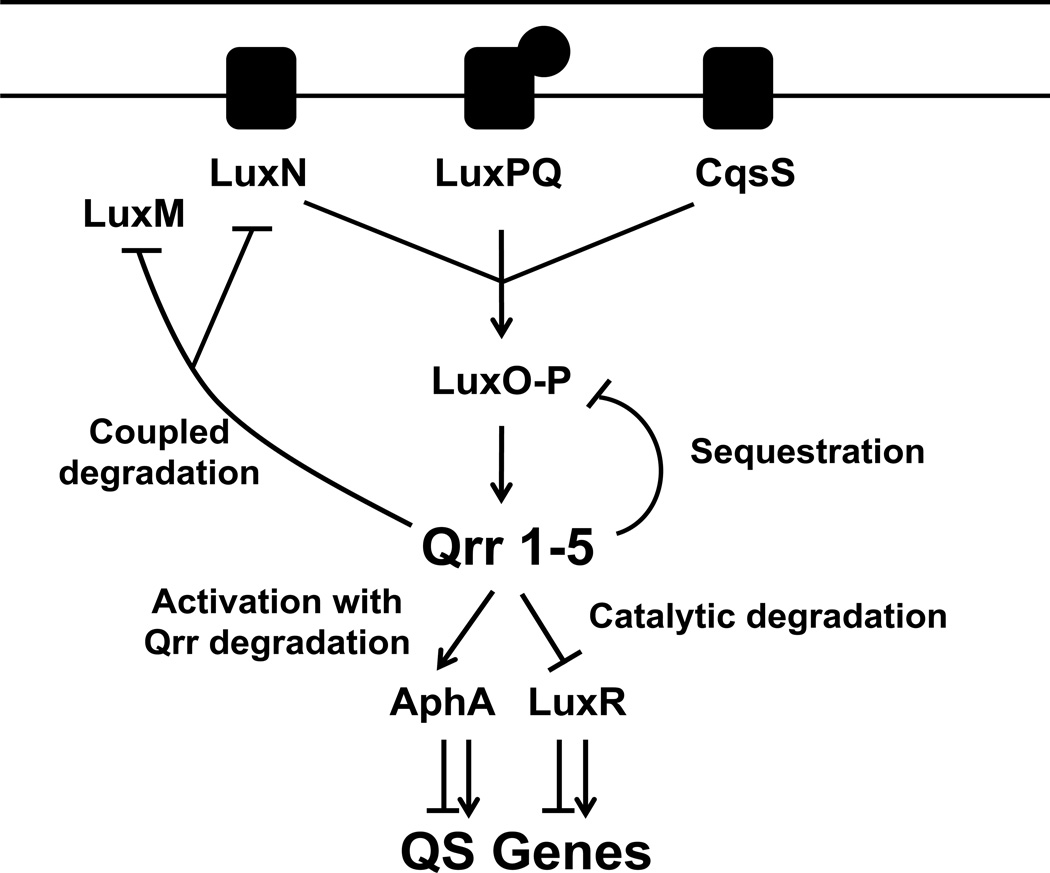
Figure 1. Schematic for how a V. harveyi quorum-sensing Qrr sRNA uses four regulatory mechanisms to control target mRNAs.
At low cell density, the three quorum-sensing receptors LuxN, LuxPQ, and CqsS transfer phosphate through LuxU (not shown) to LuxO. Phosphorylated LuxO activates transcription of genes encoding five sRNAs called Qrr1-5. Using Qrr3 as the representative quorum-sensing regulatory sRNA, we show that the Qrr sRNA catalytically represses the high-cell-density master regulator luxR. The Qrr sRNA represses luxO through sequestration. The Qrr sRNA represses the luxMN operon through coupled degradation (luxM encodes the synthase that produces the ligand for LuxN). The Qrr sRNA also activates translation of the low-cell-density master regulator aphA; base-pairing with the aphA mRNA leads to Qrr degradation.
To monitor Qrr preference, we first measured GFP fluorescence from the target mRNA in the absence of both Qrr3 and competitor mRNA to determine the basal expression level of the mRNA target. Next, we measured target GFP fluorescence when Qrr3 was induced to determine the level of target mRNA repression by Qrr3. Finally, we measured GFP fluorescence from the target mRNA when Qrr3 was induced and, additionally, the competitor mRNA was induced to different levels. This third measurement allowed us to assess the ability of different competitor mRNAs to compete with the target mRNA for regulation by Qrr3. Our expectation was that, in the case of a Qrr3 repressed target mRNA, if competition occurred, the target mRNA-GFP fluorescence should increase when we induced expression of the competitor mRNA because the amount of Qrr3 available to regulate the target mRNA would decrease. By contrast, if the target mRNA-GFP level did not change when we induced expression of the competitor mRNA, we would infer that the competitor mRNA did not compete with the target mRNA for regulation by Qrr3.
Our dual reporter system allowed us to simultaneously measure mCherry production from the competitor mRNA. A change in mCherry level following induction of Qrr3 was useful to verify that the competitor was indeed being regulated by Qrr3. We also determined the expression level and the half-life of Qrr3 in the absence and presence of the mRNA targets. Likewise, we measured the expression level of the target mRNAs in the absence and presence of Qrr3. These final measurements allowed us to discover the fates of Qrr3 and the target mRNA, and thus the mechanism used by Qrr3 to control each target – catalytic, coupled degradation, sequestration, or mRNA activation with concomitant sRNA degradation.
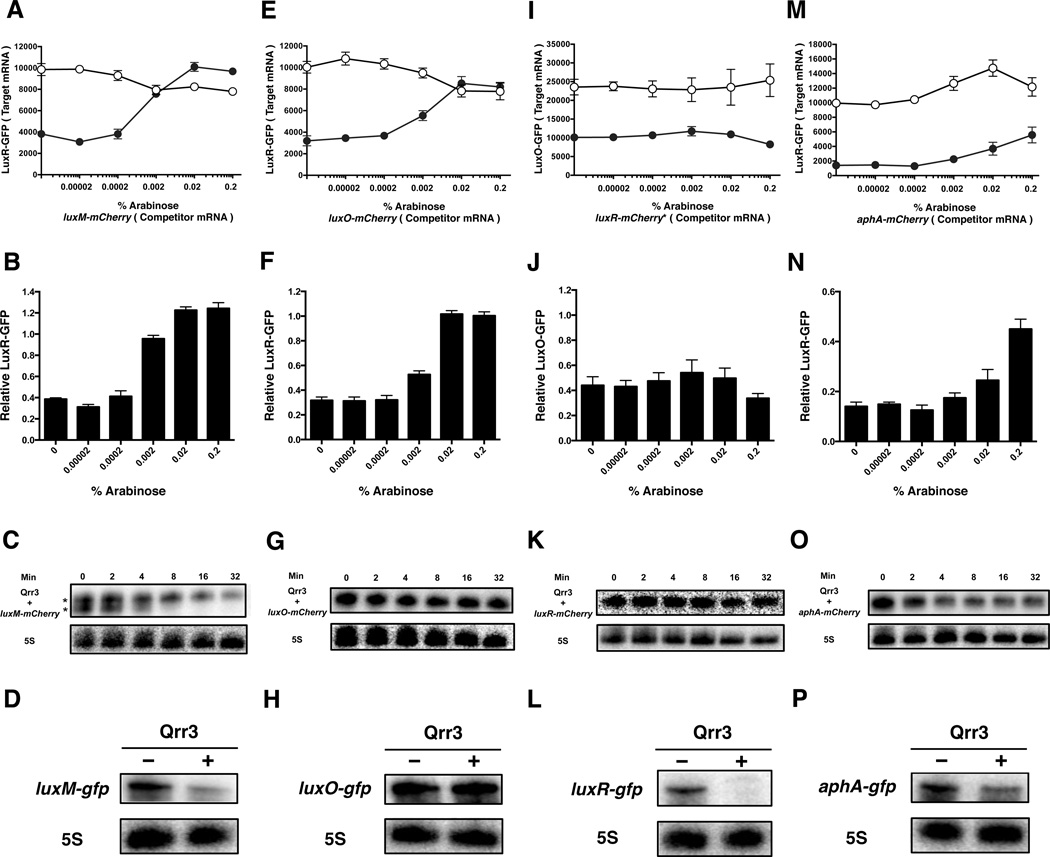
Using this strategy, we first investigated whether luxM mRNA competes with luxR mRNA for regulation by Qrr3. As a control, we show that, in the absence of Qrr3, increasing the luxM-mCherry production does not significantly alter LuxR-GFP production (Figure 2A open circles). When Qrr3 is present, LuxR-GFP is repressed 2.5-fold in the absence of luxM-mCherry (Figure 2A filled circles, Figure 2B; no arabinose). Inducing luxM-mCherry expression causes an increase in LuxR-GFP (Figure 2A filled circles, 2B). This result shows that luxM mRNA successfully competes with luxR mRNA for regulation by Qrr3. We confirmed that in the absence of Qrr3, increasing the arabinose inducer results in increasing production of luxM-mCherry (Figure S1B open circles). At low arabinose concentration and thus low luxM-mCherry levels, LuxM-mCherry is repressed 2.5-fold in the presence of Qrr3 (Figure S1C). By contrast, at high arabinose, and thus high luxM-mCherry levels, neither LuxM-mCherry nor LuxR-GFP is repressed by Qrr3 (Figures S1C, 2B). Presumably, under the latter condition, Qrr3 is saturated by the competitor mRNA or degraded. In analogous experiments, we measured the ability of luxM-mCherry to compete with luxO-gfp and with luxM-gfp for regulation by Qrr3 and obtained similar results (Figure S3A,B). Thus, luxM mRNA can successfully compete for regulation by Qrr3 against luxM (itself), luxO, and luxR mRNA.
Figure 2. mRNAs have different abilities to compete for Qrr sRNA regulation.
First row: Competition between different Qrr sRNA target mRNAs. Fluorescence from a plasmid-borne luxR-gfp ((A) (E) (M)) or luxO-gfp (I) translational fusion was measured in E. coli. Arabinose was added to drive production of the competitor mRNA luxM-mCherry (A), luxO-mCherry (E), luxR-mCherry* (I) and aphA-mCherry (M). Experiments were performed in the absence of Qrr3 (open circles) and in the presence of Qrr3 (filled circles). Means and SEMs for triplicate cultures are shown. Second row: Quantification of the fractional expression of LuxR-GFP ((B) (F) (N)) or LuxO-GFP (J) from panels A, E, M, and I, respectively. GFP fluorescence in the presence of Qrr3 was normalized to that in its absence. Means and SEMs for triplicate cultures are shown. Third row: Half-life of Qrr3 in the presence of luxM-mCherry (C), luxO-mCherry (G), luxR-mCherry (K), and aphA-mCherry (O). Fourth row: Northern blots of luxM-gfp (D), luxO-gfp (H), luxR-gfp (L), and aphA-gfp (P) translational fusions in the absence (−) and presence (+) of Qrr3. For all Northern blots, results are representative of two independent experiments and 5S rRNA was used as the loading control.
To assess what becomes of Qrr3 when regulating luxM-mCherry target mRNA, we measured Qrr3 levels in the absence and presence of luxM-mCherry mRNA. When no luxM-mCherry mRNA is present, Qrr3 appears as one band on a Northern blot (Figure S2A, left lane). In the presence of luxM-mCherry mRNA, two Qrr3 bands appear, suggesting that Qrr3 is processed (Figure S2A right lane). In the absence of any mRNA target, the Qrr3 half-life is over 32 minutes (Figure S2B). In the presence of luxM-mCherry mRNA, the half-lives of both Qrr3 RNA bands decrease, with the processed Qrr3 product exhibiting the most dramatic decline (to T1/2 <8 minutes, Figure 2C). Together, these data explain how luxM mRNA competes with luxR mRNA for regulation by Qrr3: the presence of luxM mRNA causes Qrr3 degradation, decreasing the amount of Qrr3 available to regulate luxR mRNA. We also examined what becomes of the luxM mRNA during regulation by Qrr3. We used gfp fusions to measure target mRNA levels. Both the luxM-gfp mRNA (Figure 2D) and the LuxM-GFP protein (Figure S1D) decreased in the presence of Qrr3, indicating that luxM mRNA is degraded during regulation. We therefore conclude that Qrr3 regulates luxM mRNA through a coupled degradation mechanism: when base-paired, both the luxM mRNA target and the Qrr3 sRNA are subject to degradation.
We next investigated whether luxO mRNA can compete with other mRNA targets for Qrr3 regulation. We again used luxR-gfp mRNA as the “target” for which we show data, but we note that the results are the same when luxM-gfp or luxO-gfp mRNAs act as the “target” (data not shown). Figures 2E,F and S1E,F show that luxO mRNA can indeed compete with luxR mRNA for Qrr3 regulation, similar to what we found above when luxM-mCherry acts as the competitor. However, unlike when luxM-mCherry was the competitor, the level and half-life of Qrr3 are identical in the absence and presence of luxO-mCherry competitor mRNA (Figures S2C, 2G, S2B) and no processed Qrr3 RNA band was detected (Figure S2C, 2G). Furthermore, the presence of Qrr3 does not alter luxO-gfp mRNA levels (Figure 2H), however, LuxO-GFP protein production is repressed ∼15-fold in the presence of Qrr3 (Figure S1G). Based on these data, we propose that Qrr3 and luxO mRNA sequester one another when base-paired.
We likewise tested whether luxR mRNA could act as a competitor. Induction of high luxR-mCherry caused significant reductions in target-GFP levels even in the absence of Qrr3 suggesting that high levels of the LuxR-mCherry protein make the cells sick. To circumvent this problem, we introduced a stop codon in mCherry to abolish mCherry protein production. We call this construct luxR-mCherry*. When the luxR-mCherry* mRNA is the competitor, it does not affect Qrr regulation of luxO-gfp (Figure 2I,J) or luxR-gfp (Figure S3C,D). Importantly, Figure S1H,I show that the luxR-mCherry* construct is induced by arabinose and is fully repressed by Qrr3. Thus, even though luxR-mCherry* is capable of interacting with Qrr3, it cannot compete with luxO-gfp mRNA or luxR-gfp mRNA for Qrr-regulation. (To eliminate the possibility that this lack of competition is due to the mCherry* mutation, we inserted the same stop codon into the luxM-mCherry construct. We call this luxM-mCherry*. luxM-mCherry* mRNA remains fully capable of competing for Qrr regulation against luxO-gfp mRNA (Figure S3E,F) and luxR-gfp mRNA (Figure S3G,H). As controls, we show that the luxR-mCherry* mRNA is expressed at levels comparable to the luxM-mCherry, luxO-mCherry and luxM-mCherry* mRNAs (Figure S3I). Thus, we conclude that luxR mRNA does not compete with other mRNA targets for Qrr regulation. Figures S2D, 2K show that luxR mRNA does not affect Qrr stability because both the Qrr3 level and its half-life are identical in the absence and presence of luxR-mCherry mRNA (or luxR-mCherry* mRNA (data not shown)). By contrast, the level of luxR-gfp mRNA decreased in the presence of Qrr3 (Figure 2L), and Qrr3 repressed LuxR-GFP protein production (Figure S1J). These data indicate that Qrr3 causes degradation of luxR mRNA. However, the Qrr itself is not degraded and is thus available to regulate other targets. Therefore, we propose that Qrr3 acts catalytically on luxR mRNA.
Finally, the Qrr sRNAs post-transcriptionally activate AphA production by base-pairing to aphA mRNA (Rutherford et al., 2011; Shao and Bassler, 2012). To test if an activated target can compete for Qrr regulation, we performed our competition assay using luxR-gfp as the target mRNA and aphA-mCherry as the competitor mRNA. The endogenous expression level of aphA-mCherry mRNA is much lower than other competitor mRNA targets (data not shown). We therefore introduced an additional plasmid carrying the identical arabinose inducible aphA-mCherry construct into E. coli to boost aphA-mCherry mRNA levels. Figure 2M,N show that aphA can compete for regulation by Qrr3. Specifically, Qrr repression of LuxR-GFP decreased from seven-fold to two-fold (Figure 2N). AphA-mCherry was activated by Qrr3, indicating that aphA is regulated by Qrr3 during the competition (Figure S1K,L). Qrr3 levels declined and the Qrr3 stability dramatically decreased when aphA-mCherry mRNA was present (Figure S2E,F, 2O). Notably, Qrr3 levels reached a plateau after 4 minutes. We suspect that during the first 4 minutes there exists aphA-mCherry, which fosters Qrr3 degradation. However, after 4 minutes, there is likely little or no aphA-mCherry mRNA remaining to promote Qrr3 degradation. Thus, the Qrr3 level remains stable from that point onward. We also measured what becomes of the aphA-gfp mRNA in the absence and presence of Qrr3. Although AphA-GFP protein production is activated ∼2.5-fold by Qrr3 (Figure S1M), the full-length aphA-gfp mRNA decreased ∼2.5 fold in the presence of Qrr3 (Figure 2P). We suggest that when Qrr3 pairs with the aphA mRNA, aphA translation is activated while Qrr3 is destabilized. The fate of the aphA mRNA is unclear and is under investigation.
Together, the above experiments demonstrate that Qrr3 uses four different mechanisms to regulate its mRNA targets: Qrr3 represses luxM mRNA through coupled degradation, luxR mRNA through catalytic degradation, luxO mRNA through sequestration, and Qrr3 activates aphA mRNA translation while the Qrr is itself degraded (Figure 1).
mRNA pairing to particular sRNA stem-loops dictates the Qrr sRNA half-life
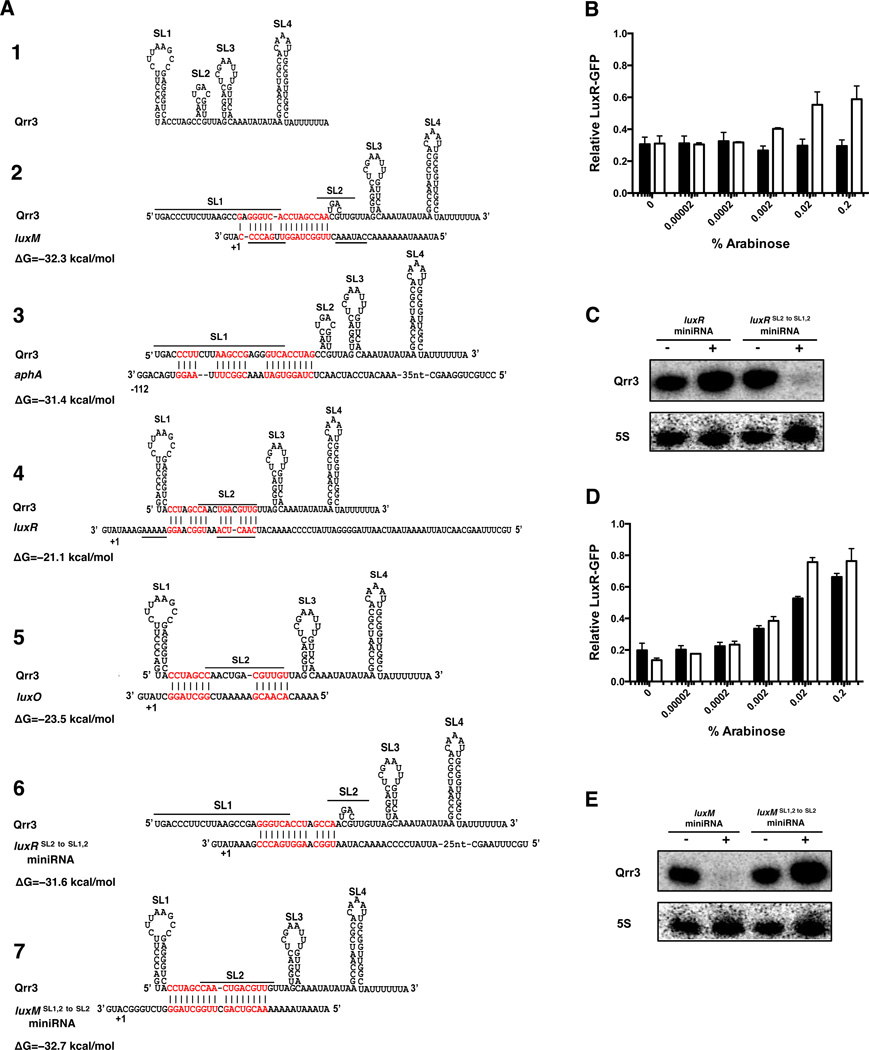
We considered what features of the Qrr-mRNA pairs dictate the Qrr fate. luxM base-pairs with the first and second stem-loops (SL1 + SL2) of the Qrr, aphA base-pairs with SL1, and luxR and luxO base-pair with SL2 (Figure 3A #1–5) (Rutherford et al., 2011; Shao and Bassler, 2012; Teng et al., 2011; Tu and Bassler, 2007; Tu et al., 2010). 5’ stem-loops commonly protect mRNAs from RppH- and RNase E-mediated degradation (Belasco, 2010). We have previously shown that this same mechanism is also used to protect the Qrr sRNAs from degradation. Deletion of SL1 or mutations that disrupt SL1 base-pairing destabilize the Qrr sRNAs, and mutations that restore SL1 base-pairing restore Qrr stability in E. coli (Shao et al., 2013) and in vivo in V. harveyi (Figure S2G). We wondered whether base-pairing to the target mRNA, if it leads to melting of SL1, causes Qrr degradation. To test this possibility, we constructed miniRNAs containing only the 5’UTRs of selected mRNA targets linked to transcription terminators. We used miniRNAs to eliminate ribosome-mediated mRNA stabilization effects. Using the above competition assay, we found that the luxR miniRNA does not affect Qrr3 repression of luxR-gfp, which is consistent with a catalytic regulatory mechanism (Figure 3B black bars). However, shifting the Qrr3-luxR base-pairing from SL2 to SL1 + SL2 (Figure 3A #6) confers competition capability to the luxR SL2 to SL1,2 miniRNA (Figure 3B white bars). The presence of the luxR miniRNA did not affect the level of Qrr3, however, the presence of the luxR SL2 to SL1,2 miniRNA caused a decrease in Qrr3 level (Figure 3C).
Figure 3. Base-pairing to the 5’ stem-loop of Qrr3 leads to Qrr degradation.
(A) Predicted secondary structure of Qrr3 and base-pairing patterns with the target mRNAs. The four predicted stem-loops of Qrr3 are labeled SL1, SL2, SL3 and SL4. Melted loops are shown with overlines. Base-pairing patterns and energies for Qrr3 and target mRNAs were predicted by RNAhybrid (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/). Nucleotides involved in base-pairing are labeled red. Nucleotides mutated to make the miniRNAs (see Supplemental Experimental Procedures) are shown with underlines. Translational start sites are denoted + 1. (B) Competition for Qrr3 regulation between the luxR miniRNA (black bars) or the luxR SL2 to SL1,2 miniRNA (white bars) and luxR-gfp. (C) Northern blot showing Qrr3 levels in the absence (−) and presence (+) of the luxR miniRNA or the luxR SL2 to SL1,2 miniRNA. (D) Competition between the luxM miniRNA (black bars) or the luxM SL1,2 to SL2 miniRNA (white bars) for Qrr3 regulation of luxR-gfp. (E) Northern blot showing Qrr3 levels in the absence (−) and presence (+) of the luxM miniRNA or the luxM SL1,2 to SL2 miniRNA. For (B) and (D), means and SEMs for triplicate cultures are shown. Normalization as in Figure 2. For (C) and (E), results are representative of two independent experiments.
We also constructed a luxM miniRNA and found that it competes with luxR-gfp for Qrr regulation identically to the full-length luxM-mCherry mRNA fusion (Figure 3D, black bars). Qrr3 was degraded in the presence of the luxM miniRNA as shown by its decreased expression level (Figure 3E). A construct that retains the number of base-pairing nucleotides but moves the base-pairing region from SL1 + SL2 to SL2 eliminated the ability of the luxM SL1,2 to SL2 miniRNA to induce Qrr degradation (Figure 3A #7,E). Together, these data suggest that pairing to, and presumably remodeling of SL1 causes Qrr degradation, while pairing to SL2 does not. As controls, we show that the luxR, luxR SL2 to SL1,2, luxM, and luxM SL1,2 to SL2 miniRNAs are expressed (Figure S4A,C, respectively), regulated by Qrr3 (Figure S4B,D, respectively), and the luxR SL2 to SL1,2 and luxM SL1,2 to SL2 miniRNAs behave identically to the luxR SL2 to SL1,2 and luxM SL1,2 to SL2 mRNAs, respectively (Figure S4E).
To map the processing site in the Qrr sRNA when base-paired to luxM, we expressed steady-state levels of Qrr3, induced target mRNA expression, and monitored the dynamics of Qrr decay. Consistent with luxM mRNA being controlled by coupled degradation and luxR mRNA being controlled by a catalytic mechanism, a Qrr sRNA degradation product appeared within ten minutes of induction of luxM mRNA (Figure S2H) whereas Qrr3 was not degraded following luxR mRNA induction (Figure S2H). Primer extension analysis revealed that processing occurred in the Qrr SL1 region after nucleotides C (position 5), U (position 7), and U (position 8) (Figure S2I red arrows), suggesting that opening the SL1 structure makes the RNase cleavage sites accessible.
We did not observe processing of the Qrr sRNA when aphA was expressed (Figure 2O, S2E,F) presumably because the cleavage sites are involved in base-pairing to the aphA mRNA (Figure 3A #3). This arrangement likely shields the sites from cleavage. We wondered whether partial opening of SL1 in a way that does not reveal the cleavage sites is sufficient to induce Qrr degradation. We constructed mutations (GAC to CUG at positions 2 to 4) to partially open SL1 (Figure S2J) and mimic when aphA is base-paired. We call this construct Qrr3mut. The Qrr3mut sRNA is unstable (Figure S2J) compared to wild-type Qrr3 (Figure S2B) and degraded without an apparent cleavage product.
The Qrr-mRNA base-pairing strength determines whether an mRNA target will sequester the Qrr sRNA
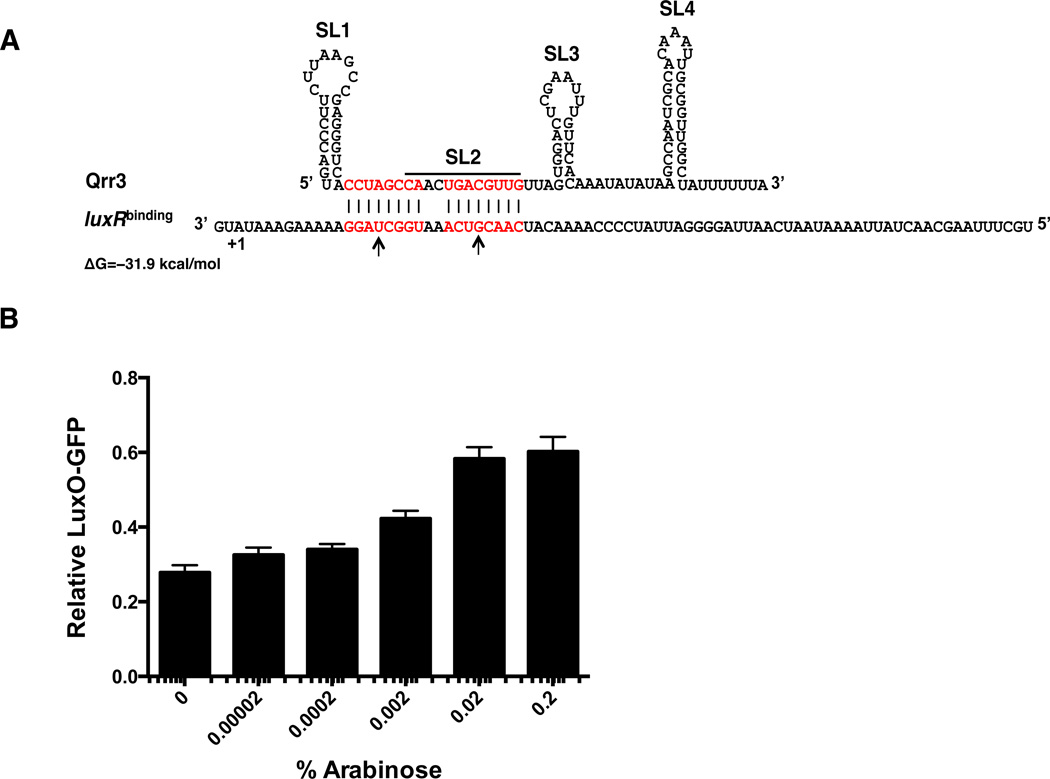
The luxM SL1,2 to SL2 miniRNA does not induce Qrr degradation (Figure 3E). Yet, unlike the luxR miniRNA, the luxM SL1,2 to SL2 miniRNA is capable of competing with luxR-gfp mRNA for Qrr regulation (Figure 3D white bars). These data suggest that the luxM SL1,2 to SL2 miniRNA competes for Qrr regulation by sequestering the Qrr sRNA. Comparison of the predicted binding energies of luxM SL1,2 to SL2 miniRNA, luxR mRNA, and luxR miniRNA (the base pairing region in the luxR mRNA and the luxR miniRNA is identical) to Qrr3 reveals that the Qrr3-luxM SL1,2 to SL2 miniRNA duplex (−32.7 kcal/mol) is more stable than the Qrr3-luxR mRNA (or luxR miniRNA) duplex (−21.1 kcal/mol) (Figure 3A #7, 4). We wondered whether mRNA targets that bind strongly to SL2 of Qrr3 sequester the Qrr sRNA, whereas targets with lower binding energy are catalytically degraded. If so, this could explain why some targets can compete for Qrr regulation while others cannot. To test this hypothesis, we introduced two point mutations into the luxR-mCherry* mRNA 5’UTR to increase its strength of binding to Qrr3 (−31.9 kcal/mol) (Figure 4A). We call this construct luxRbinding-mCherry*. Indeed, luxRbinding-mCherry* gains the capability to compete with luxO-gfp mRNA for Qrr regulation (Figure 4B, compare to Figure 2I). As a control, we show that the luxRbinding-mCherry* mRNA is made at levels comparable to that of luxR-mCherry* mRNA (Figure S3I). We suggest that the base-pairing strength between an mRNA target and the Qrr sRNA governs whether an mRNA target will sequester the Qrr sRNA and thus compete for its regulation against other target mRNAs.
Figure 4. The binding strength of the Qrr sRNA-target mRNA pair determines sequestration versus catalytic degradation.
(A) Base-pairing between Qrr3 and luxRbinding mRNA. Designations as in Figure 3A. Mutated nucleotides are labeled with arrows. (B) Competition between luxRbinding-mCherry* and LuxO-GFP for Qrr3 regulation. luxRbinding-mCherry* was driven by the arabinose promoter. Means and SEMs for triplicate cultures are shown. Normalization as in Figure 2.
The different Qrr regulatory mechanisms govern competition and potency of target control
We used mathematical modeling to explore the consequences of the distinct sRNA-mRNA interaction modes on the different RNA species and the underlying biological process being regulated. This initial modeling effort is focused on sRNA-mediated repression because sRNA-directed activation is not yet well characterized. Based on experimental evidence from previous studies, we assume that Hfq protein complexes are always close to saturated by sRNAs and mRNAs (Hussein and Lim, 2011; Moon and Gottesman, 2011), but that individual sRNA and mRNA molecules actively cycle on and off of Hfq complexes (Fender et al., 2010; Wagner, 2013).
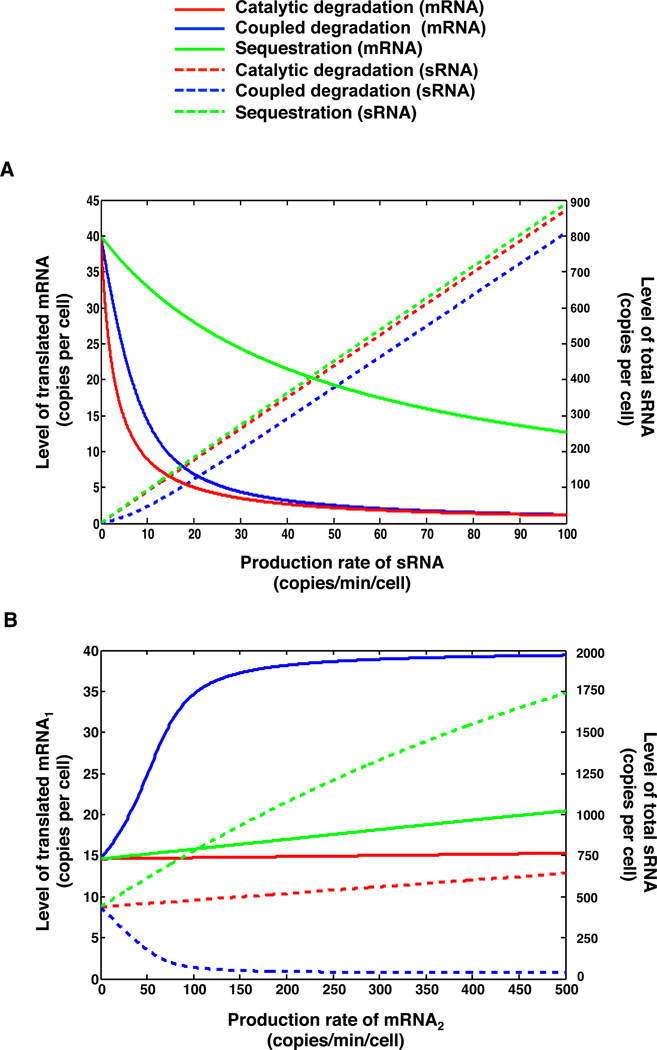
We first modeled the scenario of a single species of sRNA (e.g. a single Qrr) specifically regulating a single type of mRNA target, in the presence of a background of non-cognate sRNAs and mRNAs (see Supplemental Information). In Figure 5A we show as solid curves the copy number of mRNA molecules available for translation, which are the free mRNA molecules and the mRNA molecules bound to Hfq with non-cognate sRNAs, versus the production rate of the sRNA. We find that an mRNA target regulated by catalytic degradation (red) is the most efficiently repressed by the sRNA, followed by an mRNA regulated by coupled degradation (blue). An mRNA controlled by sequestration (green) exhibits only moderate repression by the sRNA. Moreover, mRNA targets that are regulated by coupled degradation decrease the total sRNA level (dashed blue curve). Specifically, the total sRNA level remains suppressed by coupled degradation until sRNA production exceeds the expected threshold of ∼ 7.5 copies/min/cell and then increases linearly. (The threshold occurs where the sRNA production rate/the sRNA degradation rate in the sRNA-Hfq-mRNA complex equals the mRNA production rate/the mRNA degradation rate in the sRNA-Hfq-mRNA complex). This threshold-linear behavior of total sRNA does not occur in the cases of regulation by catalytic degradation or sequestration (red and green dashed curves, respectively).
Figure 5. Modeling the strength and competition capacity of the different sRNA regulatory mechanisms.
(A) The levels of translated mRNA (solid curves) and total sRNA (dashed curves) plotted against the production rate of the sRNA, based on Equations (S1-S6). (B) The levels of translated mRNA1 (solid curves) and total sRNA (dashed curves) are plotted against the production rate of the competitor mRNA2 based on Equations (S1-S6). In (A) and (B), three different regulatory mechanisms are explored: catalytic degradation (red), coupled degradation (blue), and sequestration (green).
We next modeled the scenario in which competition occurs between different mRNA targets for sRNA regulation (See Supplemental Information). In our system, we have one target mRNA (mRNA1) and one competitor mRNA (mRNA2). Our modeling results show that coupled degradation supplies the most efficient competition: the level of translated mRNA1 increases with the production rate of mRNA2 (Figure 5B, solid blue curve), followed by sequestration (Figure 5B, solid green curve). Catalytic degradation provides only minimal competition (Figure 5B, solid red curve). This result is in good qualitative agreement with our experimental data in which luxM and luxO mRNA compete for Qrr regulation and luxR mRNA is nearly incapable of competing for Qrr regulation. We also examined the total sRNA level when we increased the production rate of the competitor mRNA2 (Figure 5B dashed curves). As expected, coupled degradation results in a significant decrease in sRNA level (Figure 5B dashed blue curve). The increase of the total sRNA level in the case of sequestration is due to the sequestering mRNA2 partially protecting the sRNA from degradation (Figure 5B dashed green curve).
The different Qrr regulatory mechanisms govern the dynamics of target mRNA expression
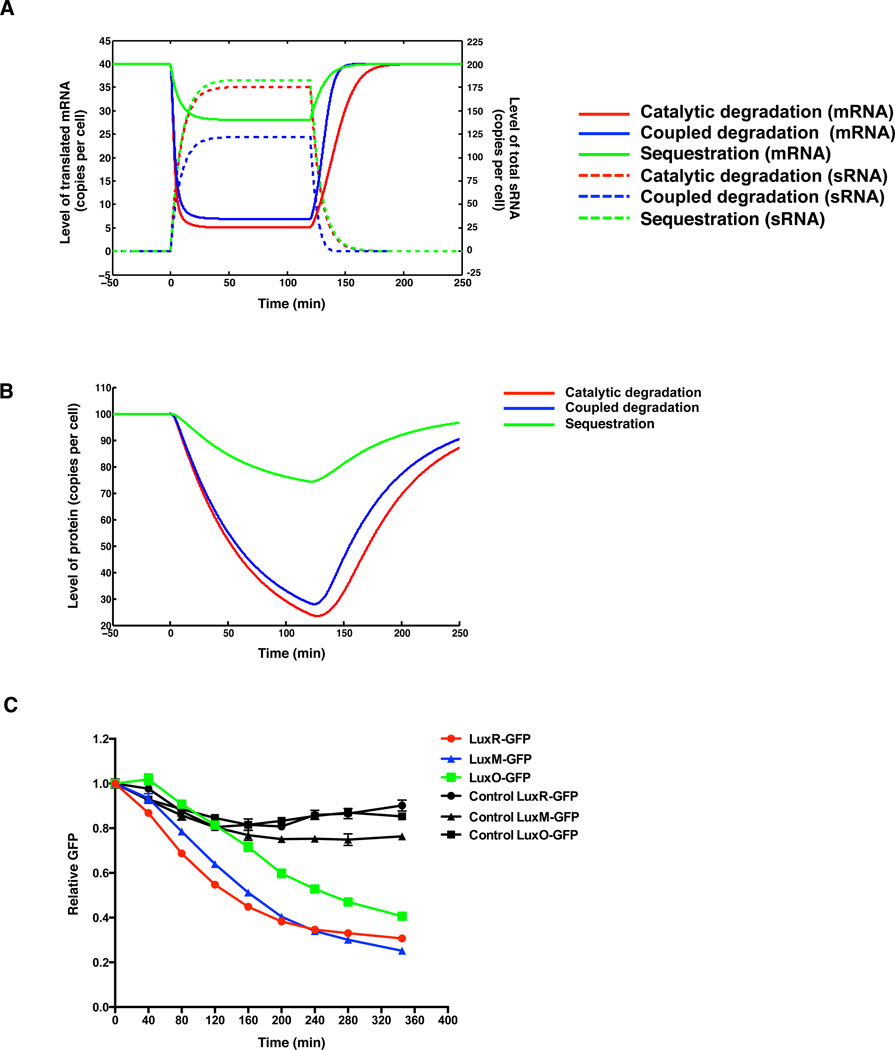
To examine the effect of different regulatory mechanisms on the dynamics of target regulation, we modeled a situation in which sRNA production is induced at time zero and then terminated after two hours. We found that when sRNA production is induced (Figure 6A dashed curves), catalytic degradation and coupled degradation provide the most rapid mRNA and protein response, while sequestration confers the slowest response (Figure 6A solid curves, 6B, and Supplemental Information). By contrast, when sRNA production is terminated, catalytic degradation yields the slowest recovery. As expected, sequestration affects the translated mRNA level but it does not affect the total mRNA level. Catalytic degradation and coupled degradation affect both mRNA translation and overall target mRNA levels (Figure S5). In all cases, repression of mRNA is rapid (∼10 minutes) (Figure 6A solid curves), whereas repression of protein is slow – approaching steady state only after hours – due to the slow dilution rate of protein (Figure 6B). These predictions are in agreement with existing experimental data (Papenfort et al., 2013; Vanderpool and Gottesman, 2004).
Figure 6. mRNA target dynamics provided by the different sRNA regulatory mechanisms.
(A) The levels of translated mRNA (solid curves) and total sRNA (dashed curves) are plotted over time based on Equations (S1-S6). (B) The level of regulated protein is plotted over time based on Equations (S1-S6 and S9). sRNA production is induced at time zero and is terminated at 120 min. Three different regulatory mechanisms are explored: catalytic degradation (red), coupled degradation (blue), and sequestration (green). (C) Repression of target mRNAs following Qrr sRNA induction. GFP fluorescence at each time-point was normalized to mCherry fluorescence, and the relative GFP levels are plotted. The results are LuxR-GFP: red and black (control) circles, LuxM-GFP: blue and black (control) triangles, LuxO-GFP: green and black (control) squares. Means and SEMs from three independent cultures are shown.
With respect to the mRNA targets studied here, the model predicts that luxR mRNA will be the most sensitive to changes in Qrr levels since it is controlled by catalytic degradation. Thus, during the high-cell-density to low-cell-density transition, when the Qrr sRNAs are produced, luxR mRNA should respond more rapidly to changes in Qrr levels than do luxM and luxO mRNA. To test the predictions of the model, we constructed three V. harveyi strains that report on target mRNA levels by integrating luxR, luxM, or a luxO 5’UTR translational GFP fusion under a constitutive promoter onto the chromosome. We used mCherry oriented in the opposite direction to normalize for cellular protein (Long et al., 2009). We measured GFP and mCherry fluorescence after we induced Qrr production by adding a quorum-sensing antagonist (Experimental Procedures) (Shao et al., 2013). Figure 6C shows that the LuxR-GFP protein is indeed the most rapid to respond to Qrr changes, followed by the LuxM-GFP protein, and finally the LuxO-GFP protein. As a control, we measured LuxR-GFP, LuxM-GFP, and LuxO-GFP levels in V. harveyi strain lacking all five Qrr sRNAs; GFP did not significantly change following addition of the antagonist.
The different Qrr regulatory mechanisms are critical for quorum-sensing circuit dynamics
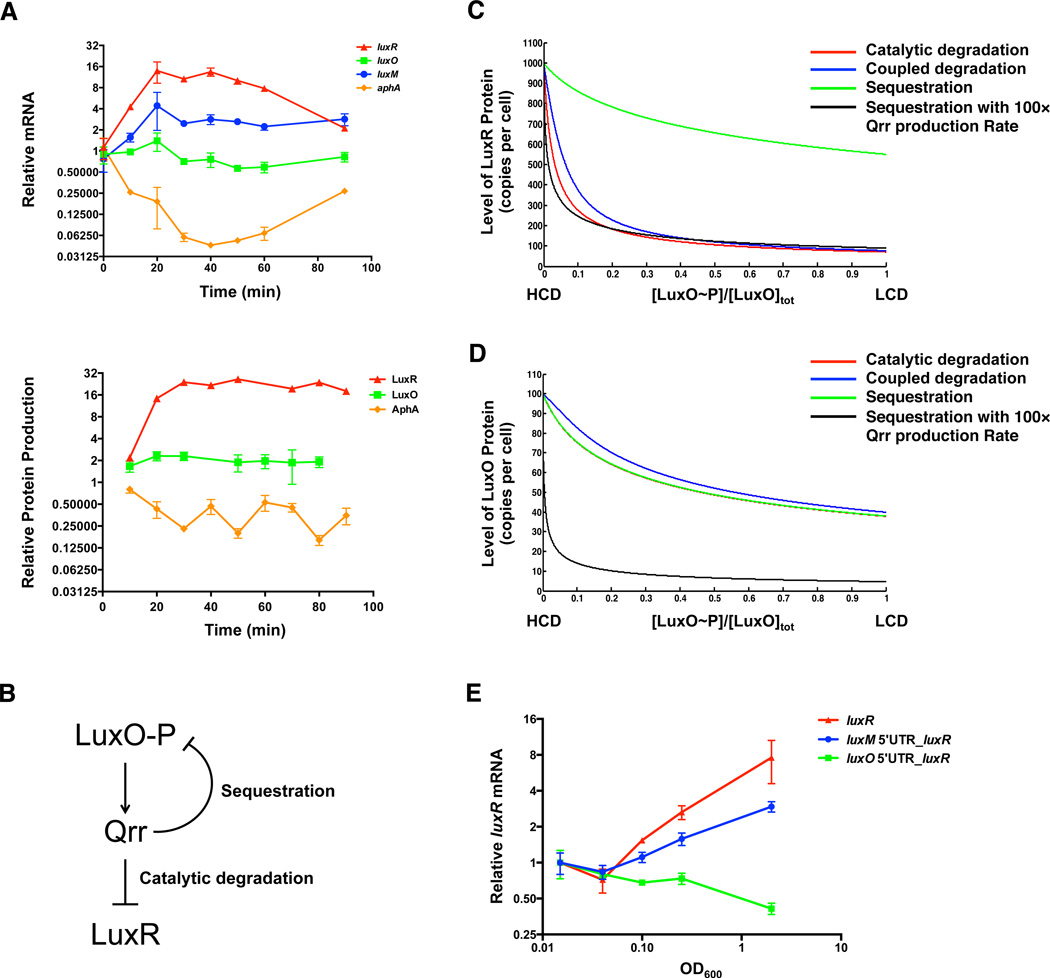
Our data and model show that the particular Qrr sRNA mechanism used to regulate each mRNA target dictates the level and dynamics of the target’s expression. We expect that these dynamics specify the quorum-sensing response timing in vivo. To investigate this notion, we measured the in vivo dynamical changes of individual quorum-sensing components that occurred in response to alterations in Qrr levels. We measured the mRNA levels and the rates of protein synthesis of LuxR, LuxO, LuxM, and AphA over a ninety-minute time period following termination of Qrr production. As controls, we show that the levels of Qrr1-4 decreased following autoinducer addition (Figure S7A). Qrr5 is not produced under our experimental conditions, so we did not measure it (Tu and Bassler, 2007). Figure 7A shows that, following the addition of the autoinducer, luxR mRNA increased ∼14 fold and the rate of LuxR protein synthesis increased ∼25 fold; luxO mRNA does not change within 90 minutes, however, the rate of LuxO protein production increased 2-fold; luxM mRNA increased ∼3 fold (we were unable to detect the LuxM protein, likely because the luxM gene is partially deleted in this locked strain); aphA mRNA decreased ∼20 fold and AphA protein synthesis decreased ∼4–6 fold. These results confirm our prediction that catalytically regulated targets (e.g., luxR) undergo larger dynamic range changes than do targets regulated by coupled degradation (e.g., luxM). Sequestered targets such as luxO are the most weakly regulated.
Figure 7. Particular Qrr regulatory mechanisms are crucial for proper quorum-sensing dynamics.
(A) qRT-PCR and BONCAT of luxR, luxO, luxM, and aphA following addition of AI-1 to TL25. Data were normalized to the first time-point in the RNA measurement. Means and SEMs from three independent cultures are shown. Relative protein synthesis rates were measured by BONCAT (Supplemental Information) and the evidence for each quantification is provided in Table S5. (B) Simplified quorum-sensing circuit used for mathematical modeling. (C) LuxR and (D) LuxO protein copy number plotted against the ratio of phosphorylated LuxO to total LuxO protein, based on Equations (S1-S6, 1–3). In (C) and (D), luxR mRNA is regulated by catalytic degradation (red), coupled degradation (blue), or sequestration (green). The black curve shows the case when luxR mRNA is regulated by sequestration but the Qrr production rate is increased 100-fold. (E) qRT-PCR of luxR mRNA from luxR R17C (red), luxM 5’UTR_luxR R17C (blue), and luxO 5’UTR_luxR R17C (green) over growth. Data from each strain were normalized to the first time-point (OD600=0.015) and plotted against OD600. Means and SEMs from four independent cultures are shown.
We next explored how important the specific mechanism is for the quorum-sensing response. We altered the regulatory mechanism by which a particular mRNA target (luxR) is controlled, and tested the effects on overall quorum-sensing dynamics. To accompany the experiment, we modeled an internal segment of the circuit in which phosphorylated LuxO activates the production of the Qrr sRNAs and the Qrr sRNAs catalytically repress luxR mRNA, whereas they sequester luxO mRNA (Figure 7B).
The subcircuit equations correspond to those used for one Qrr with two mRNA targets (Supplemental Information), with Qrr production assumed to be proportional to the level of phosphorylated LuxO. The kinetic equation for the Qrr sRNA is therefore:
| (1) |
We added two kinetic equations for LuxO and LuxR protein:
| (2) |
| (3) |
Using these equations, we explored three possible mechanisms for luxR regulation: catalytic degradation, coupled degradation, and sequestration. Our choice of parameters is based on the RNA copy numbers we measured (Figure S6) and previous measurement of LuxR protein copy number (Teng et al., 2010) (Supplemental Information).
In Figure 7C, we plot the prediction for LuxR protein copy number per cell versus the phosphorylated fraction of LuxO ([LuxO∼P]/[LuxO]). The ratio [LuxO∼P]/[LuxO] is low in the high-cell-density state and high in the low-cell-density state. Within the model, the catalytic mechanism yields the most efficient repression of LuxR protein, followed by coupled degradation, while sequestration does not achieve the experimentally observed level of LuxR repression. Can sequestration adequately repress LuxR if we allow increased Qrr production? We found that in order to achieve ∼10-fold repression of LuxR, the Qrr production rate must be increased ∼100-fold (black curve, Figure 7C). Under this condition, LuxO protein is repressed at low cell density to the unrealistically low level of fewer than five copies per cell (black curve, Figure 7D). In effect, converting regulation of LuxR from catalytic degradation to sequestration would require rewiring of much of the quorum-sensing network to achieve the same dynamics. Thus, the modeling results suggest that the particular Qrr sRNA mechanisms used to regulate specific quorum-sensing mRNA targets can be crucial for the integrated operation of the quorum-sensing circuit.
To experimentally validate the prediction from the model, we replaced the 5’UTR of luxR with that of luxM or luxO on the chromosome of a V. harveyi strain containing only Qrr3. We note that the levels of LuxR protein produced from the three endogenous 5’UTRs are different. Different levels of LuxR feedback on quorum-sensing circuit components would complicate our analysis (Chatterjee et al., 1996; Tu et al., 2008), thus we isolated LuxR production from feedback by introducing a mutation in LuxR (LuxR R17C) that eliminates DNA binding (van Kessel et al., 2013). Figure 7E shows that luxR mRNA regulated by a catalytic (red curve) or coupled degradation (blue curve) mechanism increases from low cell density to high cell density, although coupled degradation provides a smaller dynamic range (∼3.5 fold) for luxR mRNA than catalytic degradation (∼10 fold). luxR mRNA regulated by sequestration (green curve) shows no increase during growth, which agrees with our prediction. Over the course of the experiment, Qrr3 levels decrease from low cell density to high cell density and the levels of Qrr3 in the three strains are comparable (Figure S7B). These results show that the regulatory mechanism used for a particular target mRNA (luxR in this case) determines the precise timing and amplitude of the quorum-sensing response.
Discussion
sRNAs are ubiquitous regulators in bacterial genetic circuits, primarily functioning to control growth rate and in stress responses (Gottesman, 2004). Several well-characterized bacterial sRNAs, such as Spot42, RyhB, RybB, and the Qrr sRNAs each control multiple target mRNAs (Beisel and Storz, 2011; Massé and Gottesman, 2002; Papenfort et al., 2010; Shao et al., 2013; Storz et al., 2011). Here, we use the Qrr3 sRNA to show that a single sRNA can regulate its different targets by distinct mechanisms. The particular mechanism used is defined by the base-pairing strategy the Qrr sRNA employs for the particular mRNA target. Specifically, mRNA targets that base-pair with the first stem-loop of the Qrr sRNA cause Qrr degradation, and these targets include both repressed (luxM) and activated (aphA) targets. mRNA targets that base-pair with the second stem-loop of the Qrr sRNA do not cause Qrr degradation. Rather, they sequester the Qrr sRNA if the binding is strong (luxO), and they are catalytically regulated if binding is weak (luxR). Our combined mathematical modeling and experiments show that the regulatory mechanism used determines the potency of regulation, competition capability, and the temporal dynamics of each target mRNA. These distinct mechanisms are crucial to drive the overall quorum-sensing circuit dynamics.
In terms of deployment of this set of regulatory mechanisms, we suspect that mRNA targets that require complete and immediate repression will likely be regulated either catalytically or by coupled degradation. Growth and stress response mRNAs are good candidates for these modes of sRNA regulation (Beisel and Storz, 2011; Massé and Gottesman, 2002; Vanderpool and Gottesman, 2004). Catalytic degradation, because it does not alter the total sRNA pool, could be the superior mode of regulation when a target mRNA exists in high copy numbers and requires a large dynamic range for function, or when a target mRNA needs to be fully repressed even under conditions of low sRNA levels. The MicM (ChiX) target ybfM (chiP) fits this scenario. ybfM (chiP) is completely silenced by MicM (ChiX) in the absence of chitooligosaccharide inducers (Figueroa-Bossi et al., 2009; Overgaard et al., 2009).
The coupled degradation mechanism is notable because it provides a threshold-linear response: regulation depends on the relative sRNA to mRNA ratio (Levine et al., 2007). Specifically, in the low sRNA:target mRNA regime, mRNA repression is not efficient because the sRNA is degraded following base-pairing and this greatly reduces the sRNA pool. Thus, coupled degradation is an excellent mechanism for rapidly turning over, and thus eliminating, the sRNA pool once the response is complete. Good examples for this case include the sRNAs RyhB and MicM (ChiX) (Figueroa-Bossi et al., 2009; Massé et al., 2003; Overgaard et al., 2009; Plumbridge et al., 2014).
Sequestration is an excellent mechanism for regulation of target mRNAs that require modulation rather than dramatic on-off changes. In this case, the mRNA levels are not significantly affected by sRNA regulation, which has the advantage of fine-tuning target expression levels. One example to consider is the sRNA Spot42 and its target galK. Spot42-directed repression of galK by sequestration presumably fine-tunes the relative levels of the galactose catabolism proteins for optimal carbon utilization (Møller et al., 2002). We hypothesize that the propensity for a target mRNA to sequester an sRNA depends on both the binding strength of the sRNA-mRNA pair and the degradation rate of the target mRNA upon pairing. Strong sequestration will occur if the target mRNA binds tightly to the sRNA but the target mRNA is not degraded efficiently. This is the case for the luxO-Qrr pair.
Finally, we show that the Qrr sRNA is degraded following activation of the aphA mRNA. We hypothesize that this mode of action generates a negative feedback loop that has the advantage of preventing over-expression of the target mRNA when the sRNA level is low. Specifically, since the sRNA is degraded during regulation, it cannot be reused. Thus, when the sRNA:target mRNA ratio is low, the degree of target mRNA activation is limited by the concentration of the sRNA.
Here we rationalize the Qrr sRNAs use of particular regulatory mechanisms for the particular quorum-sensing target mRNAs. First, regarding luxR mRNA: LuxR is the master quorum-sensing regulator that, at high cell density, controls the bulk of genes in the quorum-sensing regulon (∼600 genes in V. harveyi) (Rutherford et al., 2011). Our measurements of luxR mRNA and LuxR protein copy numbers indicate that luxR exists in high copy numbers (at least 40 mRNA copies and ∼600 protein dimers per cell) at high cell density, presumably due to the requirement for LuxR dimers to bind their 115 DNA promoter sites (van Kessel et al., 2013; Teng et al., 2010). However, this high LuxR concentration presents a conundrum for the cell when it needs to, essentially instantaneously, transition to the low-cell-density mode upon, for example, excretion from the host or exit from a biofilm. Repression of luxR mRNA via a Qrr catalytic mechanism should be the most effective means to rapidly decrease high levels of LuxR protein to reset the quorum-sensing genetic program. Regarding luxO mRNA, which is controlled by a Qrr feedback-sequestration mechanism: The Qrr-to-luxO feedback loop acts as a rheostat to moderately adjust Qrr levels (Tu et al., 2010). We argue that using sequestration to regulate luxO mRNA prevents dramatic spikes and valleys in Qrr levels, while simultaneously buffering luxO mRNA levels from valleys and spikes due to noise associated with fluctuations in Qrr levels. With respect to the use of coupled degradation to regulate luxM mRNA: we propose that the luxM mRNA and seven other target mRNAs that are able to base-pair with SL1 of the Qrr sRNAs (Shao et al., 2013) are exploited to control Qrr turnover in order to set the appropriate concentration of the total Qrr sRNA pool under different quorum-sensing states. This feature is important for when the cells initiate the high-cell-density program and they need to eliminate the Qrr sRNAs. Finally, as we discussed above, activation of aphA with concomitant degradation of the Qrr sRNAs may prevent overproduction of the AphA protein. Our finding that AphA protein production changes only 4∼6 fold supports this hypothesis (Figure 7A). AphA fine-tunes quorum-sensing gene expression at low cell density (van Kessel et al., 2012). Keeping AphA levels in check may be critical for its subtle function. Indeed, AphA feeds back to repress Qrr transcription, which further guarantees tight control of AphA levels (Rutherford et al., 2011).
We predict that the regulatory mechanisms we discovered between Qrr3 and the target mRNAs are conserved across all five Qrr sRNAs based on their highly similar secondary structures and sequences (Tu and Bassler, 2007). The caveat is that Qrr1 lacks nine nucleotides in the first stem-loop, which makes it unable to regulate certain targets, such as aphA (Shao and Bassler, 2012). The five Qrr sRNAs are expressed at different levels and with somewhat different timing (Tu and Bassler, 2007). Thus, how the in vivo competition occurs between all five Qrr sRNAs and all 20 mRNA targets to provide a robust quorum-sensing response remains to be defined. Nonetheless, embedding the capacity for multiple regulatory mechanisms into a single sRNA is an evolutionarily economical method to endow a biological circuit with diverse dynamic behaviors. The principles underpinning the regulatory mechanisms we discovered here could be employed by other natural systems or to engineer synthetic sRNAs with numerous functions.
Experimental Procedures
Strains, plasmids and oligonucleotides used in this study are listed in Table S1, S2 and S3. Detailed protocols are described in Supplemental Experimental Procedures.
Competition assay
Overnight cultures (Table S1) were diluted 1000-fold into fresh M9 minimal medium containing 0.5% glycerol, appropriate antibiotics, 0.2 mM IPTG, 0 or 3 ng mL−1 anhydrotetracycline (aTc; Clontech) and varying amounts of arabinose. GFP and mCherry fluorescence were measured following 10 h of growth using FACS (BD Biosciences FACSAria cell sorter).
RNA expression and half-life
Qrr3 and target mRNAs were induced for 10 h and measured by Northern blot to determine expression. 250 µg mL−1 rifampicin was added to stop transcription followed by collection of RNA at different time points to determine half-life.
V. harveyi GFP assay
Overnight cultures of V. harveyi strains LF1838, LF1845, LF1848, LF2328, LF2332, LF2335 were diluted to OD600=0.0002 into fresh AB medium containing 1 µM AI-1 and grown for 6.5 h. 100 µM 3-oxo-C12-HSL (Sigma) (LuxN/AI-1 antagonist) was added to cultures. GFP and mCherry fluorescence were measured every 40 min thereafter using FACS.
qRT–PCR of luxR mRNA
Overnight cultures of V. harveyi strains LF2269, LF2246, and LF2254 were diluted into fresh LM medium to OD600=0.005 and grown for 6 h to OD600=2.0. Cultures were diluted again into fresh LM medium to OD600=0.005. Total RNA was collected at 45, 90, 135, 180 and 345 min after the dilution. 4 µg of total RNA was used for cDNA synthesis and qRT-PCR was performed as described in Supplemental Information.
qRT-PCR and Proteomics
Overnight cultures of V. harveyi strain TL25 were diluted 1:1000 fold into fresh LM medium and grown to mid-log phase. Cultures were divided in half, and one aliquot was treated with 10 µM AI-1. Total RNA was collected from both samples at 0, 10, 20, 30, 40, 50, 60 and 90 min thereafter. 4 µg of total RNA was used for cDNA synthesis and qRT-PCR was performed as described in Supplemental Information. Proteomics was performed as described in Supplemental Information.
Supplementary Material
Highlights.
The Qrr3 sRNA uses four different mechanisms to regulate target mRNAs
Base-pairing patterns determine the particular sRNA regulatory mechanism
The different regulatory mechanisms confer distinct regulatory strength and dynamics
The particular regulatory mechanisms are crucial for overall quorum-sensing dynamics
Acknowledgements
We thank Terence Hwa for generously providing the BW-RI strain and the pZA31-lucNB and pZE12G plasmids. We are indebted to members of the Bassler and Wingreen laboratories for insightful discussions and suggestions. This work was supported by the Howard Hughes Medical Institute, National Institutes of Health (NIH) Grant 5R01GM065859 and National Science Foundation (NSF) Grant MCB-0343821 to BLB NIH Grant R01GM082938 to NSW, and by NIH Grant R01GM062523 and the Institute for Collaborative Biotechnologies through grant W911NF-09-0001 from the U.S. Army Research Office to DAT. KP is supported by a post-doctoral fellowship from the Human frontiers in Science program (HFSP). STR was supported by NIH fellowship F32AI085922. JCVK is supported by funds from Indiana University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
LF, STR, KP, JDB, JCVK, DAT, NSW, and BLB designed the experiments; LF, KP, JDB and JCVK performed the experiments; LF, KP, JDB, NSW, and BLB analyzed the data; and LF, NSW, and BLB wrote the paper.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Altuvia S, Zhang A, Argaman L, Tiwari A, Storz G. The Escherichia coli OxyS regulatory RNA represses fhlA translation by blocking ribosome binding. EMBO J. 1998;17:6069–6075. doi: 10.1093/emboj/17.20.6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisel CL, Storz G. The base-pairing RNA spot 42 participates in a multioutput feedforward loop to help enact catabolite repression in Escherichia coli. Mol. Cell. 2011;41:286–297. doi: 10.1016/j.molcel.2010.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belasco JG. All things must pass: contrasts and commonalities in eukaryotic and bacterial mRNA decay. Nat. Rev. Mol. Cell Biol. 2010;11 doi: 10.1038/nrm2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee J, Miyamoto CM, Meighen EA. Autoregulation of luxR: the Vibrio harveyi lux-operon activator functions as a repressor. Mol. Microbiol. 1996;20:415–425. doi: 10.1111/j.1365-2958.1996.tb02628.x. [DOI] [PubMed] [Google Scholar]
- Fender A, Elf J, Hampel K, Zimmermann B, Wagner EGH. RNAs actively cycle on the Sm-like protein Hfq. Genes Dev. 2010;24:2621–2626. doi: 10.1101/gad.591310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa-Bossi N, Valentini M, Malleret L, Fiorini F, Bossi L. Caught at its own game: regulatory small RNA inactivated by an inducible transcript mimicking its target. Genes Dev. 2009;23:2004–2015. doi: 10.1101/gad.541609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröhlich KS, Vogel J. Activation of gene expression by small RNA. Curr. Opin. Microbiol. 2009;12:674–682. doi: 10.1016/j.mib.2009.09.009. [DOI] [PubMed] [Google Scholar]
- Gottesman S. The small RNA regulators of Escherichia coli: roles and mechanisms*. Annu. Rev. Microbiol. 2004;58:303–328. doi: 10.1146/annurev.micro.58.030603.123841. [DOI] [PubMed] [Google Scholar]
- Hussein R, Lim HN. Disruption of small RNA signaling caused by competition for Hfq. Proc. Natl. Acad. Sci. U. S. A. 2011;108:1110–1115. doi: 10.1073/pnas.1010082108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto H, Koide Y, Morita T, Aiba H. Base-pairing requirement for RNA silencing by a bacterial small RNA and acceleration of duplex formation by Hfq. Mol. Microbiol. 2006;61:1013–1022. doi: 10.1111/j.1365-2958.2006.05288.x. [DOI] [PubMed] [Google Scholar]
- Van Kessel JC, Rutherford ST, Shao Y, Utria AF, Bassler BL. The master regulators AphA and LuxR control the Vibrio harveyi quorum-sensing regulon: analysis of their individual and combined effects. J. Bacteriol. 2012;195:436–443. doi: 10.1128/JB.01998-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Kessel JC, Ulrich LE, Zhulin IB. Analysis of Activator and Repressor Functions Reveals the Requirements for Transcriptional Control by LuxR , the Master Regulator of Quorum Sensing in Vibrio harveyi. MBio. 2013;4:e00378–e00413. doi: 10.1128/mBio.00378-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, Bassler BL. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell. 2004;118:69–82. doi: 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Levine E, Zhang Z, Kuhlman T, Hwa T. Quantitative characteristics of gene regulation by small RNA. PLoS Biol. 2007;5:e229. doi: 10.1371/journal.pbio.0050229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long T, Tu KC, Wang Y, Mehta P, Ong NP, Bassler BL, Wingreen NS. Quantifying the integration of quorum-sensing signals with single-cell resolution. PLoS Biol. 2009;7:e68. doi: 10.1371/journal.pbio.1000068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majdalani N, Cunning C, Sledjeski D, Elliott T, Gottesman S. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc. Natl. Acad. Sci. U. S. A. 1998;95:12462–12467. doi: 10.1073/pnas.95.21.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massé E, Gottesman S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 2002;99:4620–4625. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massé E, Escorcia FE, Gottesman S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 2003;17:2374–2383. doi: 10.1101/gad.1127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullen CA, Benhammou JN, Majdalani N, Gottesman S. Mechanism of positive regulation by DsrA and RprA small noncoding RNAs: pairing increases translation and protects rpoS mRNA from degradation. J. Bacteriol. 2010;192:5559–5571. doi: 10.1128/JB.00464-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller T, Franch T, Udesen C, Gerdes K, Valentin-Hansen P. Spot 42 RNA mediates discoordinate expression of the E. coli galactose operon. Genes Dev. 2002;16:1696–1706. doi: 10.1101/gad.231702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon K, Gottesman S. Competition among Hfq-binding small RNAs in Escherichia coli. Mol. Microbiol. 2011;82:1545–1562. doi: 10.1111/j.1365-2958.2011.07907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overgaard M, Johansen J, Møller-Jensen J, Valentin-Hansen P. Switching off small RNA regulation with trap-mRNA. Mol. Microbiol. 2009;73:790–800. doi: 10.1111/j.1365-2958.2009.06807.x. [DOI] [PubMed] [Google Scholar]
- Papenfort K, Bouvier M, Mika F, Sharma CM, Vogel J. Evidence for an autonomous 5’ target recognition domain in an Hfq-associated small RNA. Proc. Natl. Acad. Sci. U. S. A. 2010;107:20435–20440. doi: 10.1073/pnas.1009784107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort K, Sun Y, Miyakoshi M, Vanderpool CK, Vogel J. Small RNA-mediated activation of sugar phosphatase mRNA regulates glucose homeostasis. Cell. 2013;153:426–437. doi: 10.1016/j.cell.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plumbridge J, Bossi L, Oberto J, Wade JT, Figueroa-Bossi N. Interplay of transcriptional and small RNA-dependent control mechanisms regulates chitosugar uptake in Escherichia coli and Salmonella. Mol. Microbiol. 2014 doi: 10.1111/mmi.12573. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Prévost K, Desnoyers G, Jacques J-F, Lavoie F, Massé E. Small RNA-induced mRNA degradation achieved through both translation block and activated cleavage. Genes Dev. 2011;25:385–396. doi: 10.1101/gad.2001711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford ST, Bassler BL. Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb. Perspect. Med. 2012;2:1–25. doi: 10.1101/cshperspect.a012427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford ST, van Kessel JC, Shao Y, Bassler BL. AphA and LuxR/HapR reciprocally control quorum sensing in vibrios. Genes Dev. 2011;25:397–408. doi: 10.1101/gad.2015011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y, Bassler BL. Quorum-sensing non-coding small RNAs use unique pairing regions to differentially control mRNA targets. Mol. Microbiol. 2012;83:599–611. doi: 10.1111/j.1365-2958.2011.07959.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y, Feng L, Rutherford ST, Papenfort K, Bassler BL. Functional determinants of the quorum-sensing non-coding RNAs and their roles in target regulation. EMBO J. 2013;32:2158–2171. doi: 10.1038/emboj.2013.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz G, Vogel J, Wassarman KM. Regulation by small RNAs in bacteria: expanding frontiers. Mol. Cell. 2011;43:880–891. doi: 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng S-W, Wang Y, Tu KC, Long T, Mehta P, Wingreen NS, Bassler BL, Ong NP. Measurement of the copy number of the master quorum-sensing regulator of a bacterial cell. Biophys. J. 2010;98:2024–2031. doi: 10.1016/j.bpj.2010.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng S-W, Schaffer JN, Tu KC, Mehta P, Lu W, Ong NP, Bassler BL, Wingreen NS. Active regulation of receptor ratios controls integration of quorum-sensing signals in Vibrio harveyi. Mol. Syst. Biol. 2011;7:491. doi: 10.1038/msb.2011.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu KC, Bassler BL. Multiple small RNAs act additively to integrate sensory information and control quorum sensing in Vibrio harveyi. Genes Dev. 2007;21:221–233. doi: 10.1101/gad.1502407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu KC, Waters CM, Svenningsen SL, Bassler BL. A small-RNA-mediated negative feedback loop controls quorum-sensing dynamics in Vibrio harveyi. Mol. Microbiol. 2008;70:896–907. doi: 10.1111/j.1365-2958.2008.06452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu KC, Long T, Svenningsen SL, Wingreen NS, Bassler BL. Negative feedback loops involving small regulatory RNAs precisely control the Vibrio harveyi quorum-sensing response. Mol. Cell. 2010;37:567–579. doi: 10.1016/j.molcel.2010.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udekwu KI, Darfeuille F, Vogel J, Reimegård J, Holmqvist E, Wagner EGH. Hfq-dependent regulation of OmpA synthesis is mediated by an antisense RNA. Genes Dev. 2005;19:2355–2366. doi: 10.1101/gad.354405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderpool CK, Gottesman S. Involvement of a novel transcriptional activator and small RNA in post-transcriptional regulation of the glucose phosphoenolpyruvate phosphotransferase system. Mol. Microbiol. 2004;54:1076–1089. doi: 10.1111/j.1365-2958.2004.04348.x. [DOI] [PubMed] [Google Scholar]
- Vogel J, Luisi BF. Hfq and its constellation of RNA. Nat. Rev. Microbiol. 2011;9:578–589. doi: 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner EGH. Cycling of RNAs on Hfq. RNA Biol. 2013;10:619–626. doi: 10.4161/rna.24044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.