Abstract
Sarco(endo)plasmic reticulum calcium ATPases (SERCA) are cellular pumps that transport Ca2+ into the sarcoplasmic reticulum (SR). Serca2 is the most widely expressed gene family member. The very early embryonic lethality of Serca2null mouse embryos has precluded further evaluation of loss of Serca2 function in the context of organ physiology. We have generated mice carrying a conditional Serca2flox allele which allows disruption of the Serca2 gene in an organ-specific and/or inducible manner. The model was tested by mating Serca2flox mice with MLC-2vwt/Cre mice and with αMHC-Cre transgenic mice. In heterozygous Serca2wt/flox MLC-2vwt/Cre mice, the expression of SERCA2a and SERCA2b proteins were reduced in the heart and slow skeletal muscle, in accordance with the expression pattern of the MLC-2v gene. In Serca2flox/flox Tg(αMHC-Cre) embryos with early homozygous cardiac Serca2 disruption, normal embryonic development and yolk sac circulation was maintained up to at least embryonic stage E10.5. The Serca2flox mouse is the first murine conditional gene disruption model for the SERCA family of Ca2+ ATPases, and should be a powerful tool for investigating specific physiological roles of SERCA2 function in a range of tissues and organs in vivo both in adult and embryonic stages.
Keywords: Serca2, Calcium ATPase, Endoplasmic reticulum, Sarcoplasmic reticulum, Flox, Transgenic mouse
1. Introduction
The P-type sarco(endo)plasmic reticulum (SR) Ca2+ ATPases (SERCA) are cellular ATP-driven Ca2+ pumps that transport Ca2+ into the SR. SERCA proteins are encoded by the three genes Serca1, Serca2 and Serca3 [1,2]. Serca2 is expressed in most tissues in the body, whereas Serca1 expression is restricted to fast-twitch skeletal muscle and Serca3 is expressed mainly in secretory tissues and in organs such as the kidneys and pancreas [1,2]. Splicing variants and protein isoforms with restricted expression patterns have been identified for all three gene family members. The Serca2 gene encodes two major protein isoforms. SERCA2a is expressed predominantly in cardiomyocytes and slow-twitch skeletal muscle, but has also been detected in smooth muscle cells [3], keratinocytes [4], pancreatic epithelial cells [5] and in cerebellar Purkinje cells [6]. SERCA2b has a longer C-terminal tail, and is expressed ubiquitously inmost tissues. A third inducible isoform, SERCA2c, has been identified in human monocytes and heart [7,8], but not in other species.
Much of the current understanding of SERCA2 function is derived from the heart. SERCA2 sequesters Ca2+ from the cytosol into the SR during the relaxation phase of the cardiac excitation–contraction cycle. In heart failure patients and in animal heart failure models, SERCA2 activity or expression is often reduced [9]. Decreased SERCA2 expression or function results in less efficient removal of Ca2+ from the cytosol, leading to the prolonged muscle relaxation time and reduced contractile force as found in heart failure patients and in animal models of heart failure [10–12].
Systemic disruption of Serca2 (Serca2null/null) is lethal in genetically modified mice, thus precluding further analysis of SERCA2 function both during development and in adult animals [13]. To circumvent this developmental lethality, we have created a Serca2flox mouse. In the Serca2flox allele, all SERCA2 isoforms are disrupted upon Cre recombinase activity. Here we show that tissue-specific reduction of SERCA2 expression in cardiac and slow-twitch skeletal muscle is obtained in somatic heterozygous Serca2wt/flox MLC-2vwt/Cre mice as a test of functionality of the new Serca2flox allele. Furthermore, we show that in embryos with cardiac disruption of Serca2, normal heart development and fetal yolk sac circulation is supported up to embryonic age E10.5, thus making it possible to study the role of both SERCA2-dependent and SERCA2-independent Ca2+ transport mechanisms in the early murine embryonic heart.
2. Materials and methods
2.1. Generation of Serca2flox mice
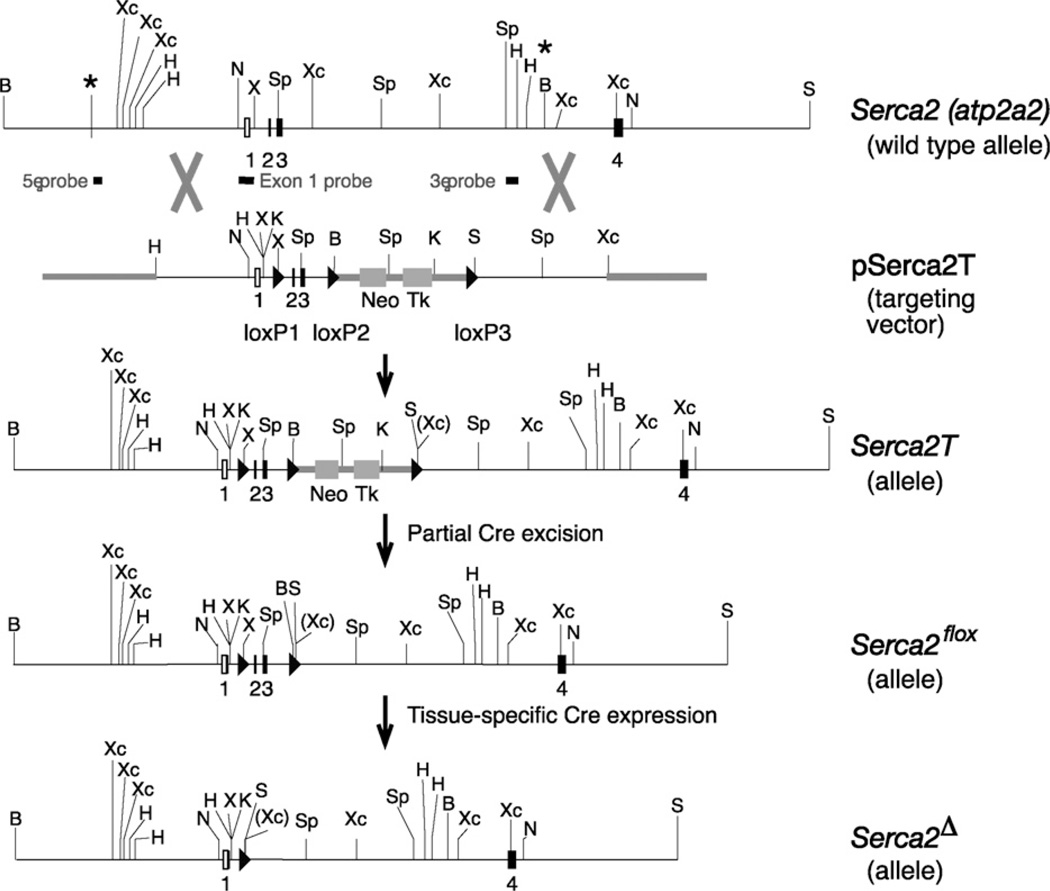
Cloning of Serca2 from a genomic library, construction of the targeting vector pSerca2T and Serca2 gene targeting in embryonic stem (ES) cells are detailed in Supporting Information Materials and methods. Correct targeting of the Serca2 gene locus (5′ and 3′ end crossover and presence of all loxP sites) was verified by Southern blotting. In the final Serca2flox allele, 172 bp and 101 bp of additional DNA including the loxP sites, remained in introns 1 and 3, respectively. Genomic maps and gene targeting strategies are shown in Fig. 1. Two independent embryonic stem cell clones with the desired floxed Serca2 gene modifications were microinjected into 3.5-day B6/Crl blastocysts (in collaboration with Marcela Pekna, University of Göteborg, Sweden) and implanted into pseudopregnant foster mothers. Chimeric males (as determined by coat color) were bred against B6/Crl (Charles River, Sweden) females. Animals with germline transmission of the Serca2flox allele were identified by PCR.
Fig. 1.
Serca2 gene and flox targeting vector design. Map of the relevant region of the Serca2 (Atp2a2) gene with non-coding exons (open boxes) and coding exons (filled boxes). Exon numbers are indicated. Non-genomic DNA is indicated by thick lines. * indicates the 5′ and 3′ ends of the cloned Serca2 genomic fragment. Restriction sites: B, BamH1; H, HindIII; K, KpnI; N, NotI; X, XhoI; Xc, XcmI; S, SalI; Sp, SpeI. Genomic probes for detection of 5′ end, exon1 and 3′ end recombination events are indicated. Triangles denote loxP recombination sites. Neo and TK, Neo and HSV-TK cassettes; wt, wild-type Serca2 gene; pSerca2T, the targeting vector; Serca2T, the allele generated by homologous recombination between the Atp2a2 gene and pSerca2T targeting vector; Serca2flox, the allele generated from Serca2T by partial Cre excision; Serca2Δ, the null allele generated from Serca2flox in cells in which Cre recombinase is expressed.
2.2. PCR detection of Serca2 and MLC-2v alleles
Different alleles of the Serca2 gene (wt, flox or deletion) and the MLC-2v gene (wt and Cre knock-in) were detected by standard PCR reactions with annealing temperature of 55 °C for 25–30 cycles and Amplitaq Gold on an ABI 9600 thermocycler (Applied Biosystems, Foster City, CA, USA). Primer sequences and PCR amplification product sizes are given in Supporting Information Table 1.
2.3. Animal husbandry
All animal experiments were performed in accordance with the Norwegian National Committee for Animal Welfare Act, which closely confirms to the NIH guidelines (NIH publication No. 85-23, revised 1996). Mice were housed in M2 or M3 cages with Bee Kay bedding (Scanbur BK, Nittedal, Norway) in 55% humidity on a 12 h light/dark cycle. Food pellets (RM1, 801151, Scanbur BK) and water were freely available.
Serca2wt/flox mice and MLC-2vwt/Cre mice [14] were backcrossed onto B6/J (n3 and n1 generations, respectively) before interbreeding to generate cardiac Serca2 heterozygous mice. Serca2wt/flox mice are maintained on the B6/J background (The Jackson Laboratory, Bar Harbor, MA, USA), and are presently at generation n > 20. Serca2flox mice and transgenic αMHC-Cre mice [15] were interbred to generate embryonic cardiac disruption of SERCA2. Genotyping was performed by PCR on 3–5 mm tail biopsies [16]. Seventeen- to 22-week-old animals of both sexes and genotypes (unless otherwise stated) were used for basic characterization. In matings of Serca2flox and Tg(αMHC-Cre) transgenic mice, live animals were genotyped as above.
2.4. Echocardiography and left ventricular pressure hemodynamics
Mice were anaesthetized, intubated and connected to a rodent ventilator using 2% isoflurane. Echocardiography was performed using a i13L 13 MHz linear array transducer (GE Healthcare Technologies, Oslo, Norway) and data were analyzed with EchoPac PC software (GE Healthcare Technologies, Oslo, Norway) as described [17]. Hemodynamic measurements were performed immediately thereafter with a 1.4 F Millar micro-tipped pressure transducer catheter (SPR-671, Millar Instruments, Houston, TX, USA) advanced from the right carotid artery into the left ventricle. Data were collected as described [17].
2.5. Tissue material
Immediately after conclusion of the in vivo measurements, the mice were sacrificed with excision of heart, lung, liver, kidney, spleen, soleus and extensor digitorum longus (EDL) muscles. Hearts and lungs were blotted dry and immediately weighed. All organs were flash frozen in liquid nitrogen. Organs were also harvested from animals not undergoing in vivo measurements.
2.6. Ca2+ ATPase content
Total Ca2+ ATPase content in mouse heart left ventricles was measured by an ATP-binding assay in tissue homogenates as described elsewhere [18,19].
2.7. Northern blotting
Poly A+ RNA was isolated as described [20]. Samples (10 µg mRNA) were size-fractionated on 1% agarose gels, transferred to nylon filters and probed with cDNA fragments for Serca2 (pRH39, EcoRI 2.2 kb insert) [3] and Gapdh (1.3 kb insert) (a kind gift from Hans Prydz, University of Oslo, Oslo, Norway).
2.8. Western blotting
Total homogenates were prepared in ice-cold buffer (210 mM sucrose, 2 mM EGTA, 40 mM NaCl, 30 mM HEPES, 5 mM EDTA, pH 7.4) supplemented with a protease inhibitor cocktail (Complete EDTA-free, Roche Diagnostics, Oslo, Norway) and 1% SDS final concentration as described [21]. Homogenization buffer was supplemented with phosphatase inhibitors (phosphatase inhibitor cocktail 1, Sigma–Aldrich, Oslo, Norway) where needed. Protein content was measured by a Micro BCA assay (Pierce 23235, Pierce Biotechnology, Rockford, IL, USA) using BSA as standard protein. Samples (left ventricular myocardium 15 µg/lane; kidney, liver and lung samples, 40 µg/lane) were electrophoresed in SDS-PAGE gels (10%) and transferred to 0.45 µm PVDF membranes (GE Healthcare Amersham Biosciences) by standard procedures. Membranes were blocked in 5% skimmed milk in TBS-T. Antibodies for detection were mouse monoclonal anti-SERCA2a (MA3-919, Affinity Bioreagents) and rabbit polyclonal anti-SERCA2b antiserum [22] (kind gift from Frank Wuytack, Katholieke Universiteit Leuven, Leuven, Belgium). Blots were incubated with appropriate HRP-conjugated sheep anti-mouse IgG or donkey anti-rabbit IgG secondary antibodies diluted in 5% skimmed milk in TBS-T (GE Healthcare Amersham Biosciences) and developed with ECL or ECLplus reagents (GE Healthcare Amersham Biosciences). Images were acquired in a LAS-1000 CCD detection system (Fuji Photo Film, Tokyo, Japan). All membranes were stained after immunoblotting with Coomassie Brilliant Blue R250 (Sigma–Aldrich) and scanned for verification of equal protein loading and membrane transfer of samples.
2.9. Basic characterization of embryos with early cardiac disruption of Serca2
In timed matings between Serca2flox/flox and Serca2flox/wt Tg(αMHC-Cre) mice, the uterine horns were removed from pregnant females at appropriate time points and blotted dry. The uterine wall was transected and embryos with intact yolk sacs were carefully removed and washed in oxygenated Tyrodes solution with low Ca2+ (136 mM NaCl, 4.5 mM KCl, 0.1 mM CaCl2, 0.33 mM NaH2PO4, 1 mM MgCl2, 10 mM HEPES, 10 mM glucose, pH 7.3). Embryos were transferred to oxygenated Tyrodes solution containing 1.8 mM CaCl2 at 37 °C to observe heart beat and yolk sac circulation. Embryos were imaged in a Leica MZ FLIII stereomicrosope with a DFC300 FX digital camera (Leica Microsystems, Heerbrugg, Switzerland). Genotyping of embryos was performed on embryonic yolk sacs or embryos proper.
2.10. Statistical analysis
The data are expressed as mean values ± SEM unless otherwise noted. Statistical significance was calculated using two-tailed Student’s unpaired t-test (Prism 4.01, Graphpad Software, San Diego, USA). p values <0.05 were considered significant. The statistical significance of genotype distributions in live mice was calculated using the χ2 distribution.
3. Results
3.1. Generation of Serca2flox targeted mice
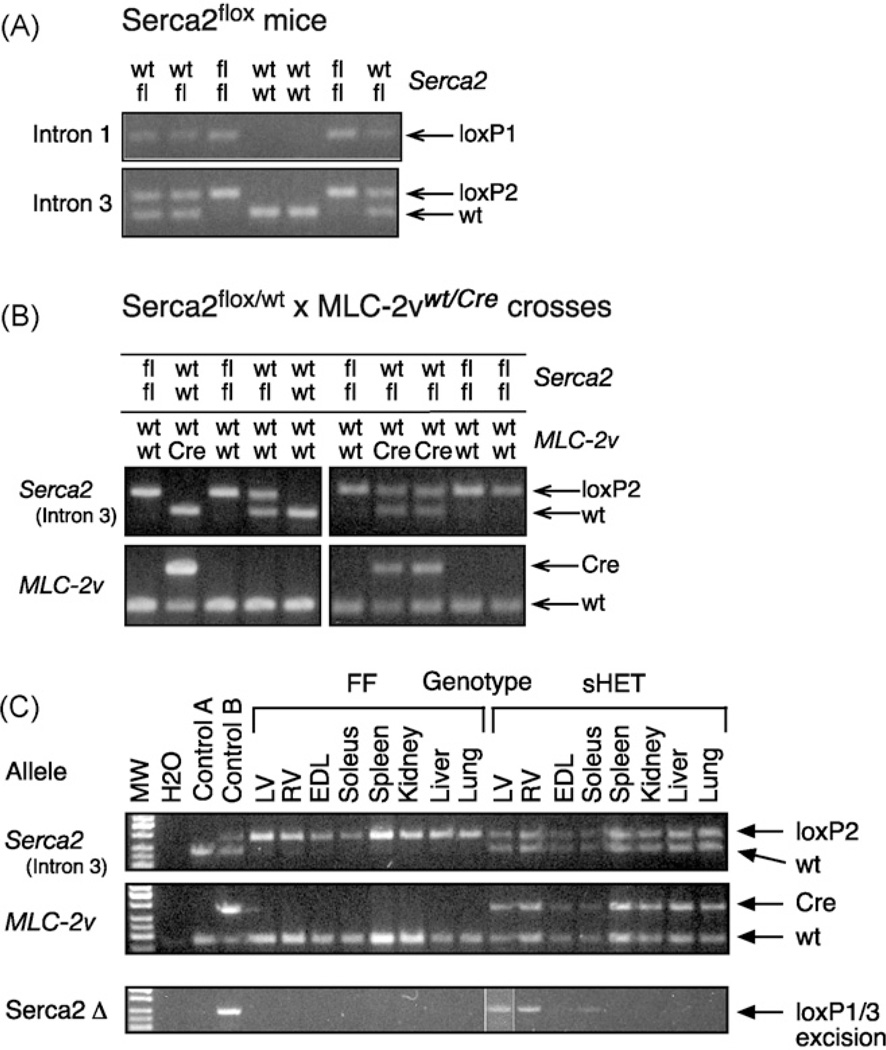
The partial structure of the Serca2 (Atp2a2) gene and gene targeting strategy is outlined in Fig. 1. The wild-type Serca2 allele was converted by a two-step procedure into a Serca2flox allele in embryonic stem cells by homologous recombination (Fig. 1). LoxP sites were placed into introns 1 and 3 of the Serca2 gene, encompassing the translational start site, thus producing a Serca2 null allele upon Cre activity and recombination of the loxP sites. All matings of targeted Serca2flox animals resulted in offspring with expected Mendelian allele frequencies and normal litter sizes. Serca2wt/flox and Serca2flox/flox animals appeared normal and were indistinguishable. Verification of the presence of the loxP sites in introns 1 and 3 in offspring is shown in Fig. 2A.
Fig. 2.
Generation of Sercaflox mice and Serca2 gene disruption in somatic heterozygous SERCA2 mice. Various combinations of Serca2 and MLC-2v alleles in adult mice were detected by PCR. Primer combinations and fragment sizes are given in Supporting Information Table 1. Genotype combinations are shown above the panels. (A) Germline transmission of the Serca2flox allele and generation of Serca2flox/flox mice. PCR products for wild type (wt) and flox (fl) alleles are indicated. The presence of loxP1 and loxP2 PCR products confirmed the presence of the Serca2flox allele. (B) Generation of somatic heterozygous SERCA2 mice. Littermates from Serca2flox/flox × Serca2wt/flox MLC-2vwt/Cre crosses were genotyped for Serca2 (wt and fl) and MLC-2v (wt and Cre) alleles. (C) Specificity of Serca2 gene deletion in somatic heterozygous SERCA2 (sHET) (Serca2wt/flox MLC-2vwt/Cre) and control FF (Serca2flox/flox) mice. Genomic DNA was extracted from the indicated tissues and analyzed by PCR. Control A, wt ES cells (ES14.1a); Control B, upper panel: Serca2wt/flox ES cells, middle panel: left ventricle MLC-2vwt/Cre mice, lower panel: Serca2wt/Δ ES cells; LV, heart left ventricle; RV, heart right ventricle; EDL, extensor digitorum longus muscle; Soleus, soleus muscle; other tissues as indicated. The PCR product for LV was from an independent PCR run.
3.2. Test of functionality in adult animals—tissue-specific heterozygous SERCA2 mice
To validate a functional Serca2flox allele and disruption of the Serca2 gene upon tissue-specific Cre activity, we mated Serca2flox/flox mice with MLC-2vwt/Cre “knock-in” mice [14,23], in which the Cre recombinase is expressed in the heart ventricles and in slow-twitch skeletal muscle with no detectable expression in other tissues. In crosses between Serca2flox/flox and MLC-2vwt/Cre mice, the Serca2 and MLC-2v alleles segregated normall2y (Fig. 2B). However, we failed to generate somatic Serca2 knockout mice (Serca2flox/flox MLC-2vwt/Cre) in crosses between Serca2flox/flox and Serca2wt/flox MLC-2vwt/Cre mice (n > 100 offspring, data not shown). This was due to the close proximity of the Serca2 (Atp2a2) and MLC-2v (Myl2) loci on mouse chromosome 5 (data not shown). For practical purposes, the genetic linkage between Serca2 and MLC-2v is 100%.
In somatic heterozygous SERCA2 mice (Serca2wt/flox MLC-2vwt/Cre), deletion of one copy of the Serca2 gene was detected in the heart (right and left ventricles) and in slow-twitch skeletal muscle (soleus), but not in fast-twitch skeletal muscle (EDL), liver, lung, kidney or spleen, nor in any tissue in floxed Serca2 littermates (FF, Serca2flox/flox MLC-2vwt/wt) (Fig. 2C). This is in accordance with the Cre recombinase being expressed in cardiac and slow-twitch skeletal muscle from within MLC-2v gene locus [14]. All subsequent analyses were performed on somatic heterozygous SERCA2 and control FF mice.
3.3. Serca2 expression in somatic heterozygous SERCA mice
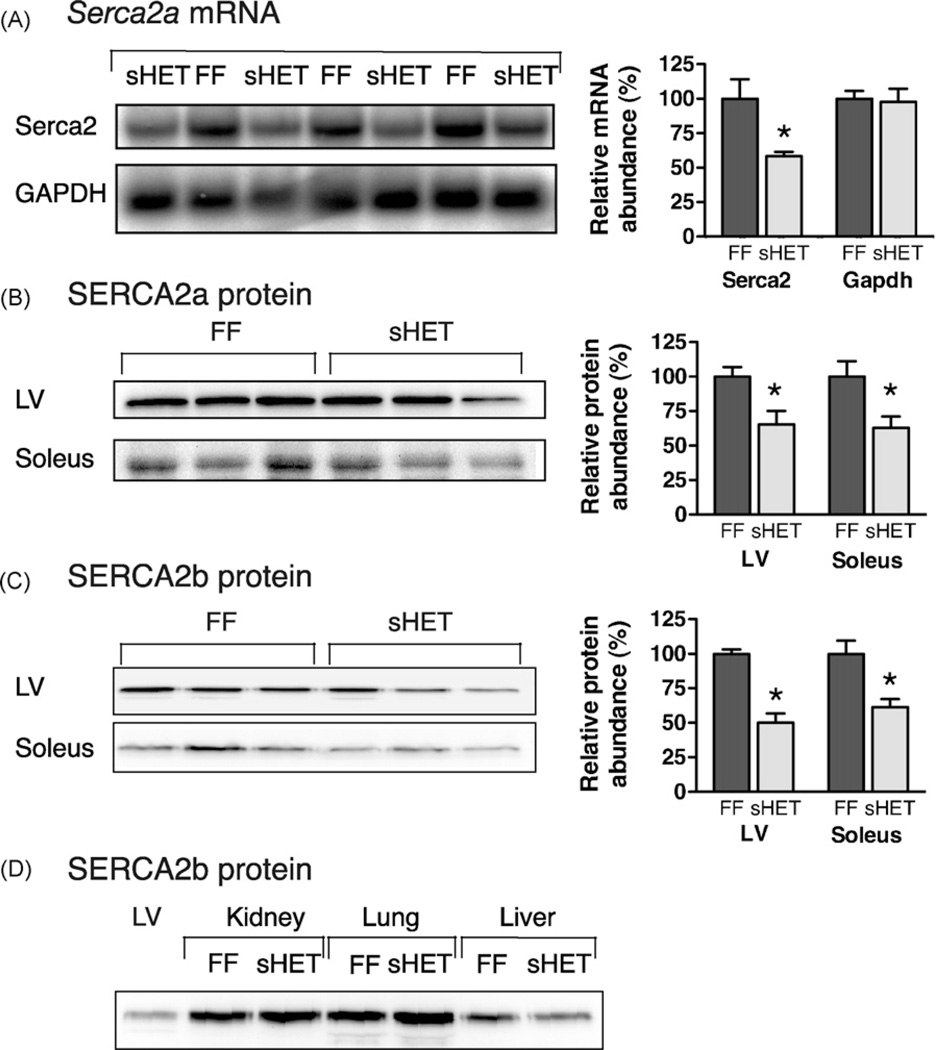
Serca2 mRNA was significantly reduced by 42% in the left ventricles of somatic heterozygous SERCA2 mice compared to control FF mice (Fig. 3A). There was no difference in Serca2 mRNA expression between Serca2wt/flox and Serca2flox/flox mice (data not shown), suggesting that the flox sites inserted into the Serca2 gene did not affect Serca2 mRNA expression.
Fig. 3.
Reduced expression of Serca2 mRNA and SERCA2 proteins in somatic heterozygous SERCA2 mice. (A) Serca2 mRNA in left ventricles of somatic heterozygous SERCA2 (sHET) and control FF mice. Northern blots sequentially probed with [32P]-dCTP-labelled probes for Serca2 and Gapdh. Right: Quantification of mRNA abundance by phosphorimager analysis. (B) Quantification of SERCA2a protein in tissues from somatic heterozygous SERCA2 and FF mice by Western blotting. Heart left ventricle (LV, 20 µg/lane); soleus and other tissues, 40 µg/lane. Blots were probed with SERCA2a-specific antibody. Right: SERCA2a quantification summary. (C) Western blots probed with SERCA2b-specific antiserum. Right: SERCA2b quantification summary. (D) Expression of SERCA2b in kidney, lung and liver. *p < 0.05.
SERCA2a protein was reduced by 35% in the left ventricles and by 37% in the soleus muscles of somatic heterozygous SERCA2 mice compared with FF control littermates (Fig. 3B) (p < 0.05). SERCA2b protein was reduced by 35% in the left ventricle and by 38% in the soleus muscle of somatic heterozygous SERCA2 animals compared to FF control littermates (Fig. 3C) (p < 0.05). Overall, half the gene dosage reduced both the SERCA2a and SERCA2b protein abundance in the heart and soleus muscle of somatic heterozygous SERCA2 mice by 35% and 38% of that in control animals. There was no significant difference in SERCA2b protein expression between the somatic heterozygous SERCA2 and FF mice in lung, spleen, liver or kidney (Fig. 3D). In somatic heterozygous SERCA2 mice, the total Ca2+ ATPase content in the heart was reduced by 28% in 19-week-old mice (23 ±1 nmol Ca2+ ATPase/g protein, n = 5) compared with FF control littermates (32 ± 3 nmol Ca2+ ATPase/g protein, n = 5) (p < 0.05).
3.4. Overall phenotype of somatic heterozygous SERCA2 mice
There was no significant difference between somatic heterozygous SERCA2 and FF control animals of the same sex with regard to size, heart or lung weights as absolute values or normalized to body weight (Supporting Information Table 2). Heart function in vivo was assessed by echocardiography (Supporting Information Table 3). We found no significant difference between somatic heterozygous SERCA2 and control FF mice in overall heart and aorta dimensions, flow rates or cardiac output as determined by echocardiography, nor dilatation of the left atrium, which is an early discriminative parameter of heart failure in post-infarction mice [17]. Moreover, the left ventricular end-diastolic pressure and the maximal positive (LVdP/dtmax) and negative derivatives (LVdP/dtmin) of the left ventricular pressure were not significantly different in somatic heterozygous SERCA2 and control FF mice (Supporting Information Table 3).
3.5. Test of functionality—cardiac embryonic disruption of SERCA2
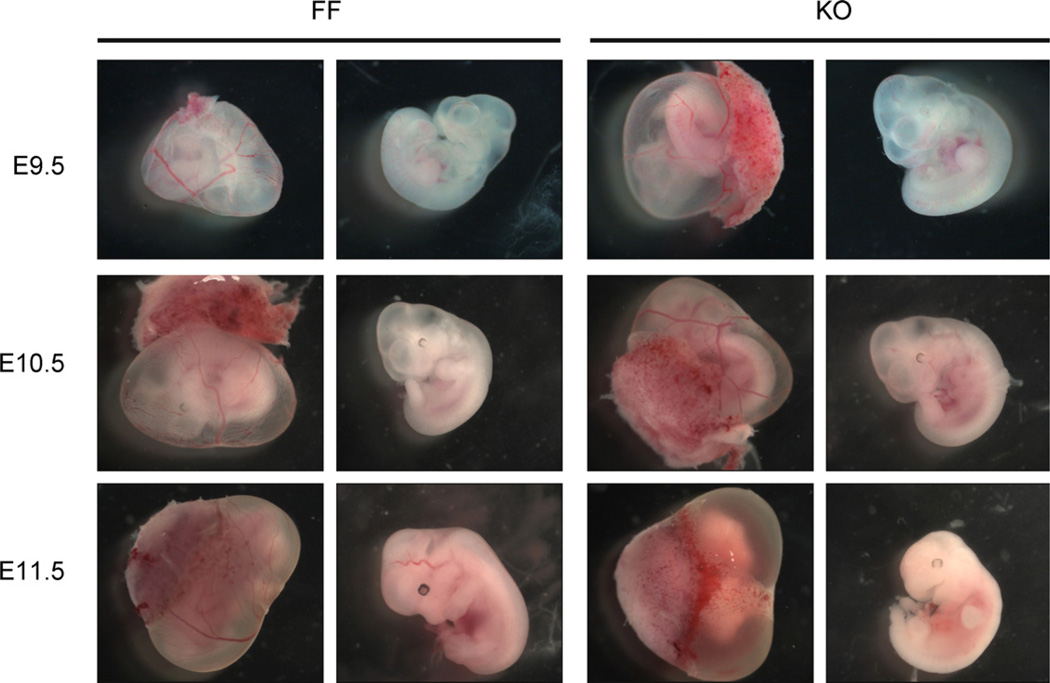
The αMHC-Cre transgene is active in the embryonic heart from day E8 [24]. We therefore mated Serca2flox/flox mice with Serca2flox/wt Tg(αMHC-Cre) carrying the αMHC-Cre transgene to generate mice with disruption of Serca2 in the heart [15]. We could not find live mice with cardiac disruption of the Serca2 gene (Supporting Information Table 4). In contrast, embryos with all four expected genotypes were identified up to E14.5 in timed matings (Supporting Information Table 5). Yolk sac circulation and heartbeat were readily observed in homozygous cardiac Serca2-disrupted embryos up to E10.5 (Fig. 4 and Supporting Information Table 5). There was no indication of pericardial enlargement as a sign of cardiac dysfunction at these stages. However, embryos at E11.5 were dead with no visible yolk sac circulation (Fig. 4), and embryos at E14.5 were heavily decomposed. Thus, cardiac Serca2-disrupted embryos may be studied in developmental window up to embryonic stage E10.5.
Fig. 4.
Heart development in embryos with homozygous cardiac disruption of the Serca2 gene. Examples of embryos of FF (Serca2flox/flox) and KO (Serca2flox/flox Tg(αMHC-Cre)) genotypes at the Theiler stages as indicated. Yolk sac (left) and released embryo (right) within each genotype, respectively.
4. Discussion
We report here the first conditional gene inactivation mouse model for the SERCA family of Ca2+ ATPases. Mice carrying the new Serca2flox allele allow disruption of the Serca2 gene in a tissue-specific and/or inducible fashion by mating with appropriate mice expressing Cre recombinase. Efficient tissue-specific disruption of the Serca2flox allele was demonstrated in adult mice by mating Serca2flox/flox mice with MLC-2vwt/Cre mice [14]. Somatic heterozygous SERCA2 mice (Serca2flox/flox MLC-2vwt/Cre) were phenotypically normal, but with reduced SERCA2 content in the heart and slow-twitch skeletal muscle. The magnitude of decrease in Serca2 mRNA, protein and Ca2+ ATPase activity in the hearts of somatic heterozygous SERCA2 mice was similar to that found in systemic heterozygous Serca2wt/null mice [13,25]. Reduction of SERCA2 expression in the heart may be compensated at several levels. One possibility is to increase the production of the higher Ca2+ affinity SERCA2b protein isoform, as seen in a Serca2 splicing site mutant mouse (Serca2b/b, Serca2anull) [26]. However, both SERCA2a and SERCA2b proteins were reduced to similar extents in the heart and in soleus muscle of somatic heterozygous SERCA2 mice.
It has been suggested that decreased SERCA2 expression or function in the heart is an important contributor to the development of heart failure [10]. Yet none of the three mouse models (Serca2wt/null, Serca2b/b i.e. Serca2anull and tissue-specific Serca2wt/null) developed severe heart failure under basal conditions. However, systemic Serca2wt/null mice responded with accelerated heart failure development when challenged with pressure overload [27] and were more sensitive to ischemia/reperfusion injury [28]. Serca2b/b splicing site mutant mice developed mild cardiac hypertrophy [26]. We did not find indications of depressed cardiac function in somatic heterozygous SERCA2 mice. Darier’s disease patients have mutations in the Serca2 gene and are functional Serca2 heterozygotes. Cardiac dysfunction was not found at rest or during exercise in these patients [29]. A possible explanation for the lack of overt heart failure in these animal models and in Darier’s patients is that the remaining SERCA2 function may be sufficient for maintaining Ca2+ transport during the cardiac contractile cycle.
We have recently shown that in an another application of the Serca2flox allele, in which tamoxifen induces Cre-dependent cardiomyocyte-specific homozygous disruption of the Serca2flox allele results in cardiac dysfunction and death, but also an unexpected time window of several weeks where cardiac function was maintained with a small reduction in cardiac output despite more than 95% loss of SERCA2 protein [30]. Furthermore, the reduction of SERCA2 was compensated by multiple mechanisms, among them augmented Ca2+ cycling over the cardiomyocyte membrane, increased adrenergic drive and enhanced myofilament responsiveness.
In embryos with homozygous disruption of Serca2 during early cardiac development (Serca2flox/flox Tg(αMHC-Cre), we found that heart development appeared normal and embryo-derived yolk sac circulation was supported up to embryonic age 10.5. Disruption of Serca2 would occur at E7.5–E8, when the αMHC promoter becomes transcriptionally active, and SERCA2 protein should rapidly decrease. Thus, it should be possible to examine cardiac function in a time window with a reduction in SERCA2 in mouse embryos.
The functional role of Serca2 in the endoplasmic reticulum (ER) of cells in other organs than the heart is poorly understood, and Serca2flox mice may be useful for such purposes. It has been proposed that SERCA proteins play a diverse role in multiple processes such as ER luminal protein processing, intracellular Ca2+ signaling and cell death, in addition to a more general ER housekeeping function [31,32]. Darier’s disease is a heritable disorder of epithelial keratinization. It was therefore surprising that mutations in these patients were mapped to the Serca2 gene [33]. It has been suggested that the reduced SERCA2 function in Darier’s keratinocytes interferes with ER protein synthesis and trafficking [34]. SERCA2 clearly serves an important function in Ca2+ homeostasis in keratinized cells, even though the exact mechanism is currently not understood. Aged systemic heterozygous Serca2wt/null mice have an unexpected high frequency of squamous cell tumors in keratinized epithelial cells in the skin, forestomach, oral cavity and esophagus [35]. In human oral squamous cell carcinomas, SERCA2 protein expression was found to be reduced [36]. These findings suggest that SERCA2 may also play a role in cancer development and our model may provide new information in this context.
The diverse role of SERCA2 in Ca2+ handling and physiological adaptability is also illustrated in the pancreatic acinar cells of heterozygous systemic heterozygous Serca2wt/null mice. Exocytosis was normal in Serca2wt/null pancreas, even though the Ca2+ oscillations were reduced by 50% [37]. Moreover, calcium homeostasis is important for cell survival in the neuronal cells [38]. In the brain, Serca2 mRNA exists as several splice variants with regional transcript expression patterns [39]. A recent report has shown that plasma membrane components such as NCX1 and sodium pumps may form microdomains with SERCA2 and other ER proteins in astrocytes [40]. However, specialized regional SERCA2 function(s) in the CNS have not yet been demonstrated in vivo.
The new Serca2flox mouse allows tissue and/or time-specific inactivation of the Serca2 gene in embryos or in adult mice, as specified by the control of Cre recombinase expression. The high efficiency of Serca2 disruption, short Serca2 mRNA and protein half-live as demonstrated in the heart [30], provides a time window to study loss or reduction of Serca2 function and biological responses and compensations which is not possible to study using acute application of pharmacological inhibitors. Cells from Serca2flox/flox mice in combination with transfection techniques may also be used to generate Serca2-deficient primary cell cultures. Serca2flox mice should be useful in unraveling multiple aspects of SERCA2 function in multiple physiological settings in vivo.
Supplementary Material
Acknowledgements
We gratefully acknowledge Johannes Wilbertz and the KTCC (Karolinska Institute, Stockholm, Sweden) for help with ES cell lines, Marcela Pekna, University of Göteborg for blastocyst injections and excellent animal husbandry, Dag Markus Eide and Kari G. Løken at the National Institute of Public Health, Oslo for excellent animal husbandry and breeding, Line Solberg and Morten Eriksen for animal care, and Tævje Strømme for technical assistance with the hemodynamic measurements. We would also like to thank Frank Wuytak (KU Leuven) for the gift of polyclonal SERCA2b-specific antibody and Franziska Wiebel for the pIC-Cre-Pac plasmid. We also gratefully acknowledge Nils Göran Larsson and Anna Wredenberg (Karolinska Institute) for the transgenic αMHC-Cre mice, Nigel Brown (University of London) for invaluable advice on mouse embryos, and Sigurd From (University of Oslo) for use of the microscope.
This work has been supported by the Norwegian Research Council, Anders Jahre’s Fund for the Promotion of Science, the Ullevål University Hospital Fund (K.B.A.), a University of Oslo EMBIO senior research fellowship (K.B.A.), an Eastern Norway Regional Health Authority senior fellow grant (K.B.A.) and a Norwegian Research Council Research fellowship (A.V.F).
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.ceca.2009.07.004.
Footnotes
Conflict of interest
Patent EP1699289 (WO2005063007) (K.B.A., G.C.).
References
- 1.Loukianov E, Ji Y, Grupp IL, et al. Enhanced myocardial contractility and increased Ca2+ transport function in transgenic hearts expressing the fast-twitch skeletal muscle sarcoplasmic reticulum Ca2+-ATPase. Circ. Res. 1998;83:889–897. doi: 10.1161/01.res.83.9.889. [DOI] [PubMed] [Google Scholar]
- 2.Wuytack F, Raeymaekers L, Missiaen L. Molecular physiology of the SERCA and SPCA pumps. Cell Calcium. 2002;32:279–305. doi: 10.1016/s0143416002001847. [DOI] [PubMed] [Google Scholar]
- 3.Lompre AM, de la Bastie D, Boheler KR, et al. Characterization and expression of the rat heart sarcoplasmic reticulum Ca2+-ATPase mRNA. FEBS Lett. 1989;249:35–41. doi: 10.1016/0014-5793(89)80010-9. [DOI] [PubMed] [Google Scholar]
- 4.Sakuntabhai A, Ruiz-Perez V, Carter S, et al. Mutations in ATP2A2, encoding a Ca2+ pump, cause Darier disease. Nat. Genet. 1999;21:271–277. doi: 10.1038/6784. [DOI] [PubMed] [Google Scholar]
- 5.Lee MG, Xu X, Zeng W, et al. Polarized expression of Ca2+ pumps in pancreatic and salivary gland cells. Role in initiation and propagation of [Ca2+]i waves. J. Biol. Chem. 1997;272:15771–15776. doi: 10.1074/jbc.272.25.15771. [DOI] [PubMed] [Google Scholar]
- 6.Baba-Aissa F, Raeymaekers L, Wuytack F, et al. Distribution of the organellar Ca2+ transport ATPase SERCA2 isoforms in the cat brain. Brain Res. 1996;743:141–153. doi: 10.1016/s0006-8993(96)01037-2. [DOI] [PubMed] [Google Scholar]
- 7.Gelebart P, Martin V, Enouf J, et al. Identification of a new SERCA2 splice variant regulated during monocytic differentiation. Biochem. Biophys. Res. Commun. 2003;303:676–684. doi: 10.1016/s0006-291x(03)00405-4. [DOI] [PubMed] [Google Scholar]
- 8.Dally S, Bredoux R, Corvazier E, et al. Ca2+-ATPases in non-failing and failing heart: evidence for a novel cardiac sarco/endoplasmic reticulum Ca2+-ATPase 2 isoform (SERCA2c) Biochem. J. 2006;395:249–258. doi: 10.1042/BJ20051427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colucci WS, Braunwald E. In: Braunwald’s Heart Disease: a Textbook of Cardiovascular Medicine. Zipes DP, Bonow RO, Braunwald E, editors. Saunders: Elsevier; 2005. pp. 519–520. [Google Scholar]
- 10.Hasenfuss G, Pieske B. Calcium cycling in congestive heart failure. J. Mol. Cell Cardiol. 2002;34:951–969. doi: 10.1006/jmcc.2002.2037. [DOI] [PubMed] [Google Scholar]
- 11.Frank KF, Bolck B, Erdmann E, et al. Sarcoplasmic reticulum Ca2+-ATPase modulates cardiac contraction and relaxation. Cardiovasc. Res. 2003;57:20–27. doi: 10.1016/s0008-6363(02)00694-6. [DOI] [PubMed] [Google Scholar]
- 12.Sande JB, Sjaastad I, Hoen IB, et al. Reduced level of serine(16) phosphorylated phospholamban in the failing rat myocardium: a major contributor to reduced SERCA2 activity. Cardiovasc. Res. 2002;53:382–391. doi: 10.1016/s0008-6363(01)00489-8. [DOI] [PubMed] [Google Scholar]
- 13.Periasamy M, Reed TD, Liu LH, et al. Impaired cardiac performance in heterozygous mice with a null mutation in the sarco(endo)plasmic reticulum Ca2+-ATPase isoform 2 (SERCA2) gene. J. Biol. Chem. 1999;274:2556–2562. doi: 10.1074/jbc.274.4.2556. [DOI] [PubMed] [Google Scholar]
- 14.Chen J, Kubalak SW, Minamisawa S, et al. Selective requirement of myosin light chain 2v in embryonic heart function. J. Biol. Chem. 1998;273:1252–1256. doi: 10.1074/jbc.273.2.1252. [DOI] [PubMed] [Google Scholar]
- 15.Wredenberg A, Wibom R, Wilhelmsson H, et al. Increased mitochondrial mass in mitochondrial myopathy mice. Proc. Natl. Acad. Sci. U.S.A. 2002;99:15066–15071. doi: 10.1073/pnas.232591499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Torres RM, Kühn R. Laboratory Protocols for Conditional Gene Targeting. Oxford, NY: Oxford University Press; 1997. [Google Scholar]
- 17.Finsen AV, Christensen G, Sjaastad I. Echocardiographic parameters discriminating myocardial infarction with pulmonary congestion from myocardial infarction without congestion in the mouse. J. Appl. Physiol. 2005;98:680–689. doi: 10.1152/japplphysiol.00924.2004. [DOI] [PubMed] [Google Scholar]
- 18.Everts ME, Andersen JP, Clausen T, et al. Quantitative determination of Ca2+-dependent Mg2+-ATPase from sarcoplasmic reticulum in muscle biopsies. Biochem. J. 1989;260:443–448. doi: 10.1042/bj2600443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holt E, Lunde PK, Sejersted OM, et al. Electrical stimulation of adult rat cardiomyocytes in culture improves contractile properties and is associated with altered calcium handling. Basic Res. Cardiol. 1997;92:289–298. doi: 10.1007/BF00788941. [DOI] [PubMed] [Google Scholar]
- 20.Andersson KB, Florholmen G, Winer LH, et al. Regulation of neuronal type genes in congestive heart failure rats. Acta Physiol. (Oxf.) 2006;186:17–27. doi: 10.1111/j.1748-1716.2005.01503.x. [DOI] [PubMed] [Google Scholar]
- 21.Ploug T, Wojtaszewski J, Kristiansen S, et al. Glucose transport and transporters in muscle giant vesicles: differential effects of insulin and contractions. Am. J. Physiol. 1993;264:E270–E278. doi: 10.1152/ajpendo.1993.264.2.E270. [DOI] [PubMed] [Google Scholar]
- 22.Campbell AM, Wuytack F, Fambrough DM. Differential distribution of the alternative forms of the sarcoplasmic/endoplasmic reticulum Ca(2+)-ATPase, SERCA2b and SERCA2a, in the avian brain. Brain Res. 1993;605:67–76. doi: 10.1016/0006-8993(93)91357-x. [DOI] [PubMed] [Google Scholar]
- 23.Minamisawa S, Gu Y, Ross J, Jr, et al. A post-transcriptional compensatory pathway in heterozygous ventricular myosin light chain 2-deficient mice results in lack of gene dosage effect during normal cardiac growth or hypertrophy. J. Biol. Chem. 1999;274:10066–10070. doi: 10.1074/jbc.274.15.10066. [DOI] [PubMed] [Google Scholar]
- 24.Lyons GE, Schiaffino S, Sassoon D, et al. Developmental regulation of myosin gene expression in mouse cardiac muscle. J. Cell Biol. 1990;111:2427–2436. doi: 10.1083/jcb.111.6.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ji Y, Loukianov E, Periasamy M. Analysis of sarcoplasmic reticulum Ca2+ transport and Ca2+ ATPase enzymatic properties using mouse cardiac tissue homogenates. Anal. Biochem. 1999;269:236–244. doi: 10.1006/abio.1999.4059. [DOI] [PubMed] [Google Scholar]
- 26.VerHeyen M, Heymans S, Antoons G, et al. Replacement of the muscle-specific sarcoplasmic reticulum Ca(2+)-ATPase isoform SERCA2a by the nonmuscle SERCA2b homologue causes mild concentric hypertrophy and impairs contraction–relaxation of the heart. Circ. Res. 2001;89:838–846. doi: 10.1161/hh2101.098466. [DOI] [PubMed] [Google Scholar]
- 27.Schultz Jel J, Glascock BJ, Witt SA, et al. Accelerated onset of heart failure in mice during pressure overload with chronically decreased SERCA2 calcium pump activity. Am. J. Physiol. Heart Circ. Physiol. 2004;286:H1146–H1153. doi: 10.1152/ajpheart.00720.2003. [DOI] [PubMed] [Google Scholar]
- 28.Talukder MA, Kalyanasundaram A, Zuo L, et al. Is reduced SERCA2a expression detrimental or beneficial to postischemic cardiac function and injury? Evidence from heterozygous SERCA2a knockout mice. Am. J. Physiol. Heart Circ. Physiol. 2008;294:H1426–H1434. doi: 10.1152/ajpheart.01016.2007. [DOI] [PubMed] [Google Scholar]
- 29.Mayosi BM, Kardos A, Davies CH, et al. Heterozygous disruption of SERCA2a is not associated with impairment of cardiac performance in humans: implications for SERCA2a as a therapeutic target in heart failure. Heart. 2006;92:105–109. doi: 10.1136/hrt.2004.051037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andersson KB, Birkeland JA, Finsen AV, et al. Moderate heart dysfunction in mice with inducible cardiomyocyte-specific excision of the Serca2 gene. J. Mol. Cell Cardiol. 2009;47:180–187. doi: 10.1016/j.yjmcc.2009.03.013. [DOI] [PubMed] [Google Scholar]
- 31.Burdakov D, Petersen OH, Verkhratsky A. Intraluminal calcium as a primary regulator of endoplasmic reticulum function. Cell Calcium. 2005;38:303–310. doi: 10.1016/j.ceca.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 32.Petersen OH, Michalak M, Verkhratsky A. Calcium signalling: past, present and future. Cell Calcium. 2005;38:161–169. doi: 10.1016/j.ceca.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 33.Hovnanian A. Darier’s disease: from dyskeratosis to endoplasmic reticulum calcium ATPase deficiency. Biochem. Biophys. Res. Commun. 2004;322:1237–1244. doi: 10.1016/j.bbrc.2004.08.067. [DOI] [PubMed] [Google Scholar]
- 34.Dhitavat J, Cobbold C, Leslie N, et al. Impaired trafficking of the desmoplakins in cultured Darier’s disease keratinocytes. J. Invest. Dermatol. 2003;121:1349–1355. doi: 10.1046/j.1523-1747.2003.12557.x. [DOI] [PubMed] [Google Scholar]
- 35.Prasad V, Boivin GP, Miller ML, et al. Haploinsufficiency of Atp2a2, encoding the sarco(endo)plasmic reticulum Ca2+-ATPase isoform 2 Ca2+ pump, predisposes mice to squamous cell tumors via a novel mode of cancer susceptibility. Cancer Res. 2005;65:8655–8661. doi: 10.1158/0008-5472.CAN-05-0026. [DOI] [PubMed] [Google Scholar]
- 36.Endo Y, Uzawa K, Mochida Y, et al. Sarcoendoplasmic reticulum Ca(2+) ATPase type 2 downregulated in human oral squamous cell carcinoma. Int. J. Cancer. 2004;110:225–231. doi: 10.1002/ijc.20118. [DOI] [PubMed] [Google Scholar]
- 37.Zhao XS, Shin DM, Liu LH, et al. Plasticity and adaptation of Ca2+ signaling and Ca2+-dependent exocytosis in SERCA2(±) mice. EMBO J. 2001;20:2680–2689. doi: 10.1093/emboj/20.11.2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verkhratsky A, Toescu EC. Endoplasmic reticulum Ca(2+) homeostasis and neuronal death. J. Cell. Mol. Med. 2003;7:351–361. doi: 10.1111/j.1582-4934.2003.tb00238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baba-Aissa F, Raeymaekers L, Wuytack F, et al. Distribution and isoform diversity of the organellar Ca2+ pumps in the brain. Mol. Chem. Neuropathol. 1998;33:199–208. doi: 10.1007/BF02815182. [DOI] [PubMed] [Google Scholar]
- 40.Lencesova L, O’Neill A, Resneck WG, et al. Plasma membrane–cytoskeleton–endoplasmic reticulum complexes in neurons and astrocytes. J. Biol. Chem. 2004;279:2885–2893. doi: 10.1074/jbc.M310365200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.