Abstract
A selected group of oral bacteria commonly associated with dental health is capable of producing alkali via the arginine deiminase system (ADS), which has a profound impact on the pH of human oral biofilms. An increased risk for dental caries has been associated with reduced ADS activity of the bacteria in oral biofilms. Arginolytic bacterial strains from dental plaque samples of caries-free (CF) and caries-active (CA) adults were isolated and characterized to investigate the basis for differences in plaque ADS activity between individuals. Fifty-six ADS-positive bacterial strains were identified by 16S rRNA gene sequencing and their ADS activity levels were compared under standard growth conditions. The spectrum of bacterial ADS activity ranged from 45.2 to 688.0 units (mg protein)−1. Although Streptococcus sanguinis was the most prevalent species, other Streptococcus were also represented. Biochemical assays carried out using twenty-seven ADS-positive strains under conditions known to induce or repress ADS gene expression, showed substantial variation in arginolytic activity in response to pH, oxygen, and the availability of carbohydrate or arginine. This study reveals that the basis for the wide spectrum of arginolytic expression observed among clinical strains is, at least in part, attributable to differences in the regulation of the ADS within and between species. The results provide insights into the microbiological basis for inter-subject differences in ADS activity in oral biofilms and enhance our understanding of dental caries as an ecologically-driven disease in which arginine metabolism moderates plaque pH and promotes dental health.
Keywords: Arginine deiminase, biofilm, plaque, microflora, caries, pH
INTRODUCTION
Oral biofilms, often called dental plaque, constantly form and grow on all tooth surfaces. When conditions are favorable, dental biofilms is able to produce sufficient acids from carbohydrate fermentation to demineralize the tooth and leading to the formation of dental caries, the most prevalent infectious disease affecting humans. The transition from dental health to dental caries is characterized by compositional and metabolic changes in the complex microbial communities of oral biofilms. In particular, it is recognized that increases in the proportions of aciduric organisms appear to occur at the expense of species that are less acid tolerant [Aas et al., 2008; Becker et al., 2002; Corby et al., 2005]. Importantly, a subset of abundant bacteria in dental biofilms derives protection from acidic conditions from the metabolism of substrates that yield alkaline products; and these species often shows a strong positive correlation with dental health [Burne and Marquis, 2000b].
The hydrolysis of urea and the catabolism of arginine are the primary sources of bacterially-generated alkali in dental biofilms. Urea is present in relatively high concentrations (3–10 mM) in human saliva and in gingival crevicular fluid [Golub et al., 1971; Kopstein and Wrong, 1977], and is rapidly hydrolyzed to two molecules of ammonia and one of CO2 by bacterial ureases. Although the anti-caries effect of urea have been demonstrated in a rat model [Clancy et al., 2000], a variety of problems associated with urea have limited its utility as an anti-caries agent in humans. Arginine is found free in saliva in micromolar concentrations [Van Wuyckhuyse et al., 1995], but is abundant in salivary peptides and proteins. Arginine is primarily catabolized by the arginine deiminase system (ADS) of oral bacteria into ornithine, ammonia and CO2, with the concomitant generation of ATP [Burne and Marquis, 2000b; Clancy et al., 2000; Margolis et al., 1988b; Van Wuyckhuyse et al., 1995]. Hence, the ADS serves key physiological functions in bacteria, providing protection from the deleterious effects of low pH while generating ATP for growth and maintenance [Burne, 2010; Casiano-Colon and Marquis, 1988; Liu and Burne, 2009; Liu et al., 2008; Marquis et al., 1987b; Vander Wauven et al., 1984]. Importantly, the ADS activity in oral biofilms can have major impacts on the ecology of oral microbial communities by moderating the pH through ammonia production [Casiano-Colon and Marquis, 1988; Liu and Burne, 2009; Liu et al., 2008; Huang et al., 2012; Marquis et al., 1987b; Vander Wauven et al., 1984]. Inclusion of arginine in dentifrices or confections [Acevedo et al., 2005; Acevedo et al., 2008] was demonstrated to have significant effects in inhibiting caries, and the potential for the cost-effective use of this technology to reduce caries incidence and severity appears high. More recently, a number of studies, including two-year randomized clinical trials, have shown that a dentifrice with arginine and fluoride was substantially better at preventing the initiation and progression of caries than fluoride alone [Kraivaphan et al., 2013; Srisilapanan et al., 2013; Yin et al., 2013a; Yin et al., 2013b].
A variety of bacteria that are present in significant proportions in biofilms on the teeth and soft tissues or the oral cavity can express the ADS, including Streptococcus sanguinis, Streptococcus gordonii, Streptococcus parasanguis, Streptococcus mitis, Streptococcus oralis, Streptococcus rattus, certain Lactobacillus species and a few spirochetes [Burne and Marquis, 2000a; Marquis et al., 1987a; Rogers, 1990]. Knowledge derived from previous studies using laboratory strains of oral streptococci indicate that the expression of ADS genes is substrate inducible, sensitive to carbon catabolite repression (CCR), and requires low pH and anaerobic conditions for optimal expression [Dong et al., 2004; Liu and Burne, 2009; Liu et al., 2008]]. Specific and global transcriptional regulators, multiple two-component systems (TCS) and other factors have been shown to regulate ADS activity through transcriptional and post-transcriptional mechanisms [Burne, 1991; Dong et al., 2004; Liu and Burne, 2009; Liu et al., 2008]].
Evidence continues to accumulate from in vitro and clinical observations that supports a significant influence of alkali generation on oral ecology and the inhibition of dental caries [Dawes and Dibdin, 2001; Margolis et al., 1988a; Nascimento et al., 2009b; Peterson et al., 1985; Shu et al., 2007b; Wijeyeweera and Kleinberg, 1989b]. A positive correlation between oral arginine metabolism and absence of caries activity has been demonstrated in adults [Nascimento et al., 2009b], and more recently in children [Nascimento et al., 2012]. Specifically, oral bacteria from dental plaque of caries-free subjects present higher ADS activity compared with those from caries-active subjects. From these studies, it was also evident that there is an exceptionally high degree of variability in the rate of ammonia production from arginine among individuals, in some cases greater than 1000-fold. Importantly, an in vitro study showed that as little as a five-fold decrease in the ammonia-generating capacity of a genetically-modified strain of the caries pathogen Streptococcus mutans resulted in the loss of ability to offset environmental acidification by glycolysis [Clancy et al., 2000]. Therefore, many individuals may lack sufficient ADS activity to counteract dental plaque acidification. It is clear, then, that the ADS activity of plaque bacteria may greatly impact the pH profiles of resting and carbohydrate-challenged plaque, and therefore, the risk for caries development.
Differences in t he microbial composition of oral biofilms and differential expression of the ADS are the most likely factors that affect the capacity of oral samples from different individuals to metabolize arginine. The use of qPCR in a previous clinical study [Nascimento et al., 2009b] did not reveal a statistically significant association between the proportions of two recognized arginolytic species, S. sanguinis and S. gordonii, and the caries-status of adults. These results suggested that the diminution in ADS activity associated with caries experience may not be due simply to lower proportions of known ADS-positive bacteria, and also raised the possibility that species other than those examined may be contribute to overall oral ADS activity. It is also possible that environmental conditions and host factors encourage differential expression of the ADS in caries-active versus caries-free subjects. Thus, there is a critical need to characterize more thoroughly the organisms that contribute to arginolysis in the oral cavity. To begin to understand the fundamental microbiology and ecology of oral arginolytic bacterial communities and their relationship to dental health and dental caries, the goals of the present study were to isolate and characterize arginolytic bacterial strains from supragingival dental plaque of caries-free and caries-active adult subjects, and to explore the responses of these strains to environmental stimuli.
MATERIALS AND METHODS
Isolation of bacterial strains
Supragingival dental plaque was collected from 11 caries-free (CF) subjects with no clinical or reported evidence of present or past caries experience [decayed, missing and filled teeth (DMFT) = 0] and 3 caries-active (CA) subjects with at least four active, cavitated (dentin level) and unrestored caries lesions (DT ≥4, MFT ≥ 0), as described elsewhere [Nascimento et al., 2009a; Schulte et al., 2009]. The activity of caries lesions was determined by clinical appearance, plaque stagnation, and tactile sensation. Informed consent was obtained from all participating subjects under a protocol reviewed and approved by the Institutional Review Board of the University of Florida Health Science Center. The criterion for collecting plaque samples was the same as described elsewhere [Nascimento et al., 2009a]. To acquire a variety of cultivable microflora, plaque samples were dispersed by external sonication (W375, Sonicator Heat Systems-Ultrasonics Inc, Farmingdale, NY, USA) for 2 cycles of 15 seconds, with cooling on ice during the interval. Samples were then serially diluted in 10 mM sodium phosphate buffer (pH 7.0) and 100 µl of the 10−4 to 10−7 diluted samples were cultured on Sheep Blood Agar plates (Columbia Agar containing 5% V/V of anti-coagulated sheep blood, Difco Laboratories, Detroit, MI, USA) and on Brain-Heart Infusion (BHI) Agar plates (Difco Laboratories, Detroit, MI, USA). Plates were incubated at 37°C in anaerobic jars (BBL GasPak™ Systems, BD, Sparks, MD, USA) for 3 days with subsequent aerobic incubation at 37°C in 5% CO2 for 2 days. After the incubation period, colonies of clinical strains representing all morphological types were subcultured on the same media until pure colonies were obtained.
Screening of ADS-positive strains
Bacterial strains were screened for the potential to liberate ammonia from arginine in a microtiter-based assay [Schulte et al., 2009]. Briefly, strains were grown in clear polystyrene microtiter-plates (Fisher Scientific Inc., USA) containing tryptone-vitamin (TV)-based broth [Burne et al., 1999] with 0.2% galactose and 10 mM arginine. The plates were incubated under anaerobic conditions (85% N2, 5% CO2, 10% H2, 80% relative humidity) at 37°C for 48 hours. Bacterial cells were collected by centrifuging the plates for 3 min at 10,000× g in a refrigerated microcentrifuge, washed once with 10 mM Tris-maleate (pH 7.0) and resuspended in 100 µl of 50 mM Tris-maleate buffer (pH 6.0). The ADS-positive phenotype was identified by detecting the ammonia generated from the incubation of bacteria in the presence of 50 mM arginine-HCl for 2 hours at 37°C using the Nessler´s reagent (Sigma-Aldrich Inc., USA). Controls for background and interference were routinely included in each reaction. The library of the ADS-positive strains was stored at −80°C for further analysis. From this library, fifty-six ADS-positive strains were randomly selected from the plaque of the various CF and CA subjects to be identified by 16S rRNA gene sequencing and characterized in this study.
Amplification and sequencing of 16S rRNA genes by PCR
An optimized colony PCR reaction was used to amplify 16S rRNA genes. Colonies were picked up with a sterilized pipette tip and directly transferred to the tubes for the PCR reaction. Sequences of the 16S rRNA genes were amplified under standardized conditions using a universal primer set (Forward: 5’-AGA GTT TGA TCC TGG CTC AG-3’, Reverse: 5’-TAC GGG TAC CTT GTT ACG ACT-3) [Aas et al., 2005; Paster et al., 2001]. DNA purification was carried out using the QIAprep spin miniprep kit purchased from Qiagen, Inc. (Valencia, CA). Purified PCR-products of 16S rRNA were sequenced using an ABI Prism cycle sequencing kit and the primers and conditions and the data analyzed as previously described [Aas et al., 2005; Paster et al., 2001]. DNA sequences were compared to the 16S rRNA sequences deposited at the Human Oral Microbiome Database (HOMD), Ribosomal Database Project, and the GenBank database. The complete 16S rRNA gene sequences generated in this study will be available for electronic retrieval from the EMBL, GenBank, and DDBJ nucleotide sequence databases.
ADS activity, growth conditions and reagents
ADS activity of bacterial strains was measured by monitoring citrulline production from arginine using protocols validated by our group [Liu et al., 2008]. Bacterial strains were maintained on fresh BHI agar and inoculated into tryptone-yeast (TY) extract broth containing 25 mM galactose and 10 mM arginine for overnight growth in a 5% CO2 aerobic atmosphere at 37°C. ADS activity of the 56 selected ADS-positive strains was determined under the following standard growth conditions: overnight cultures of each strain were diluted (1:20 dilution) into fresh TY medium containing 25 mM galactose and 10 mM arginine and incubated as above until the OD600 reached 0.5–0.6 [Liu et al., 2008]. The cells were then permeabilized in toluene-acetone (1:9) prior to determination of their ADS activity. The concentration of protein in the permeabilized cell preparations was determined using a Pierce BCA protein assay kit (Waltham, MA, USA) with bovine serum albumin as the standard. ADS activity levels of bacterial strains were normalized to protein content and defined as nmol of citrulline generated [minute × (mg protein)]−1.
To monitor ADS expression as a function of environmental conditions known to induce or repress the ADS, 27 representatives of different bacterial species were grown in TY base medium containing: 25 mM galactose with or without 10 mM arginine; 25 mM galactose and 10 mM arginine that had been acidified to pH 5.7 with HCl or buffered at pH 7.0 with 50 mM potassium phosphate buffer (TY/KPB); 10 mM arginine and 25 mM galactose or 25 mM glucose; or 25 mM galactose and 10 mM arginine with the cultures incubated under aerobic or anaerobic conditions. For aerobic growth, the cells were inoculated into a 250 ml conical flask containing 40 ml of TY medium supplemented with galactose and arginine and grown on a rotary shaker (50 rpm) at 37°C. For anaerobic growth, cultures were similarly diluted and incubated in an anaerobic chamber at 5%CO2, 10% H2, 85% N2 and 37°C. All the cells were collected at OD600 = 0.5–0.6 for the detection of ADS activity.
Statistical analysis
For descriptive analysis, distribution of percentages and means were calculated when appropriate. Student’s t-test or ANOVA were used to test the differences of continuous variables; and chi-square test was used for categorical variables. The correlation between the proportions of ADS-positive bacterial st rains and total cultivable organisms with the subjects’ caries status was analyzed using the Two Proportions Z-test. The level of significance was determined at p < 0.05.
RESULTS
Arginolytic Bacterial Strains of Oral Biofilms
A total of 2328 bacterial strains were isolated from plaque samples of the 14 participating subjects (11 CF and 3 CA; ratio of 166.3 strains isolated per subject), and screened for arginolytic capacity by detection of ADS activity. Of these 2328 strains, 288 were ADS-positive, which represents a ratio of 20.5 strains per subject, or 15.8 strains per CF subject (minimum of 5 ADS+ and maximum of 51 ADS+ strains within this caries group) and 38 strains per CA subject (minimum of 6 ADS+ and maximum of 84 ADS+ strains within this caries group). Despite considerable variation among the number of ADS-positive strains identified within subjects and within the caries groups, there was a fair or unbiased distribution of the strains tested across subjects. There was no significant correlation between the proportions of ADS-positive strains in the total cultivable flora with the caries status of the subjects.
Table 1 shows the diversity of arginolytic species isolated from supragingival dental plaque and identified by 16S rRNA gene sequencing. All 56 ADS-positive strains identified had greater than 99% sequence similarity with their assigned bacterial taxa. A total of 11 different bacterial taxa of Streptococcus representing the bacterial phyla Firmicutes were detected: S. sanguinis (67.9%), S. gordonii (8.9%), Streptococcus intermedius (8.9%), Streptococcus cristatus (8.9%), Streptococcus australis (3.6%) and S. parasanguinis (1.8%). The spectrum of bacterial ADS activity ranged from 45.2 to 688.0 units (mg protein)−1 when bacterial cells were incubated under standard growth conditions. The ADS activity of clinical strains of S. sanguinis varied from 45.2 to 263.4 units (mg protein)−1. Of note, higher ADS activity was observed among strains of S. parasanguinis, S. intermedius, S. gordonii and S. australis, when compared to strains of S. sanguinis and S. cristatus. There was no statistical difference between the average of bacterial ADS activity among the caries groups, despite the fact that, on average, arginolytic strains of CF subjects [200.2 units (mg protein)−1] presented slightly higher activity compared to those of CA subjects [188.1 units (mg protein)−1)] under standard growth conditions.
Table 1.
Identity and ADS activity levels of plaque bacterial strains.
| STUDY CODE | ACCESSION NUMBER |
HOT NO.▫ |
CLOSEST RELATIVE | SOURCE | ADS ACTIVITY (Mean ±SD) |
|---|---|---|---|---|---|
| Streptococcus gordonii DL1 | Lab strain | 339.3±33.0 | |||
| arcA-deficient strains of S. gordonii | Lab strain | 0 | |||
| Streptococcus parasanguinis A1 | KF733681.1 | 721 | Streptococcus parasanguinis ChDC B356 | CF | 688.0±57.1* |
| Streptococcus intermedius A2 | KF733728.1 | 644 | Streptococcus intermedius ChDC B589 | CF | 390.1±17.3* |
| Streptococcus intermedius A3 | KF733728.1 | 644 | Streptococcus intermedius ChDC B589 | CF | 476.9±43.8* |
| Streptococcus intermedius A4 | KF733728.1 | 644 | Streptococcus intermedius ChDC B589 | CF | 233.1±15.7 |
| Streptococcus intermedius A5 | KF733728.1 | 644 | Streptococcus intermedius ChDC B589 | CF | 252.85±61.79 |
| Streptococcus intermedius A6 | KF733728.1 | 644 | Streptococcus intermedius ChDC B589 | CF | 237.5±11.5 |
| Streptococcus gordonii A7 | CP000725.1 | 622 | Streptococcus gordonii str. Challis substr. CH1◆ | CA | 283.3±5.2 |
| Streptococcus gordonii A8 | NR_074516.1 | 622 | Streptococcus gordonii str. Challis substr. CH1 | CF | 431.9±15.4* |
| Streptococcus gordonii A9 | NR_115242.1 | 622 | Streptococcus gordonii ATCC 10558 | CA | 244.8±10.7 |
| Streptococcus gordonii A10 | NR_115242.1 | 622 | Streptococcus gordonii ATCC 10558 | CA | 241.3±15.9 |
| Streptococcus gordonii A11 | NR_115242.1 | 622 | Streptococcus gordonii ATCC 10558 | CF | 354.8±20.9* |
| Streptococcus australis A12 | NR_036936.1 | 073 | Streptococcus australis AI-1▫ | CF | 309.2±1.4 |
| Streptococcus australis A13 | NR_036936.1 | 073 | Streptococcus australis AI-1▫ | CF | 287.3±12.7 |
| Streptococcus sanguinis A14 | CP000387.1 | 758 | Streptococcus sanguinis SK36 | CF | 129.0±5.6 |
| Streptococcus sanguinis A15 | CP000387.1 | 758 | Streptococcus sanguinis SK36 | CF | 119.7±3.7 |
| Streptococcus sanguinis A16 | CP000387.1 | 758 | Streptococcus sanguinis SK36 | CF | 129.1±10.4 |
| Streptococcus sanguinis A17 | CP000387.1 | 758 | Streptococcus sanguinis SK36 | CF | 94.1±4.3 |
| Streptococcus sanguinis A18 | CP000387.1 | 758 | Streptococcus sanguinis SK36 | CF | 107.1±9.1 |
| Streptococcus sanguinis A19 | CP000387.1 | 758 | Streptococcus sanguinis SK36 | CF | 98.0±5.7 |
| Streptococcus sanguinis A20 | CP000387.1 | 758 | Streptococcus sanguinis SK36 | CF | 93.1±1.4 |
| Streptococcus sanguinis A21 | CP000387.1 | 758 | Streptococcus sanguinis SK36 | CF | 116.8±14.0 |
| Streptococcus sanguinis A22 | CP000387.1 | 758 | Streptococcus sanguinis SK36 | CF | 88.9±10.0 |
| Streptococcus sanguinis A23 | NR_113260.1 | 758 | Streptococcus sanguinis SK1284_K2-1 | CA | 127.6±1.1 |
| Streptococcus sanguinis A24 | NR_113260.1 | 758 | Streptococcus sanguinis JCM 5708 | CF | 45.2±6.0 |
| Streptococcus sanguinis A25 | NR_113260.1 | 758 | Streptococcus sanguinis JCM 5708 | CF | 50.2±3.4 |
| Streptococcus sanguinis A26 | NR_113260.1 | 758 | Streptococcus sanguinis JCM 5708 | CF | 46.0±0.3 |
| Streptococcus sanguinis A27 | NR_113260.1 | 758 | Streptococcus sanguinis JCM 5708 | CF | 63.4±0.0 |
| Streptococcus sanguinis A28 | NR_113260.1 | 758 | Streptococcus sanguinis JCM 5708 | CF | 56.4±13.0 |
| Streptococcus sanguinis A29 | NR_113260.1 | 758 | Streptococcus sanguinis JCM 5708 | CF | 187.1±20.3 |
| Streptococcus sanguinis A30 | NR_113260.1 | 758 | Streptococcus sanguinis JCM 5708 | CF | 173.3±4.0 |
| Streptococcus sanguinis A31 | NR_113260.1 | 758 | Streptococcus sanguinis JCM 5708 | CF | 246.2±2.4 |
| Streptococcus sanguinis A32 | NR_113260.1 | 758 | Streptococcus sanguinis JCM 5708 | CF | 227.3±0.0 |
| Streptococcus sanguinis A33 | NR_113260.1 | 758 | Streptococcus sanguinis JCM 5708 | CF | 199.2±8.3 |
| Streptococcus sanguinis A34 | NR_113260.1 | 758 | Streptococcus sanguinis JCM 5708 | CF | 200.6±14.6 |
| Streptococcus sanguinis A35 | NR_113260.1 | 758 | Streptococcus sanguinis JCM 5708 | CF | 201.9±15.2 |
| Streptococcus sanguinis A36 | NR_113260.1 | 758 | Streptococcus sanguinis JCM 5708 | CF | 263.4±29.9 |
| Streptococcus sanguinis A37 | NR_113260.1 | 758 | Streptococcus sanguinis JCM 5708 | CF | 227.9±89.7 |
| Streptococcus sanguinis A38 | NR_113260.1 | 758 | Streptococcus sanguinis JCM 5708 | CF | 167.7±9.9 |
| Streptococcus sanguinis A39 | NR_113260.1 | 758 | Streptococcus sanguinis JCM 5708 | CF | 198.1±5.1 |
| Streptococcus sanguinis A40 | NR_113260.1 | 758 | Streptococcus sanguinis JCM 5708 | CF | 212.5±0.6 |
| Streptococcus sanguinis A41 | NR_113260.1 | 758 | Streptococcus sanguinis JCM 5708 | CF | 144.0±10.0 |
| Streptococcus sanguinis A42 | NR_113260.1 | 758 | Streptococcus sanguinis JCM 5708 | CF | 190.1±10.6 |
| Streptococcus sanguinis A43 | NR_113260.1 | 758 | Streptococcus sanguinis JCM 5708 | CF | 106.4±4.0 |
| Streptococcus sanguinis A44 | NR_113260.1 | 758 | Streptococcus sanguinis JCM 5708 | CF | 104.3±4.1 |
| Streptococcus sanguinis A45 | NR_113260.1 | 758 | Streptococcus sanguinis JCM 5708 | CA | 161.3±3.3 |
| Streptococcus sanguinis A46 | NR_113260.1 | 758 | Streptococcus sanguinis JCM 5708 | CA | 169.8±6.4 |
| Streptococcus sanguinis A47 | NR_113260.1 | 758 | Streptococcus sanguinis JCM 5708 | CA | 221.3±4.8 |
| Streptococcus sanguinis A48 | NR_113260.1 | 758 | Streptococcus sanguinis JCM 5708 | CA | 136.9±4.3 |
| Streptococcus sanguinis A49 | NR_113260.1 | 758 | Streptococcus sanguinis JCM 5708 | CA | 238.2±31.1 |
| Streptococcus sanguinis A50 | NR_113260.1 | 758 | Streptococcus sanguinis JCM 5708 | CA | 182.0±39.3 |
| Streptococcus sanguinis A51 | KF733682.1 | 758 | Streptococcus sanguinis ChDC B357 | CF | 250.8±1.3 |
| Streptococcus cristatus A52 | NR_115274.1 | 578 | Streptococcus cristatus ATCC 51100 | CA | 187.5±41.4 |
| Streptococcus cristatus A53 | NR_115274.1 | 578 | Streptococcus cristatus ATCC 51100 | CA | 129.2±31.6 |
| Streptococcus cristatus A54 | GU470899.1 | 578 | Streptococcus cristatus F0329 | CA | 160.0±26.2 |
| Streptococcus cristatus A55 | GU470899.1 | 578 | Streptococcus cristatus F0329 | CA | 159.0±9.1 |
| Streptococcus cristatus A56 | GU470899.1 | 578 | Streptococcus cristatus F0329 | CA | 185.9±41.8 |
The 56 ADS-positive strains identified had greater than 99% sequence similarity with their assigned bacterial taxa, as derived from their database accession numbers.
Human Oral Taxon ID (HOT) from the Human Oral Microbiome Database (HOMD). ADS activity was expressed as nmol of citrulline generated [minute × (mg of protein)]−1.
An asterisk (*) indicates that the bacterial strain expressed higher ADS than S. gordonii DL1. CF: caries-free and CA: caries-active subjects; SD: standard deviation.
The closest relative strains to S. gordonii A7 are Streptococcus sp. JCM 5703 and S. gordonii str. Challis substr. CH1.
The closest relative strains to S. australis A12 and A13 are S. australis AI-1, but also Streptococcus rubneri LMG 27207 (accession number: NR_109720.1). Because A12 and A13 were collected from dental plaque and are ADS-positive, whereas the S. rubneri strain in the database is an ADS-negative throat isolate, A12 and A13 will be considered herein as strains of S. australis.
ADS express ion as a function of environmental stimuli
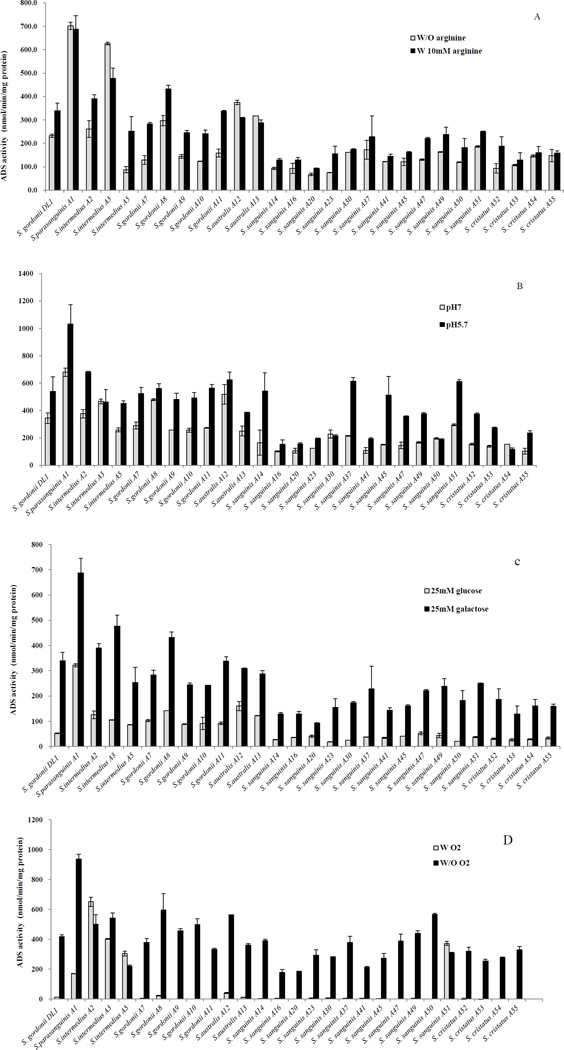
A substantial variation in ADS expression patterns was observed in response to pH, oxygen, and the availability of arginine and carbohydrate as illustrated in Figures 1 and 2. Figure 1A shows that for most strains, including the laboratory strain S. gordonii DL1, optimal expression of ADS was strongly dependent on the presence of arginine. However, strains such as S. parasanguinis A1, S. intermedius A2 and A3, S. gordonii A8 and S. australis A12 and A13 demonstrated higher ADS expression compared to S. gordonii DL1, even in the absence of arginine. Also of note, a number of the strains presented high levels of ADS in the absence of added arginine, including S. parasanguinis A1, S. intermedius A3, and S. australis A12 and A13.
Figure 1. ADS activity levels of S. gordonii DL1 and arginolytic clinical strains under different environmental conditions.
Cultures of TY medium containing: (A) 25 mM galactose with (w/) or without (w/o) 10 mM arginine, (B) 25 mM galactose and 10 mM arginine that had been acidified to pH 5.7 with HCl or buffered at pH 7.0, (C) 10 mM arginine and 25 mM galactose or 25 mM glucose, and (D) 25 mM galactose and 10 mM arginine and the cells were cultured under aerobic (w/O2) or anaerobic (w/o O2) conditions. Results represent the mean and standard deviations (error bars) of three independent experiments.
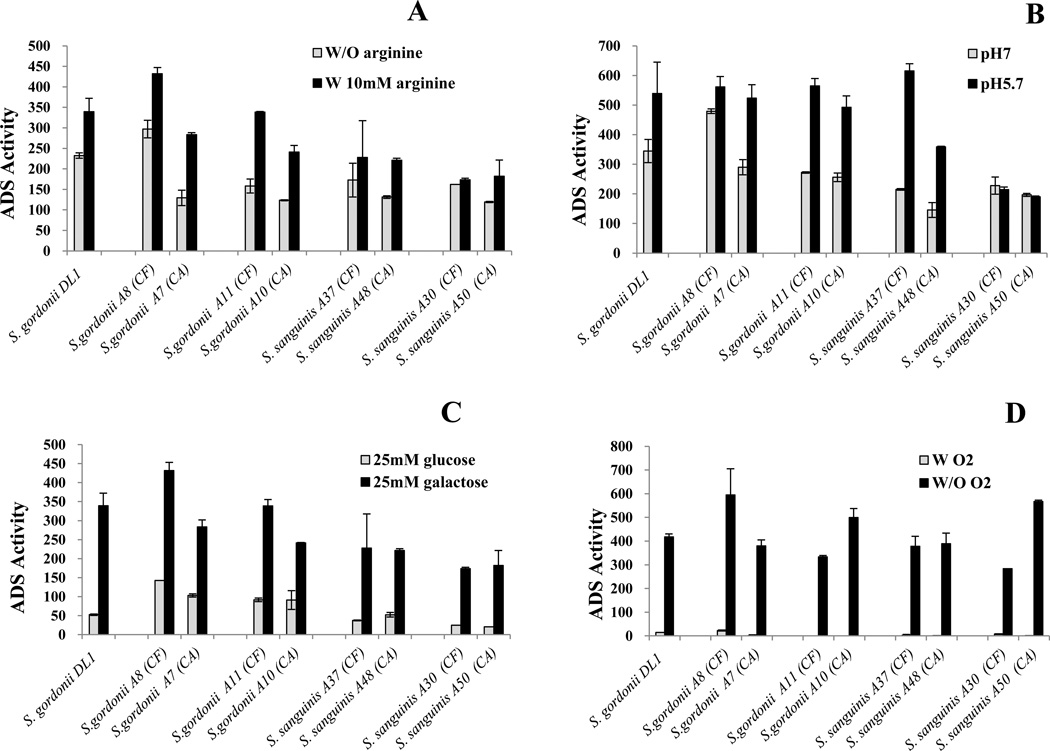
Figure 2. Comparison of the ADS activity levels of arginolytic strains from caries-active and caries-free subjects grown under different environmental conditions.
Cultures of TY medium containing: (A) 25 mM galactose with (w/) or without (w/o) 10 mM arginine, ((B) 25 mM galactose and 10 mM arginine that had been acidified to pH 5.7 with HCl or buffered at pH 7.0, (C) 10 mM arginine and 25 mM galactose or 25 mM glucose, and (D) 25 mM galactose and 10 mM arginine and the cells were cultured under aerobic (w/ O2) or anaerobic (w/o O2) conditions. CA: caries-active and CF: caries-free subjects. Results represent the mean and standard deviations (error bars) of three independent experiments.
A low pH environment is known to enhance ADS activity in S. gordonii DL1, however, S. intermedius A3, S. gordonii A8 and S. australis A12 expressed significantly higher levels of ADS activity than S. gordonii when cells were cultured at neutral pH (Fig. 1B) and did not show evidence of induction at pH 5.7. In contrast, S. parasanguinis A1 showed very high levels of ADS activity at pH and sigificant induction at pH 5.7 was observed.
Expression of the genes for the ADS of S. gordonii DL1 is also very sensitive to CCR [Dong et al., 2004], with growth in glucose resulting in 5-fold lower ADS activity compared to cells cultivated in galactose [Dong et al., 2004], which is less effective at eliciting CCR than glucose. Similarly, glucose could lower ADS activity of all the clinical strains to varying degrees (Fig. 1C). Yet strains such as S. parasanguinis A1, S. intermedius A2, A3 and A5, S. gordonii A7, A8, A9, A10 and A11, and S. australis A12 and A13, exhibited ADS activity that was higher than that expressed by S. gordonii DL1 when cells were grown in the presence of 25 mM glucose.
Figure 1D confirms that expression of ADS activity by S. gordonii DL1 was highly repressed in cells growing under aerobic conditions and similar repression by oxygen was noted in the majority of clinical strains. However, ADS activity in S. intermedius A2, A3 and A5, and in S. sanguinis A51 were either far less sensitive to, or resistant to the repressive effects of growth in aerated conditions.
Intraspecies heterogeneity was also highlighted by further phenotypic characterization of the clinical strains studied here (data not shown). For example, 16S sequencing revealed that the closest relative strains in the database to S. gordonii A7 are Streptococcus sp. JCM 5703 and S. gordonii str. Challis substr. CH1. However, phenotypically, the A7 strain is able to produce H202 from lactate; a property that is also found in A9, A10, and A11 strains, which are most similar at the 16S level to S. gordonii 10558. Unlike the A7, A9, A10 and A11 strains, the A8 strain, which at the 16S level is most similar to S. gordonii str. Challis substr. CH1, does not exhibit the capacity of producing H202 from lactate.
ADS expression and caries status
As shown in Table 1, different levels of ADS activity were observed among strains of the same species isolated from plaque of CF and CA subjects. For example, the strains S. sanguinis A48 [136.9 units (mg protein)−1] and S.gordonii A7 [283.3 units (mg protein)−1]) from CA subjects and S. sanguinis A36 [263.4 units (mg protein)−1] and S.gordonii A8 [431.9 units (mg protein)−1] from CF subjects presented different ADS expression under standard growth conditions, respectively. To further explore whether the arginolytic capacity of oral bacteria was related to the caries status of the subjects, we compared ADS expression in response to different environmental conditions for clinical strains of same species isolated from the different caries groups (Fig. 2). The selected strains included those with highest 16S rRNA sequence similarity to S. gordonii Challis substr. CH1 (A7 and A8), S. gordonii ATCC 10558 (A10 and A11), S. sanguinis JCM 5708 (A37 and A38), and S. sanguinis JCM 5708 (A30 and A50).
Figure 2A shows that most strains of the same species presented comparable differences in ADS expression in response to arginine independently of the subjects’ caries status. For example, both S. gordonii A10 from a CA subject and S. gordonii A11 from a CF subject showed about twice as high as ADS activity in the presence of arginine compared to growth in the absence of arginine. The exception was with the S. sanguinis A30 and A50 strains. While the expression levels of ADS were generally similar between the two strains, ADS expression in S. sanguinis A30 from a CF subject did not require supplemental arginine for optimal expression of the ADS, whereas S. sanguinis A50 did.
Similar patterns for ADS expression and induction by acidic pH were observed among strains of S. gordonii A7 and A8, S. gordonii A10 and A11, and S. sanguinis A37 and A38 from both caries groups (Fig. 2B). The exception was again for the S. sanguinis A30 and A50 strains, where S. sanguinis A30 from a CF subject and S. sanguinis A50 from a CA subject showed no induction of ADS by low pH, compared to neutral pH. In contrast to effects of arginine and pH, all strains of S. gordonii and S. sanguinis from both caries groups showed a similar pattern of ADS repression by glucose (Fig. 2C) and ADS repression by oxygen (Fig. 2D).
DISCUSSION
Arginine metabolism in oral biofilms offers the opportunity for the development of novel anti-caries approaches that may be beneficial for short-term moderation of acid challenges to teeth and long-term effects on the persistence of desirable bacteria in dental plaque. However, if arginolysis is to be used in the development of strategies to assess caries risk and to control caries, there is a need to fully understand the distribution, regulation and function of the ADS in oral biofilms in health and disease. Although genome sequencing and other molecular techniques have revealed new levels of complexity in the cariogenic microflora and in the nature of individual bacterial species [Aas et al., 2008; Corby et al., 2005; Crielaard et al., 2011; Mager et al., 2003; Russell, 2008], limited attempts [Sissons et al., 1988a; Sissons et al., 1988b; Sissons et al., 1994] have been made to identify and characterize the clinically-relevant oral organisms capable of producing alkali that can potentially diminish the cariogenicity of oral biofilms. In this study, a rapid and simple protocol was developed for screening of cultivable arginolytic bacteria isolated from dental plaque samples. Even though the majority of the ADS-positive bacterial species identified were strains of S. sanguinis, additional cultivable species including S. gordonii, S. intermedius, S. cristatus, S. australis and S. parasanguinis were shown to be present and to have the potential to contribute to different extents to total oral ADS activity. In conjunction with previous and extensive microbiological studies demonstrating the abundance in human oral biofilms of the commensal streptococci that were identified here [Aas et al., 2008; Aas et al., 2005; Corby et al., 2005; Crielaard et al., 2011; Dewhirst et al., 2010; Gross et al., 2010; Mager et al., 2003], our results reveal that these abundant streptococci likely have a dominant influence on the arginolytic capacity of human oral biofilms. Importantly, this study clearly demonstrates that the ADS of clinical strains is in fact regulated in response to those specific environmental factors that have the greatest impact on the composition and biochemical activities of supragingival biofilms; e.g. availability and source of carbohydrate, low pH and oxygen are all well-recognized environmental factors that can influence the development of caries.
The diversity of the oral microbiota associated with health and disease is only beginning to be described by high-throughput methodologies [Aas et al., 2008; Corby et al., 2005; Crielaard et al., 2011; Gross et al., 2010; Mager et al., 2003]. While this species- or taxa-level identification is tremendously valuable [Dewhirst et al., 2010], it does not address the fact that there is significant heterogeneity within given species of oral bacteria. Based on current sequencing efforts, the majority of ADS-positive oral bacterial species are cultivable, and mostly consist of abundant oral streptococci. Although uncultivable organisms may contribute to the total arginolytic activity measured in oral biofilms, organisms that were unlikely to grow under the conditions we used to cultivate plaque samples generally do not possess the ADS (Nascimento and Burne, Unpublished observation) or are not well represented in supragingival plaque, e.g. certain spirochetes. Further, many uncultivable bacterial species are represented in plaque in far lower proportions than cultivable species [Aas et al., 2008; Corby et al., 2005; Crielaard et al., 2011; Gross et al., 2010; Mager et al., 2003]. Thus, one would expect that the contribuition to the total ADS of uncultivable organisms is minor at best. Importantly, the present study enhances ongoing oral microbiome efforts by highlighting the phenotypic heterogeneity of the more abundant species in the oral cavity in the context of their abilities to modulate the pH, and thus the cariogenic potential, of oral biofilms. This study also presents novel theoretical concepts - the molecular basis for heterogeneity in alkali production - while concurrently generating knowledge, strains, probes and reagents that will advance existing methodologies for evaluating and understanding the pathogenic potential of the oral microbiome.
Markedly less is known about the production of alkali than is known about sugar metabolism in oral biofilms. The causal relationship between bacterial sugar metabolism and acid production by a mixed population of plaque bacteria was first described by Stephan [Stephan, 1940]. Stephan also pointed out that the drop in plaque pH detected after sugar challenge is followed by a gradual rise in plaque pH that eventually reaches a plateau. Later, the plateau - or resting pH - of caries-active plaque was found to be more acidic than that of caries-free plaque [Margolis et al., 1988b], further supporting a correlation between acid production and dental caries. Subsequent studies showed that the rise in plaque pH is largely due to ammonia production from arginine or urea by a subset of acid-sensitive organisms present in saliva and plaque [Wijeyeweera and Kleinberg, 1989a]. Marquis suggested [Marquis, 1995] that the buffering capacity from ammonia production in oral biofilms moderates the speed of the pH drop and allows time for the base-producing bacteria to adjust their physiology for survival. Kleinberg showed that carbohydrate starved plaque was more alkaline than the saliva bathing the plaque, mainly in regions of greater saliva flow, so it was suggested that plaque bacteria generate ammonia from salivary substrates more rapidly than the forces of diffusion can clear them from dental plaque [Kleinberg and Jenkins, 1964]. Kleinberg also foretold [Kleinberg, 1970] that the plaque pH would be determined by the acid-base metabolism of plaque organisms, which in turn could be affected by plaque thickness, the proportions of acid- and base-producing organisms in plaque, and the relative availability of nitrogenous and carbohydrates substrates.
Our clinical studies to date support that caries susceptibility involves a deficiency in alkali production and not solely acid production, as has been traditionally assumed [Nascimento et al., 2009a; Nascimento et al., 2012; Shu et al., 2007a]. Here, we began to examine whether the heterogeneity of oral bacterial strains, the constitutional difference in the ADS genes expression levels, and/or differential sensitivity of the ADS genes to induction or repression by environmental factors, could account for the high degree of variability in alkali production detected in dental health and when caries activity is evident. Although ADS-positive strains from caries-free subjects showed slightly higher levels of ADS activity than those isolated from caries-active subjects, there was no significant correlation between levels of bacterial ADS activity and the caries status of the host. Yet, this study examining a collection of arginolytic plaque bacteria, or more specifically, the ADS activity in closely-related commensal streptococci, revealed a considerable spectrum of responses of the ADS to multiple environmental stimuli. While control of expression and activity of the ADS in vivo may be more complex and involve more inputs than tested here, the results clearly show that differences in the regulation of the ADS and absolute levels of AD activity exist in the most abundant members of the commensal flora. Thus, it is possible that the basis for differences in arginolysis observed between caries-free and caries-active subjects can be associated with: (i) the carriage in oral biofilms of strains that have inherent differences in the regulation of the ADS by environmental factors, and/or (ii) host and biofilm micro-environmental factors that influence ADS expression in vivo. For example, the biofilms of caries-active subjects may not be conducive to high ADS expression or may produce inhibitory factors that decrease ADS gene expression or activity. Clearly, arginolytic clinical strains with constitutionally high ADS-expressing phenotypes and those in which ADS expression is insensitive to conditions known to cause dental caries - sugar availability and acidic environment - may have great potential in probiotic therapies to prevent and control dental caries.
In conclusion, this study reveals that the microbial basis for intra-subject variation in oral arginolysis is more complex than previously appreciated; not only may the arginolytic potential of oral biofilms be associated with the carriage of certain strains of bacteria, but also arginolytic species display a range of ADS activity as a function of environmental variables. The results are significant in the context of understanding caries as an ecologically-driven disease by supporting that high ADS-expressing strains could positively affect plaque ecology in a synergistic manner, i.e. by moderating plaque pH and reducing the risk for caries. This study has expanded knowledge on the diversity of the oral alkali-generating bacteria and, potentially on their role in oral health and disease. Future studies will assess whether there are associations of particular strains possessing beneficial properties with dental health, or if particular strains with undesirable properties – high aciduricity, low arginolysis, poor antagonism against acidogenic organisms – are associated with disease. Undoubtedly, a better understanding of the microbial composition of plaque and its pH responses are needed in order to efficiently reduce the cariogenicity of oral biofilms.
ACKNOWLEDGEMENTS
We thank Yaling Liu and Roshan Kalra for their contributions to the development and refinement of some of the methodologies utilized in this study. This work was supported by Colgate-Palmolive and NIH/NIDCR R01-DE10362, K23-DE023579, and R90-DE022530.
Footnotes
DECLARATION OF INTERESTS
The authors declare that there are no conflicts of interest.
AUTHOR CONTRIBUTIONS
M.M. Nascimento and R.A. Burne designed the study. M.M. Nascimento performed the clinical examinations for recruitment of research subjects and collected dental plaque samples from the eligible subjects. R.M. Schulte isolated the bacterial strains from the plaque samples. X. Huang performed all the biochemical and molecular biology assays of the study and analyzed the raw data. X. Huang, R.A. Burne and M.M. Nascimento evaluated and interpreted the data, and wrote the manuscript.
REFERENCES
- Aas JA, Griffen AL, Dardis SR, Lee AM, Olsen I, Dewhirst FE, Leys EJ, Paster BJ. Bacteria of dental caries in primary and permanent teeth in children and young adults. J Clin Microbiol. 2008;46:1407–1417. doi: 10.1128/JCM.01410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43:5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acevedo AM, Machado C, Rivera LE, Wolff M, Kleinberg I. The inhibitory effect of an arginine bicarbonate/calcium carbonate cavistat-containing dentifrice on the development of dental caries in venezuelan school children. J Clin Dent. 2005;16:63–70. [PubMed] [Google Scholar]
- Acevedo AM, Montero M, Rojas-Sanchez F, Machado C, Rivera LE, Wolff M, Kleinberg I. Clinical evaluation of the ability of cavistat in a mint confection to inhibit the development of dental caries in children. J Clin Dent. 2008;19:1–8. [PubMed] [Google Scholar]
- Becker MR, Paster BJ, Leys EJ, Moeschberger ML, Kenyon SG, Galvin JL, Boches SK, Dewhirst FE, Griffen AL. Molecular analysis of bacterial species associated with childhood caries. J Clin Microbiol. 2002;40:1001–1009. doi: 10.1128/JCM.40.3.1001-1009.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burne R, Liu Y, Zeng L. Acid tolerance strategies of commensal and pathogenic oral streptococci. Society for General Microbiology Autumn 2010 Meeting; 2010; Nottingham, UK. [Google Scholar]
- Burne RA, Marquis RE. Alkali production by oral bacteria and protection against dental caries. FEMS Microbiol Lett. 2000;193:1–6. doi: 10.1111/j.1574-6968.2000.tb09393.x. [DOI] [PubMed] [Google Scholar]
- Burne RA, Parsons DT, Marquis RE. Environmental variables affecting arginine deiminase expression in oral streptococci; in Genetics and molecular biology of streptococci, lactococci, and enterococci. American Society for Microbiology; 1991; Washington DC. [Google Scholar]
- Burne RA, Wen ZT, Chen YY, Penders JE. Regulation of expression of the fructan hydrolase gene of streptococcus mutans gs-5 by induction and carbon catabolite repression. J Bacteriol. 1999;181:2863–2871. doi: 10.1128/jb.181.9.2863-2871.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casiano-Colon A, Marquis RE. Role of the arginine deiminase system in protecting oral bacteria and an enzymatic basis for acid tolerance. Appl Environ Microbiol. 1988;54:1318–1324. doi: 10.1128/aem.54.6.1318-1324.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy KA, Pearson S, Bowen WH, Burne RA. Characterization of recombinant, ureolytic Streptococcus mutans demonstrates an inverse relationship between dental plaque ureolytic capacity and cariogenicity. Infection and immunity. 2000;68:2621–2629. doi: 10.1128/iai.68.5.2621-2629.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corby PM, Lyons-Weiler J, Bretz WA, Hart TC, Aas JA, Boumenna T, Goss J, Corby AL, Junior HM, Weyant RJ, Paster BJ. Microbial risk indicators of early childhood caries. J Clin Microbiol. 2005;43:5753–5759. doi: 10.1128/JCM.43.11.5753-5759.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crielaard W, Zaura E, Schuller AA, Huse SM, Montijn RC, Keijser BJ. Exploring the oral microbiota of children at various developmental stages of their dentition in the relation to their oral health. BMC Med Genomics. 2011;4:22. doi: 10.1186/1755-8794-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawes C, Dibdin GH. Salivary concentrations of urea released from a chewing gum containing urea and how these affect the urea content of gel-stabilized plaques and their pH after exposure to sucrose. Caries Res. 2001;35:344–353. doi: 10.1159/000047473. [DOI] [PubMed] [Google Scholar]
- Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, Lakshmanan A, Wade WG. The human oral microbiome. J Bacteriol. 2010;192:5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Chen YY, Burne RA. Control of expression of the arginine deiminase operon of Streptococcus gordonii by Ccpa and Flp. J Bacteriol. 2004;186:2511–2514. doi: 10.1128/JB.186.8.2511-2514.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golub LM, Borden SM, Kleinberg I. Urea content of gingival crevicular fluid and its relation to periodontal diseases in humans. J Periodont Res. 1971;6:243–251. doi: 10.1111/j.1600-0765.1971.tb00615.x. [DOI] [PubMed] [Google Scholar]
- Gross EL, Leys EJ, Gasparovich SR, Firestone ND, Schwartzbaum JA, Janies DA, Asnani K, Griffen AL. Bacterial 16s sequence analysis of severe caries in young permanent teeth. J Clin Microbiol. 2010;48:4121–4128. doi: 10.1128/JCM.01232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Exterkate RA, Ten Cate JM. Factors associated with alkali production from arginine in dental biofilms. J Dent Res. 2012;91:1130–1134. doi: 10.1177/0022034512461652. [DOI] [PubMed] [Google Scholar]
- Kleinberg I. Biochemistry of the dental plaque. Advances in oral biology. 1970;4:43–90. doi: 10.1016/b978-0-12-030504-9.50010-x. [DOI] [PubMed] [Google Scholar]
- Kleinberg I, Jenkins GN. The pH of dental plaques in the different areas of the mouth before and after meals and their relationship to the pH**rate of flow of resting saliva. Arch Oral Biol. 1964;72:493–516. doi: 10.1016/0003-9969(64)90015-9. [DOI] [PubMed] [Google Scholar]
- Kopstein J, Wrong OM. The origin and fate of salivary urea and ammonia in man. Clin Sci Mol Med. 1977;52:9–17. doi: 10.1042/cs0520009. [DOI] [PubMed] [Google Scholar]
- Kraivaphan P, Amornchat C, Triratana T, Mateo LR, Ellwood R, Cummins D, Devizio W, Zhang YP. Two-Year caries clinical study of the efficacy of novel dentifrices containing 1.5% arginine, an insoluble calcium compound and 1,450 ppm fluoride. Caries Res. 2013;47:582–590. doi: 10.1159/000353183. [DOI] [PubMed] [Google Scholar]
- Liu Y, Burne RA. Multiple two-component systems modulate alkali generation in Streptococcus gordonii in response to environmental stresses. J Bacteriol. 2009;191:7353–7362. doi: 10.1128/JB.01053-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Burne RA. The major autolysin of Streptococcus gordonii is subject to complex regulation and modulates stress tolerance, biofilm formation, and extracellular-DNA release. J Bacteriol. 2011;193:2826–2837. doi: 10.1128/JB.00056-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Dong Y, Chen YY, Burne RA. Environmental and growth phase regulation of the Streptococcus gordonii arginine deiminase genes. Appl Environ Microbiol. 2008;74:5023–5030. doi: 10.1128/AEM.00556-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mager DL, Ximenez-Fyvie LA, Haffajee AD, Socransky SS. Distribution of selected bacterial species on intraoral surfaces. J Clin Periodontol. 2003;30:644–654. doi: 10.1034/j.1600-051x.2003.00376.x. [DOI] [PubMed] [Google Scholar]
- Margolis HC, Duckworth JH, Moreno EC. Composition and buffer capacity of pooled starved plaque fluid from caries-free and caries-susceptible individuals. J Dent Res. 1988;67:1476–1482. doi: 10.1177/00220345880670120701. [DOI] [PubMed] [Google Scholar]
- Marquis RE. Oxygen metabolism, oxidative stress and acid-base physiology of dental plaque biofilms. J Ind Microbiol. 1995;15:198–207. doi: 10.1007/BF01569826. [DOI] [PubMed] [Google Scholar]
- Marquis RE, Bender GR, Murray DR, Wong A. Arginine deiminase system and bacterial adaptation to acid environments. Appl Environ Microbiol. 1987;53:198–200. doi: 10.1128/aem.53.1.198-200.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento MM, Gordan VV, Garvan CW, Browngardt CM, Burne RA. Correlations of oral bacterial arginine and urea catabolism with caries experience. Oral Microbiol Immunol. 2009;24:89–95. doi: 10.1111/j.1399-302X.2008.00477.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento MM, Liu Y, Kalra R, Perez S, Adewumi A, Xu X, Burne RA. Arginine metabolism may confer caries resistance in children. J Dent Res. 2012;91(Spec Iss A) doi: 10.1177/0022034513487907. Abst.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, Levanos VA, Sahasrabudhe A, Dewhirst FE. Bacterial diversity in human subgingival plaque. J Bacteriol. 2001;183:3770–3783. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson S, Woodhead J, Crall J. Caries resistance in children with chronic renal failure: Plaque pH, salivary pH, and salivary composition. Pediatr Res. 1985;19:796–799. doi: 10.1203/00006450-198508000-00003. [DOI] [PubMed] [Google Scholar]
- Rogers AH. Utilization of nitrogenous compounds by oral bacteria. Aust Dent J. 1990;35:468–471. doi: 10.1111/j.1834-7819.1990.tb05432.x. [DOI] [PubMed] [Google Scholar]
- Russell RR. How has genomics altered our view of caries microbiology? Caries Res. 2008;42:319–327. doi: 10.1159/000151326. [DOI] [PubMed] [Google Scholar]
- Schulte R, Burne RA, Gordan VV, Nascimento MM. Alkali generation capacity of oral bacteria. J Dent Res. 2009;88(Spec Iss A) Abst. 113. [Google Scholar]
- Shu M, Morou-Bermudez E, Suarez-Perez E, Rivera-Miranda C, Browngardt CM, Chen YY, Magnusson I, Burne RA. The relationship between dental caries status and dental plaque urease activity. Oral Microbiol Immunol. 2007;22:61–66. doi: 10.1111/j.1399-302X.2007.00325.x. [DOI] [PubMed] [Google Scholar]
- Sissons CH, Hancock EM, Cutress TW. The source of variation in ureolysis in artificial plaques cultured from human salivary bacteria. Arch Oral Biol. 1988a;33:721–726. doi: 10.1016/0003-9969(88)90005-2. [DOI] [PubMed] [Google Scholar]
- Sissons CH, Hancock EM, Perinpanayagam HE, Cutress TW. The bacteria responsible for ureolysis in artificial dental plaque. Arch Oral Biol. 1988b;33:727–733. doi: 10.1016/0003-9969(88)90006-4. [DOI] [PubMed] [Google Scholar]
- Sissons CH, Wong L, Hancock EM, Cutress TW. The pH response to urea and the effect of liquid flow in 'artificial mouth' microcosm plaques. Arch Oral Biol. 1994;39:497–505. doi: 10.1016/0003-9969(94)90146-5. [DOI] [PubMed] [Google Scholar]
- Srisilapanan P, Korwanich N, Yin W, Chuensuwonkul C, Mateo LR, Zhang YP, Cummins D, Ellwood RP. Comparison of the efficacy of a dentifrice containing 1.5% arginine and 1450 ppm fluoride to a dentifrice containing 1450 ppm fluoride alone in the management of early coronal caries as assessed using Quantitative Light-induced Fluorescence. J Dent. 2013;41(2):29–34. doi: 10.1016/j.jdent.2010.04.005. [DOI] [PubMed] [Google Scholar]
- Stephan RM. Changes in hydrogen-ion concentration on tooth surfaces and in carious lesions. J Am Dent Assoc. 1940;27:718–723. [Google Scholar]
- Van Wuyckhuyse BC, Perinpanayagam HE, Bevacqua D, Raubertas RF, Billings RJ, Bowen WH, Tabak LA. Association of free arginine and lysine concentrations in human parotid saliva with caries experience. J Dent Res. 1995;74:686–690. doi: 10.1177/00220345950740021001. [DOI] [PubMed] [Google Scholar]
- Vander Wauven C, Pierard A, Kley-Raymann M, Haas D. Pseudomonas aeruginosa mutants affected in anaerobic growth on arginine: Evidence for a four-gene cluster encoding the arginine deiminase pathway. J Bacteriol. 1984;160:928–934. doi: 10.1128/jb.160.3.928-934.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijeyeweera RL, Kleinberg I. Arginolytic and ureolytic activities of pure cultures of human oral bacteria and their effects on the pH response of salivary sediment and dental plaque in vitro. Arch Oral Biol. 1989;34:43–53. doi: 10.1016/0003-9969(89)90045-9. [DOI] [PubMed] [Google Scholar]
- Yin W, Hu DY, Fan X, Feng Y, Zhang YP, Cummins D, Mateo LR, Pretty IA, Ellwood RP. A clinical investigation using quantitative light-induced fluorescence (QLF) of the anticaries efficacy of a dentifrice containing 1.5% arginine and 1450 ppm fluoride as sodium monofluorophosphate. J Clin Dent. 2013;24(A):15–22. [PubMed] [Google Scholar]
- Yin W, Hu DY, Li X, Fan X, Zhang YP, Pretty IA, Mateo LR, Cummins D, Ellwood RP. The anti-caries efficacy of a dentifrice containing 1.5% arginine and 1450 ppm fluoride as sodium monofluorophosphate asse ssed using Quantitative Light-induced Fluorescence (QLF) J Dent. 2013;41(2):22–28. doi: 10.1016/j.jdent.2010.04.004. [DOI] [PubMed] [Google Scholar]