Abstract
Autophagy is a catabolic recycling pathway triggered by various intra- or extracellular stimuli that is conserved from yeast to mammals. During autophagy diverse cytosolic constituents are enveloped by double-membrane vesicles, autophagosomes, which later fuse with lysosomes or the vacuole in order to degrade their cargo. Dysregulation in autophagy is associated with a diverse range of pathologies including cardiovascular disease, the leading cause of death in the world. As such, there is great interest in identifying novel mechanisms that govern the cardiovascular response to disease-related stress. First described in failing hearts, autophagy within the cardiovascular system has been widely characterized in cardiomyocytes, cardiac fibroblasts, endothelial cells and vascular smooth muscle cells. In all cases, a window of optimal autophagic activity appears to be critical to the maintenance of cardiovascular homeostasis and function; excessive or insufficient levels of autophagic flux can each contribute to heart disease pathogenesis. Here we review the molecular mechanisms that govern autophagosome formation and analyze the link between autophagy and cardiovascular disease.
Keywords: atherosclerosis, autophagy, blood vessels, cardiac hypertrophy, cardiomyocyte, cardiovascular diseases, cardiovascular system, cell signaling, endothelial cell, fibroblast, heart, heart failure, myocardial infarction, vascular smooth muscle
Introduction
Autophagy is a key catabolic process for cell survival against different types of stress. During macroautophagy the cell carries out the sequestration of different substrates such as invading pathogens, proteins, lipids, or even damaged or superfluous organelles inside double-membrane vesicles termed autophagosomes. In order to degrade their cargo, autophagosomes fuse with the lysosome/vacuole, which contains various hydrolases able to break down the sequestered substrates. Following this event, the basic components obtained from cargo degradation are released into the cytoplasm for recycling.2 Autophagy is triggered by different stimuli such as hypoxia,3 oxidative stress,4 pathogen infection,5 endoplasmic reticulum (ER) stress,6 and most notably nutrient starvation.
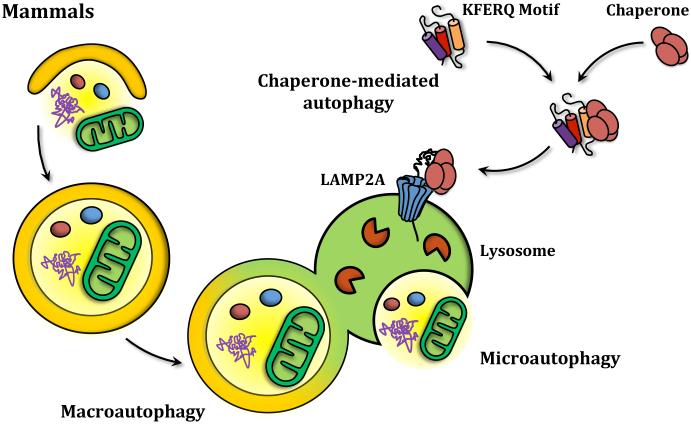
Autophagy can be divided into three main types: Macroautophagy, microautophagy and chaperone-mediated autophagy. Whereas microautophagy and macroautophagy can be selective or nonselective processes found in yeast and higher eukaryotes, chaperone-mediated autophagy is a selective process that has only been described in mammalian cells. During chaperone-mediated autophagy, specific protein substrates containing the amino acid sequence KFERQ are recognized by chaperones, unfolded and translocated into the lysosome through the lysosomal membrane protein LAMP2A (Figure 1A). In microautophagy uptake also occurs directly at the limiting membrane of the lysosome/vacuole. In this case, however, the process operates by directly sequestering the substrates through invagination of the vacuole/lysosome membrane.7 Of the three types mentioned, macroautophagy is clearly the most well studied process (Figure 1B). Multiple autophagy-related (ATG) genes and proteins have been described as being involved in the different stages of autophagy, comprising what is now known as the core autophagy machinery that is required for autophagosome formation, and additional proteins that act in making the process selective, or in stages other than autophagosome biogenesis. Accordingly, macroautophagy (hereafter referred to as autophagy) can be dissected into different steps based on the proteins involved, including: Induction, nucleation of the autophagosome precursor (termed the phagophore), membrane expansion and maturation of the autophagosome, fusion with the lysosome/vacuole, and recycling of the degraded cargo.
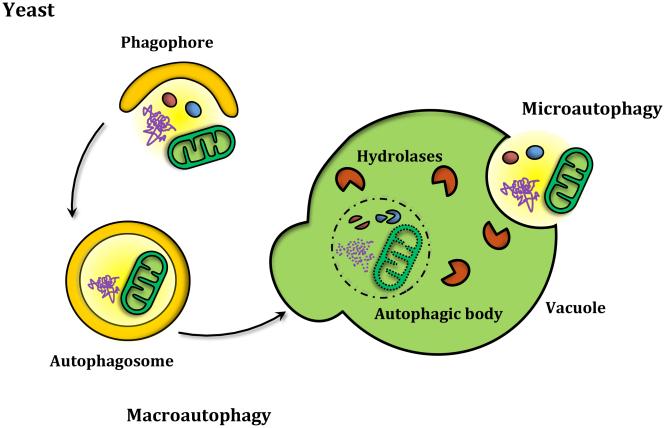
Figure 1. The three main types of autophagy.
(A) Yeast cells carry out both macroautophagy and microautophagy. While macroautophagy consists of the bulk degradation of cytoplasmic material that is sequestered inside double-membrane autophagosomes that then fuse with the vacuole, microautophagy works by directly taking up the substrates through invagination of the vacuole. (B) In mammals along with microautophagy and macroautophagy, chaperone-mediated autophagy enables the degradation of specific protein substrates that contain a KFERQ motif that is recognized by chaperones that mediate the translocation of the protein into the lysosome through LAMP2A.
Autophagy induction in yeast
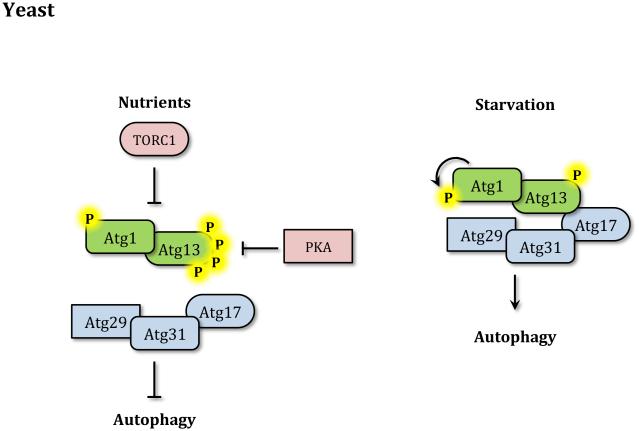
During growth in nutrient-rich conditions autophagy activity is kept to a minimum by different nutrient sensing pathways including those regulated by the target of rapamycin (TOR) kinase and cAMP-dependent protein kinase A (PKA). The ability of TOR to sense nutrient levels, in particular amino acids, makes it a critical negative regulator of autophagy. The rapamycin-sensitive TOR complex 1 (TORC1) inhibits autophagy in part by preventing the activation of the Atg1 kinase complex.8 In yeast the Atg1 kinase complex is formed by the Ser/Thr kinase Atg1, the regulatory subunit protein Atg13, and the Atg17-Atg31-Atg29 complex, which is thought to function as a scaffold.8,9 Although specific substrates of Atg1 may remain to be discovered, the Atg1 kinase complex plays an essential role in autophagy induction by recruiting other Atg proteins to what is known as the phagophore assembly site (PAS), a perivacuolar location found in yeast that is proposed to be the organizing center for phagophore formation.10-13 Upon nutrient starvation or rapamycin treatment TORC1 activity is inhibited and Atg13 is rapidly but partially dephosphorylated leading to the activation of Atg1.14 Autophosphorylation of Atg1 within its activation loop is also important for activating its kinase activity and inducing autophagy (Figure 2A).15 Atg13 dephosphorylation was linked to an increased interaction with Atg1 leading to a model in which starvation increased affinity between the two proteins; however, recent data as well as interaction studies in other model organisms support the idea that Atg1 and Atg13 interact independently of nutrient conditions.16 In addition to Atg13, TORC1 may also inhibit autophagy by directly phosphorylating Atg1.17
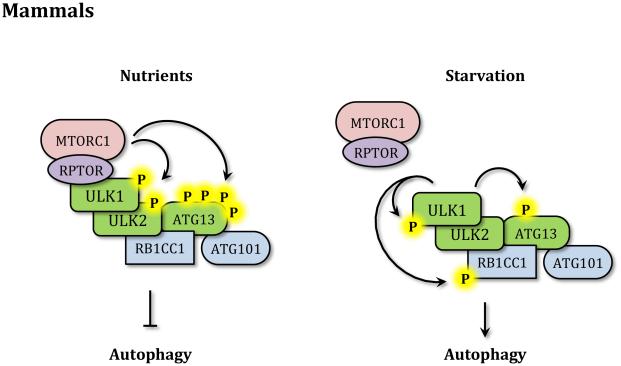
Figure 2. Autophagy induction.
(A) In yeast in nutrient-rich conditions TORC1 and PKA inhibit autophagy by phosphorylating Atg1 and Atg13. During starvation the Atg1 kinase complex is no longer repressed, Atg13 is partially dephosphorylated and Atg1 is activated. Atg1 then phosphorylates itself and other targets to induce autophagy. (B) In mammals in nutrient-rich conditions MTORC1 directly binds ULK1 through RPTOR and inhibits ULK1/2 and ATG13 by phosphorylation. Upon starvation MTORC1 dissociates from the ULK1 kinase complex, allowing ATG13 dephosphorylation and activating ULK1/2 that then phosphorylates members of the complex and other targets to induce autophagy.
PKA is also a negative regulator of autophagy. PKA suppresses autophagy by phosphorylating both Atg1 and Atg13.18-20 While the two pathways target similar proteins, TOR and PKA seem to work largely independent from one another by targeting different phosphorylation sites.20 Other nutrient sensing kinases involved in autophagy induction are Snf1 and Gcn2. While the former corresponds to the yeast homolog of 5’-AMP-activated protein kinase (AMPK), which will be discussed below, Gcn2 promotes autophagy during amino acid starvation by phosphorylating Sui2/eIF2α. Phosphorylation of Sui2 blocks general protein synthesis and specifically activates the translation of the transcription factor Gcn4, which in turn induces the transcription of various ATG genes.21, 22
Autophagy induction in mammals
In contrast to yeast, mammalian cells have multiple Atg1 homologs, and the ones most relevant to autophagy are ULK1 (unc-51 like autophagy activating kinase 1) and ULK2.23, 24 Thus, in mammals the Atg1 kinase complex is known as the ULK kinase complex and is formed by ULK1/2, the mammalian homolog of Atg13 (ATG13), the functional homolog of Atg17 (RB1CC1) and the ATG13 stabilizing protein ATG101, which has no yeast counterpart. All members of the ULK kinase complex are required for autophagy induction in mammalian cells.25-27 As mentioned above, in mammalian cells the interaction between the members of the ULK kinase complex does not depend on nutrient conditions.28 While some studies indicate that ATG13 mediates the interaction between RB1CC1 and ULK,26 others have reported that all members of the complex can interact independently.28 Similar to the yeast Atg1 complex, regulation of the ULK kinase complex depends on MTORC1. During nutrient-rich conditions MTORC1 interacts directly with ULK1 through the scaffold protein RPTOR and inhibits its kinase activity by phosphorylating both ATG13 and ULK1/2.26, 29 Upon nutrient starvation or rapamycin treatment MTORC1 is released from the ULK kinase complex leading to the dephosphorylation of both proteins and the activation of ULK kinase activity.26,28,29 Once activated, ULK1 phosphorylates ATG13, RB1CC1 and itself, stabilizing its enzymatic activity and inducing the autophagic process (Figure 2B).26,29,30
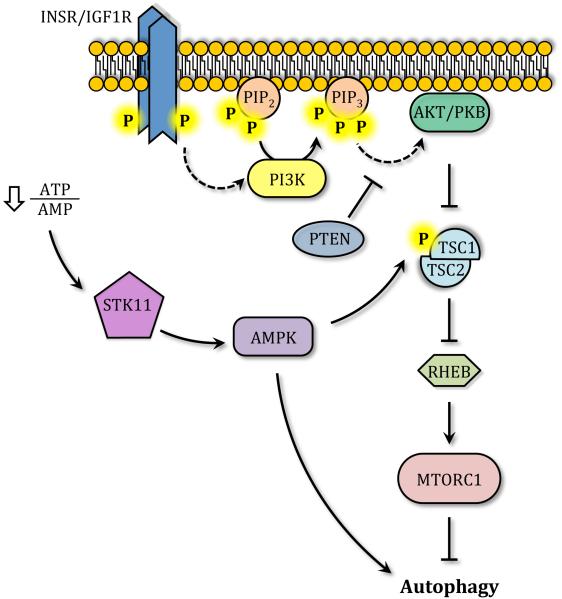
Another protein capable of sensing energy levels that is involved in autophagy regulation is AMPK. Through the upstream kinase STK11/LKB1, AMPK is able to sense decreases in the cellular ATP/AMP ratio leading to its activation and autophagy induction.31 During glucose deprivation AMPK phosphorylates and activates the tuberous sclerosis complex, TSC1/2, which in turn inactivates the GTPase activating protein RHEB, leading to MTORC1 inhibition and the release of the ULK kinase complex (Figure 3).32, 33 Once MTORC1 leaves the ULK complex, AMPK directly phosphorylates ULK1, stimulating its catalytic activity and inducing autophagy.31 Interestingly, the ULK kinase complex also phosphorylates and inactivates AMPK, through a mechanism that has been described as an inhibitory feedback loop.34
Figure 3. Autophagy regulation.
Through STK11/LKB1, AMPK senses decreases in the ATP/AMP ratio and phosphorylates TSC1-TSC2, which then targets RHEB, leading to MTORC1 inhibition and autophagy activation. INSR/IGF1R triggers the activation of the class I PI3K, inducing the formation of phosphatidylinositol(3,4,5)triphosphate (PIP3) and AKT/PKB activation; AKT can inhibit TSC1/TSC2, blocking autophagy. PTEN works as a PIP3 phosphatase generating phosphatidylinositol(4,5)bisphosphate (PIP2) and inducing autophagy.
Although functioning in part in a hormone-sensing pathway, AKT/PKB can also regulate autophagy by controlling MTORC1 activation. Upon ligand binding, dimerization, autophosphorylation and activation of INSR (insulin receptor) or IGF1R (insulin-like growth factor 1 receptor), the class I phosphoinositide 3-kinase (PI3K) is recruited to the plasma membrane and activated.35 PI3K catalyzes the phosphorylation of phosphatidylinositol(4,5)bisphosphate (PIP2) generating the lipid second messenger phosphatidylinositol(3,4,5)trisphosphate (PIP3) which in turn recruits AKT to the plasma membrane where it is activated via phosphorylation by PDPK1 and MTORC2.35,36 AKT-dependent phosphorylation of TSC2 prevents RHEB inhibition, leading to MTORC1 activation and autophagy inhibition.37, 38 As a consequence, the tumor suppressor and lipid phosphatase PTEN can induce autophagy by dephosphorylating PIP3 and downregulating the AKT-PI3K pathway (Figure 3).39
Membrane nucleation and source
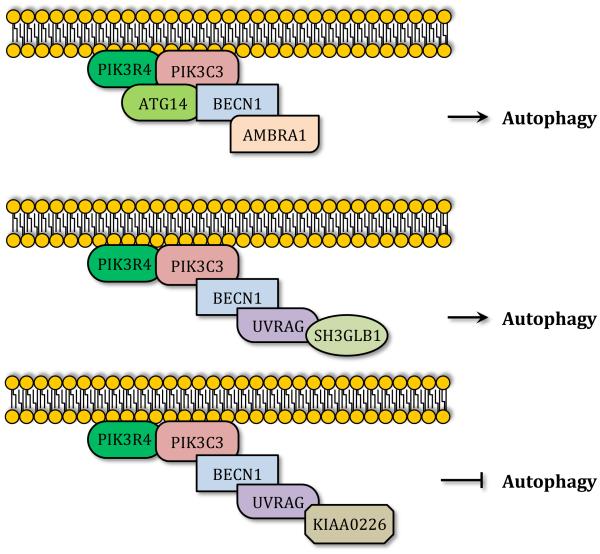
Once autophagy is induced, assembly of the phagophore is initiated by membrane nucleation. As mentioned above, in yeast the PAS corresponds to the location at which several Atg proteins are recruited to assemble the phagophore. In contrast, mammalian cells lack a single defined PAS, and autophagosome formation seems to be initiated at different locations inside the cell. In both yeast and mammals the class III phosphatidylinositol 3-kinase (PtdIns3K) catalyzes the nucleation of the phagophore by generating phosphatydilinositol 3-phosphate (PtdIns3P) and inducing the recruitment of PtdIns3P binding proteins.38 In yeast the PtdIns3K is formed by the regulatory subunit Vps15, the catalytic subunit Vps34, Vps30/Atg6, Atg14 and Atg38, all of which are essential for autophagy.40-42 Similarly, the core mammalian PtdIns3K is composed of the Vps15 homolog PIK3R4, the Vps34 homolog PIK3C3, and the Vps30/Atg6 homolog BECN1.38,43 While these three proteins constitute the core machinery of the mammalian PtdIns3K, distinct interactions with specific proteins lead to the formation of at least three different PtdIns3K complexes that play different roles in autophagy.44-46 One of these complexes is formed by the interaction of the PtdIns3K core complex with the mammalian Atg14 homolog (ATG14) and AMBRA1.47, 48 The ATG14-containing PtdIns3K complex is thought to positively regulate autophagy by promoting translocation of the complex to the phagophore and inducing the generation of PtdIns3P.45,48,49 The other two PtdIns3K complexes contain the BECN1-interacting protein UVRAG as a common component. Whereas the PtdIns3K complex formed by UVRAG and SH3GLB1/Bif-1 promotes autophagosome formation,49,50 the PtdIns3K complex formed by UVRAG and KIAA0226/RUBICON downregulates autophagy by impairing autophagosome maturation (Figure 4).45, 46 Other BECN1-interacting proteins include the anti-apoptotic protein BCL2, which inhibits the PtdIns3K complex by sequestering BECN1 under nutrient-rich conditions.51 Besides alterations in protein interactions, BECN1 post-translational modifications also regulate PtdIns3K activity. For example, BECN1 phosphorylation by DAPK promotes dissociation of BCL2 and autophagy induction.52 ULK1-dependent phosphorylation of BECN1 activates the ATG14- and UVRAG-containing PtdIns3K complexes inducing autophagy during amino acid starvation.53 Activation of both these PtdIns3K complexes by ULK1-mediated BECN1 phosphorylation would argue for the importance of this post-translational modification for autophagosome induction and later maturation, and provides a link between the ULK kinase initiation complex and the membrane nucleation complex. Most recently, AMPK was described as regulating the activity of different PtdIns3K complexes by phosphorylating BECN1 and PIK3C3.54
Figure 4. Class III PtdIns3K complexes.
Three class III PtdIns3K complexes can be observed in mammals. All of them require PIK3C3/VPS34, PIK3R4/VPS15 and BECN1. Specific subunits regulate the function of the different complexes. Binding of ATG14 and AMBRA1 leads to autophagy induction. UVRAG and SH3GLB1 binding also activates autophagy, whereas binding to KIAA0226/RUBICON inhibits autophagosome maturation.
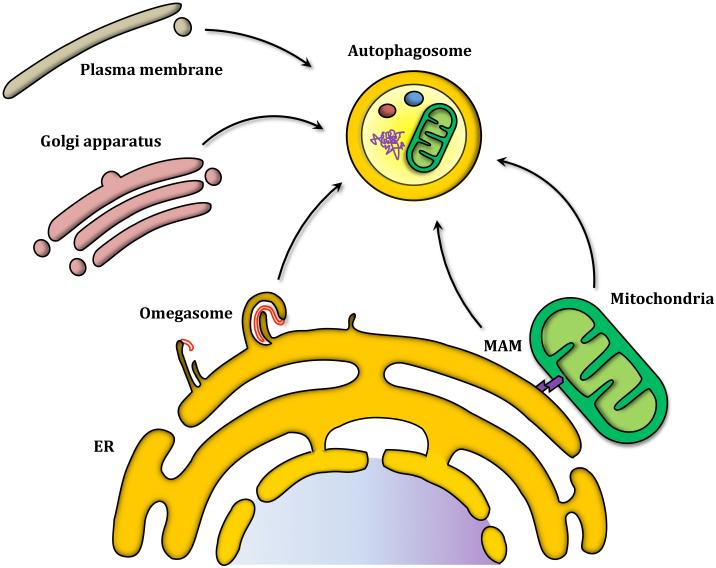
Even though membrane nucleation has been established as a key step in the autophagic process, the origin of the membrane that gives rise to the phagophore–and subsequently the autophagosome–remains an open question. Different studies have described the ER, mitochondria, plasma membrane and trans-Golgi network as possible membrane donors (Figure 5).55-57 Evidence supporting the ER as a possible membrane source include 3D tomography studies showing a connection between the phagophore and ER, as well as ATG14-containing PtdIns3K complex localization to the ER in order to initiate autophagosome formation.49, 58 Generation of PtdIns3P at the ER triggers the recruitment of the PtdIns3P binding protein ZFYVE1/DFCP1 (zinc finger, FYVE domain containing 1) and one of the mammalian homologs of Atg18, WIPI2 (WD repeat domain, phosphoinositide interacting 2). Both of these proteins have been linked to autophagosome formation from a PtdIns3P-enriched ER-associated structure termed the omegasome for its Ω-like shape.59, 60 Omegasomes have been described as platforms for autophagome formation which seem to depend on PtdIns3P, since ATG14 depletion leads to omegasome disappearance.49 Although the role of ZFYVE1 in autophagy is not well defined, WIPI2 silencing results in accumulation of omegasome structures and failure to mature into autophagosomes, suggesting WIPI2 is involved in the transition between omegasomes and autophagosomes.60 Other ATG proteins that have been associated with omegasomes include the ULK kinase complex, which localizes transiently to omegasomes in a PtdIns3P-dependent manner.59, 61 The precise mechanism by which the ER gives rise to autophagosomes via an omegasome intermediate is unknown, and several questions remain to be answered regarding the conditions, specific proteins involved and the selectivity of the process (Figure 5).62
Figure 5. Autophagosomes have a diverse range of potential membrane sources.
The trans-Golgi Network, mitochondria, mitochondrial associated membrane and ER have been postulated as membrane donors. Omegasomes have been described as the ER structures that work as a platform for autophagosome formation. The phagophore (shown in red) elongates and engulfs part of a cisternae before it buds off the ER and becomes an autophagosome.
As mentioned above, mitochondria are another organelle that has been proposed as a membrane source for the phagophore. During starvation conditions an outer mitochondria membrane fluorescent marker colocalizes with autophagosomes; mitochondrial lipids also appear to transit to autophagosomes.55 The same study showed that autophagosome formation during nutrient starvation is impaired in cells lacking the ER-mitochondria tethering protein MFN2 (mitofusin 2). Mitochondria-associated ER membrane (MAM), which are sites where the mitochondria and ER are in close proximity to each other, have been implicated in autophagosome formation. ATG14 and other autophagy markers localize to the MAM during starvation conditions. In MFN2-depleted cells, which are unable to tether the ER to the mitochondria, ATG14 localization to the MAM is impaired. Additionally, the omegasome protein marker ZFYVE1 localizes to the MAM upon starvation.63 This remarkable finding opens the possibility that the functions of omegasomes and mitochondria in autophagosome formation are essentially one and the same, unified by the association between the two.
Other studies regarding the transmembrane protein Atg9 have advanced our understanding of the membrane source from which phagophores are assembled. Atg9 has been characterized as a self-interacting protein containing 6 putative transmembrane domains, with both its carboxyl and amino termini facing the cytosol.64, 65 In yeast, Atg9 cycles from the PAS to peripheral membranes; Atg9-containing vesicles are thought to be part of the initial membranes that will generate the phagophore.66, 67 The Atg9-containing membrane reservoir appears to be composed of tubules and vesicle clusters formed through the ER-Golgi trafficking pathway;66 however, Atg9 also cycles between peri-mitochondrial sites and the PAS.68 While the precise mechanisms by which Atg9 cycling is controlled remains unknown, a number of Atg proteins are involved in the regulation of Atg9 movement. On the one hand, Atg9 anterograde transport, which is defined as movement from the peripheral sites to the PAS, depends on Atg11, Atg23 and Atg27.12, 69, 70 On the other hand, retrograde transport, that is from the PAS to the peripheral sites, is directed by the Atg1-Atg13 complex, Atg2, Atg18 and the PtdIns3K complex.12 In mammalian cells, nutrient starvation induces ATG9 redistribution from the trans-Golgi network to phagophores. Both ULK1 silencing and PtdIns3K inhibition block ATG9 trafficking to phagophores, suggesting both complexes are involved in mammalian ATG9 cycling.57, 71 The MAPK pathway is also implicated in mammalian ATG9 traffic, SUPT20H/FAM48A/p38IP [suppressor of Ty 20 homolog (S. cerevisiae)] interacts with ATG9 and induces its redistribution leading to autophagy activation. Conversely, binding between SUPT20H and MAPK14/p38α inhibits ATG9 interaction with SUPT20H, and autophagy.72
Phagophore expansion
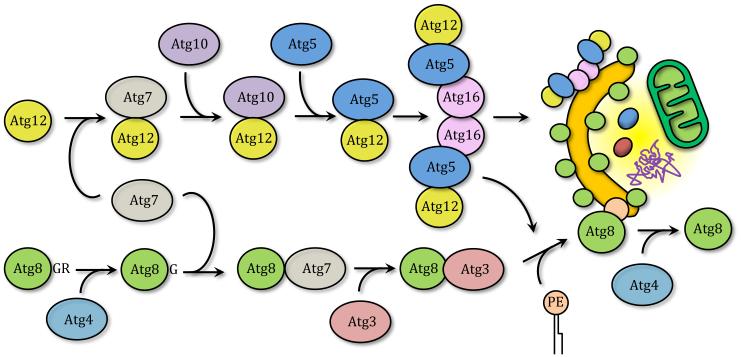
Elongation and expansion of the phagophore membrane is a key step in the autophagic process. The Atg12–Atg5-Atg16 and Atg8 conjugation systems, two interrelated ubiquitin-like (UBL) conjugation pathways, regulate this stage in both yeast and mammals. Before being covalently linked to their final substrates, both Atg12 and Atg8 go through an activation and conjugation reaction, triggered by an E1-like and an E2-like enzyme, respectively. In the Atg12–Atg5-Atg16 system, Atg12 is first activated in an ATP-dependent manner by the E1-like activating enzyme Atg7, forming a thioester bond between the two proteins.73, 74 Following this event, Atg12 is transferred to the E2-like conjugating enzyme Atg10, generating the Atg12–Atg10 intermediate through the formation of another thioester bond.75 Finally, Atg12 is covalently attached to a specific lysine residue on Atg5 in a process that, unlike ubiquitination, seems to be constitutive, irreversible and does not require an E3-like ligase enzyme.76, 77 Further interaction between Atg12–Atg5 and Atg16 leads to the formation of the Atg12–Atg5-Atg16 complex. Unlike Atg12, Atg16 is not covalently bound to Atg5 and is able to self-interact when bound to Atg12–Atg5 forming a large multimeric protein complex.78-80 The Atg12–Atg5-Atg16 complex is essential for autophagy and localizes to the phagophore.79, 81, 82 In yeast the second UBL conjugation system catalyzes the lipidation of Atg8 by covalently linking it to phosphatidylethanolamine (PE). The first event in this process corresponds to the cleavage of the carboxyl terminus of Atg8 by the cysteine protease Atg4, exposing a glycine residue.83, 84 In the next step, Atg7, again working as an E1-like enzyme, activates Atg8.85 The activated protein is then conjugated to the E2-like enzyme Atg3, before finally being linked to PE through an amide bond.85 Different studies have proposed that the E3-like enzyme that facilitates the Atg8–PE linkage is the Atg12–Atg5-Atg16 complex.86-89 While in its conjugated form, Atg8 is bound to both sides of the autophagosome membrane and thus its N terminus GFP-tagged form is widely used as an autophagosome marker.90, 91 However, Atg8 lipidation is a reversible process since Atg8–PE bound to the external autophagosome membrane can be cleaved by Atg4, releasing it from the autophagosome (Figure 6).83
Figure 6. Two ubiquitin-like conjugation systems.
Atg8 and Atg12 go through subsequent activation, mediated by Atg7 and conjugation mediated by Atg10 and Atg3, respectively, before covalently binding to PE in the case of Atg8, and Atg5 in the case of Atg12. Atg8–PE binds both the inner and outer membrane of the autophagosome, but can be deconjugated by Atg4, the same protein that removes the C-terminal arginine initially present at the Atg8 C terminus. Atg12–Atg5 bind Atg16 creating a large multimeric complex that locates to the phagophore and enhances Atg8 lipidation and membrane expansion.
Both UBL conjugation pathways are conserved and work similarly between mammals and yeast with the specific difference being that mammalian cells have several Atg8 homologs further divided into the LC3 and GABARAP subfamilies. Even though all of the homologs go through a similar conjugation process, each subfamily works at different stages of autophagy;92-94 the LC3 subfamily is involved in expansion of the phagophore and the GABARAP subfamily participates at a later stage in autophagosome maturation.95
Lysosome/vacuole fusion and recycling of the degraded cargo
The fusion of autophagosomes with lysosomes/vacuoles results in the generation of autolysosomes in higher eukaryotes and autophagic bodies in yeast. In either case, the fusion process appears to involve similar machinery that plays a role in other transport processes that terminate at these degradative organelles.96 In yeast, this machinery includes the class C Vps/HOPS complex, the SNARE family proteins Ykt6, Vti1, Vam3 and Vam7, the small GTPase Ypt7 and the proteins Mon1 and Ccz1.96–102 Fewer details are known in mammalian cells; however, the Ypt7 homolog RAB7 is required.103 One difference between yeast and mammals is that there is a clear convergence between autophagy and the endocytic pathway in the latter; autophagosomes can fuse with endosomes to form amphisomes that subsequently fuse with the lysosome.104 Once fusion occurs, the inner autophagosomal membrane and its cargo are degraded inside the lysosome/vacuole by various hydrolases. The resulting macromolecules such as amino acids that are obtained after cargo degradation are transported back into the cytoplasm for recycling. In yeast this process is regulated by protein permeases such as Atg22.105
Autophagy in the cardiovascular system
According to the World Health Organization, cardiovascular disease is the leading cause of mortality in the globalized world, accounting for 30% of all deaths.106 As expected, considerable resources have gone toward understanding the nature of cardiovascular disease and to search for possible therapeutic targets. Autophagy has been widely described in the cardiovascular system,107-112 and our understanding of the molecular machinery as described above provides the possibility for specific therapeutic intervention in treating cardiovascular disease. Autophagic activity is linked to cardiovascular development, preserving heart and vascular homeostasis, as well as in the onset and progression of several cardiovascular diseases. Interestingly, whether autophagy plays a survival role or has a deleterious effect during heart disease it is still a matter of discussion.
Autophagy and cardiovascular development
Autophagy plays a role in the regulation of mammalian cardiac development starting at a very early stage. Morpholino knockdown of atg5, atg7, and becn1 result in abnormal heart structure, including defects in cardiac looping, abnormal chamber morphology and aberrant valve development in zebrafish.113 Similarly, Atg5 knockout mice display defects in heart valve development and chamber septation indicating that autophagy regulates cardiac progenitor cell differentiation and is involved in heart development.
Autophagy in the genesis of cardiovascular diseases
Several risk factors underlie the genesis and progression of cardiovascular diseases. These factors include age, tobacco, unhealthy diet, insufficient physical activity, excessive weight/obesity, hypertension, diabetes and hyperlipidemia/hypercholesterolemia.106 Hypertension is one of the largest contributors to the worldwide burden of cardiovascular diseases, and its prevalence is close to 30% in the world population.114 The main mechanisms involved in the regulation of blood pressure are the sympathetic, parasympathetic, renin-angiotensin-aldosterone and antidiuretic hormone systems. Dysregulation of these systems, as well as obesity and diabetes, are associated with the genesis and development of hypertension.115 Because most of the peptides and hormones belonging to these systems are capable of regulating autophagy, it is possible to speculate that dysregulation of autophagy could be associated with hypertension, obesity, diabetes and end organ damage (Figure 7).
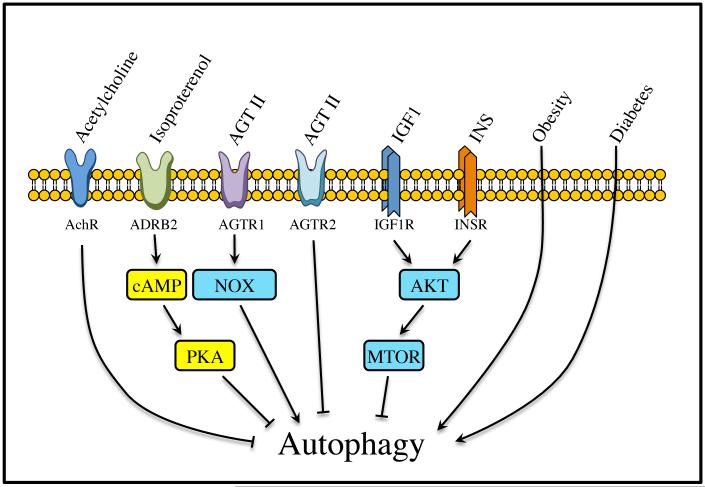
Figure 7. Autophagy regulators in cardiomyocytes.
A wide variety of stimuli can regulate autophagy in cardiomyocytes. Some of them are autophagy activators and are associated with cardiovascular diseases. However, acetylcholine, catecholamines, aging, IGF1 and INS/insulin are capable of inhibiting autophagy. INS and IGF1 are well known cardioprotective agents. AchR, acetylcholine receptor; AGT II, angiotensin II.
Sympathetic and renin-angiotensin-aldosterone systems
While there is not a lot of information regarding the parasympathetic system and autophagy, studies involving the sympathetic system have shown that autophagy protects cells against excessive β-adrenergic stimulation.116 Regarding the renin-angiotensin-aldosterone system, autophagy is stimulated by angiotensin II via the type 1 receptor (AGTR1) but diminished via AGTR2 in neonatal rat cardiomyocytes overexpressing either AGTR1, AGTR2 or both receptors after adenoviral transduction.117, 118 Cardiomyocytes derived from a genetic rat model of heart hypertrophy are more susceptible to AGTR1–induced autophagy, but also show a strong reduction of autophagy via AGTR2.117 Therefore, besides their well-known favorable hemodynamic and neurohumoral effects in the treatment of heart failure (HF), AGTR1 antagonists may also downregulate excessive autophagic cell death and consequently preserve cardiomyocytes.
Obesity
Autophagy is upregulated in adipose tissue of obese patients, correlating with the degree of obesity, visceral fat distribution, and adipocyte hypertrophy.119 Xu et al. showed that the autophagosome maturation process is involved in high-fat diet (HFD)- and AKT2 knockout-induced cardiac hypertrophy and contractile dysfunction.120 In the heart, HFD promotes the initiation and accumulation of autophagy, although it disrupts autophagosome maturation probably at the step of autophagosome-lysosome fusion. However, although AKT2 knockout does not affect the initiation of autophagy by HFD, it rescues HFD-induced disruption of the autophagosome maturation process and facilitates the transition from autophagosomes to autolysosomes, indicating a cardioprotective effect of cardiac autophagy in the presence of a HFD.120
Diabetes
Upon INS (insulin)-resistance, pancreatic β cells enhance their INS secretion to compensate for hyperglycemia. However, the progressive diminution of the number of pancreatic β cells, mainly due to apoptosis, and the decrease of their function leads to the development of type 2 diabetes mellitus (T2DM). In this model autophagy plays a protective role by limiting death of β cells. Upon exposure to a HFD, β-cell-specific Atg7-deficient mice display a decrease in the β cell number and an impairment of glucose tolerance due to a reduction in INS secretion.121-124 Genetic ablation of Atg7 in pancreatic β-cells results in degeneration of islets, impaired glucose tolerance, and reduced INS secretion.121, 125 Moreover, cardiomyocytes isolated from T2DM db/db mice and HFD-induced obese mice exhibit reduced autophagic activity.126,127 However, some recent reports conflict with these findings. Mellor et al. reported that increased myocardial autophagic flux in fructose diet-induced T2DM mice results in pathological remodeling of the heart.128 Upregulation of autophagy was also found in human T2DM pancreatic β-cells.129 These results suggest that in T2DM, increased autophagy may serve as a compensatory response to INS resistance by providing cellular components essential for maintaining normal cellular architecture and function.
Autophagy in cardiovascular diseases
In the heart, isolated cardiomyocytes, vascular epithelial cells and vascular smooth muscle cells, autophagy is strongly induced by physiological conditions, such as nutrient starvation.130-135 In those conditions, autophagy is important for the turnover of organelles and protein aggregate degradation at low basal levels under normal conditions.107,130 During conditions of cardiovascular stress, including ischemia/reperfusion (I/R) and heart failure, among others, autophagy is also activated.107,110,112,136-139 However, whether autophagy in these contexts is beneficial or detrimental is not well defined.
Ischemia/reperfusion
During ischemia, autophagy is triggered as an adaptive mechanism providing nutrients and eliminating damaged mitochondria, which could otherwise release damaging reactive oxygen species and initiate apoptosis.140 During mild ischemic stress, autophagy activation depends on AMPK-mediated inhibition of MTOR.141,142 Pharmacological inhibition of autophagy increases cardiomyocyte death, indicating that autophagy functions as a pro-survival mechanism.143 During chronic ischemia, autophagy can inhibit apoptosis and diminish tissue damage.144 During reperfusion, cardiomyocyte autophagy is upregulated dramatically in rat,145 rabbit,146 and swine144 hearts, and primary neonatal cardiomyocytes.141,143 Cardiac autophagy triggered by reperfusion can be either adaptive or detrimental and involves BECN1 activation.141 In cultured neonatal cardiomyocytes exposed to simulated I/R, inhibition of autophagy with 3-methyladenine enhances cell viability.143 In contrast, other studies describe protective actions of autophagy in simulated I/R.147,148 One possible explanation to these discrepancies is the dual role of BECN1 during I/R. While BECN1 is essential for initiation of autophagy, it can also inhibit vesicle processing late in the autophagic cascade, leading to cell death.149 Thus, BECN1 abundance can be an important determinant of autophagic activity, ensuring either survival or triggering cell death during I/R.150
Heart failure
The initial response of the heart to several stresses is hypertrophy.111 If the stresses persist, a pathological hypertrophy is developed followed by heart failure.151 In a model of pressure overload, the degree of autophagic activity correlates with the magnitude of hypertrophic growth and the rate of transition to HF.152 Cardiomyocyte-specific overexpression of BECN1 amplifies the pathological remodeling response.152,153 Conversely, BECN1 haploinsufficiency partially rescues HF.152 These data suggest that autophagy can be maladaptive under conditions of severe pressure overload. Analysis of human samples has supported additional evidence that autophagic cell death contributes to HF pathogenesis;154,155 however, complete abrogation of autophagy is similarly maladaptive and can accelerate the progression to HF. For example, inactivation of ATG5 in adult heart is sufficient to trigger rapid-onset HF,116 consistent with the notion that basal levels of cardiomyocyte autophagy are required for cellular proteostasis. Thus, whether elevated autophagy observed in human failing heart promotes cell death and contributes to the progression of heart failure, or if it represents an adaptive response of the heart to promote survival remains to be elucidated. Given these considerations, we propose a model in which titration of cardiomyocyte autophagy within an optimal, adaptive zone is a therapeutic goal of interest (Figure 8).109
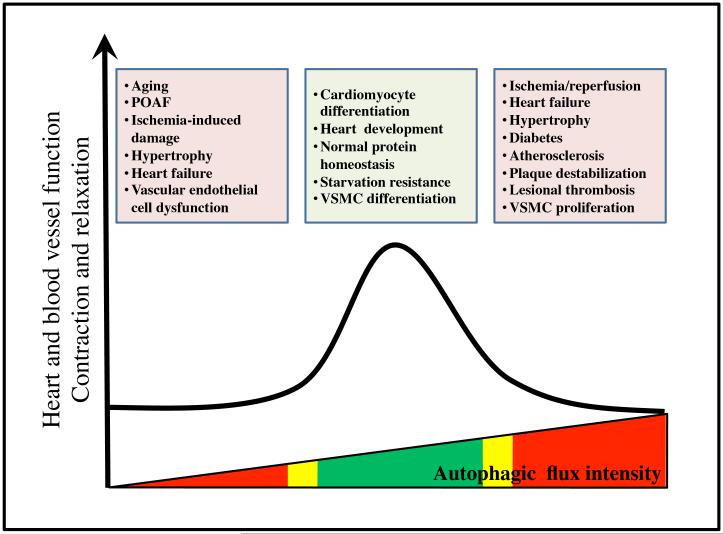
Figure 8. Autophagy in cardiovascular diseases.
The relationship between autophagy and cardiovascular diseases is complex. Although basal autophagy is critical to maintain cell homeostasis, both increases and decreases in autophagy to an excessive degree can induce alterations in normal heart and blood vessel functions. In ischemia/reperfusion, heart failure, hypertrophy, diabetes, atherosclerosis, plaque destabilization, lesional thrombosis and vascular smooth muscle cell (VSMC) proliferation, autophagic flux is abnormally elevated, contributing to cardiac and vessel dysfunction. In Atg5 and Becn1 knockout animals and during aging, autophagic activity is decreased, perturbing cellular homeostasis and contributing to cardiovascular diseases, such as post-operative atrial fibrillation (POAF), ischemia-induced damage, hypertrophy, heart failure, and vascular endothelial cell dysfunction.
Conclusions and perspectives
As one of the major catabolic pathways in the cell, autophagy has become a very important focus of research in a diverse range of biological fields. Autophagy is essential in normal cardiovascular homeostasis. However, alterations in autophagic flux are seen in all forms of cardiovascular diseases. In some instances, that response is beneficial; in other cases, it is maladaptive, promoting disease progression. Despite all knowledge accumulated so far, a substantial amount of work must be done to clarify the real contribution of autophagy in cardiovascular systems. This knowledge will not only help develop our basic understanding on how and to what end autophagy is activated, but may also give rise to potential drug treatment for several diseases. To this end, we must advance our current comprehension of the autophagic pathway and its various modulators. We are optimistic that in the near future pharmacological targeting of autophagy will emerge as a therapeutic alternative to treat cardiovascular diseases.
Acknowledgments
We would like to thank Roberto Bravo at the Advanced Center for Chronic Diseases (ACCDiS) at the University of Chile for kindly providing figure templates. This work was supported by NIH grant GM053396 (DJK), and by grants from the Comision Nacional de Investigacion Cientifica y Tecnologica (CONICYT) Chile (FONDAP 15130011 to SL and MC; FONDECYT 1140329 to MC).
Abbreviations
- AGEs
advanced glycation endproducts
- Atg
autophagy related
- ER
endoplasmic reticulum
- HF
heart failure
- HFD
high-fat diet
- I/R
ischemia/reperfusion
- MAM
mitochondria-associated ER membrane
- PAS
phagophore assembly site
- PE
phosphatidylethanolamine
- PI3K
phosphoinositide 3-kinase
- PtdIns3K
phosphatidylinositol 3-kinase
- PtdIns3P
phosphatydilinositol 3-phosphate
- T2DM
type 2 diabetes mellitus
- UBL
ubiquitin-like
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
References
- 1.Kaushik S, Cuervo AM. Chaperone-mediated autophagy: A unique way to enter the lysosome world. Trends Cell Biol. 2012;22:407–417. doi: 10.1016/j.tcb.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klionsky DJ, Baehrecke EH, Brumell JH, Chu CT, Codogno P, Cuervo AM, et al. A comprehensive glossary of autophagy-related molecules and processes. Autophagy. (2nd) 2011;7:1273–1294. doi: 10.4161/auto.7.11.17661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azad MB, Chen Y, Henson ES, Cizeau J, McMillan-Ward E, Israels SJ, Gibson SB. Hypoxia induces autophagic cell death in apoptosis-competent cells through a mechanism involving bnip3. Autophagy. 2008;4:195–204. doi: 10.4161/auto.5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Y, McMillan-Ward E, Kong J, Israels SJ, Gibson SB. Oxidative stress induces autophagic cell death independent of apoptosis in transformed and cancer cells. Cell Death Differ. 2008;15:171–182. doi: 10.1038/sj.cdd.4402233. [DOI] [PubMed] [Google Scholar]
- 5.Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting bcg and mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 6.Yorimitsu T, Nair U, Yang Z, Klionsky DJ. Endoplasmic reticulum stress triggers autophagy. J Biol Chem. 2006;281:30299–30304. doi: 10.1074/jbc.M607007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mijaljica D, Prescott M, Devenish RJ. Microautophagy in mammalian cells: Revisiting a 40-year-old conundrum. Autophagy. 2011;7:673–682. doi: 10.4161/auto.7.7.14733. [DOI] [PubMed] [Google Scholar]
- 8.Kamada Y, Yoshino K, Kondo C, Kawamata T, Oshiro N, Yonezawa K, Ohsumi Y. Tor directly controls the atg1 kinase complex to regulate autophagy. Mol Cell Biol. 2010;30:1049–1058. doi: 10.1128/MCB.01344-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kabeya Y, Noda NN, Fujioka Y, Suzuki K, Inagaki F, Ohsumi Y. Characterization of the atg17-atg29-atg31 complex specifically required for starvation-induced autophagy in saccharomyces cerevisiae. Biochem Biophys Res Commun. 2009;389:612–615. doi: 10.1016/j.bbrc.2009.09.034. [DOI] [PubMed] [Google Scholar]
- 10.Cheong H, Nair U, Geng J, Klionsky DJ. The Atg1 kinase complex is involved in the regulation of protein recruitment to initiate sequestering vesicle formation for nonspecific autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 2008;19:668–681. doi: 10.1091/mbc.E07-08-0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim J, Huang W-P, Stromhaug PE, Klionsky DJ. Convergence of multiple autophagy and cytoplasm to vacuole targeting components to a perivacuolar membrane compartment prior to de novo vesicle formation. J Biol Chem. 2002;277:763–773. doi: 10.1074/jbc.M109134200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reggiori F, Tucker KA, Stromhaug PE, Klionsky DJ. The Atg1-Atg13 complex regulates atg9 and atg23 retrieval transport from the pre-autophagosomal structure. Dev Cell. 2004;6:79–90. doi: 10.1016/s1534-5807(03)00402-7. [DOI] [PubMed] [Google Scholar]
- 13.Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–5981. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. Tor-mediated induction of autophagy via an apg1 protein kinase complex. J Cell Biol. 2000;150:1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yeh YY, Wrasman K, Herman PK. Autophosphorylation within the atg1 activation loop is required for both kinase activity and the induction of autophagy in Saccharomyces cerevisiae. Genetics. 2010;185:871–882. doi: 10.1534/genetics.110.116566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kraft C, Kijanska M, Kalie E, Siergiejuk E, Lee SS, Semplicio G, Stoffel I, Brezovich A, Verma M, Hansmann I, Ammerer G, Hofmann K, Tooze S, Peter M. Binding of the Atg1/Ulk1 kinase to the ubiquitin-like protein Atg8 regulates autophagy. EMBO J. 2012;31:3691–3703. doi: 10.1038/emboj.2012.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kijanska M, Dohnal I, Reiter W, Kaspar S, Stoffel I, Ammerer G, Kraft C, Peter M. Activation of atg1 kinase in autophagy by regulated phosphorylation. Autophagy. 2010;6:1168–1178. doi: 10.4161/auto.6.8.13849. [DOI] [PubMed] [Google Scholar]
- 18.Budovskaya YV, Stephan JS, Deminoff SJ, Herman PK. An evolutionary proteomics approach identifies substrates of the camp-dependent protein kinase. Proc Natl Acad Sci U S A. 2005;102:13933–13938. doi: 10.1073/pnas.0501046102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Budovskaya YV, Stephan JS, Reggiori F, Klionsky DJ, Herman PK. The ras/camp-dependent protein kinase signaling pathway regulates an early step of the autophagy process in Saccharomyces cerevisiae. J Biol Chem. 2004;279:20663–20671. doi: 10.1074/jbc.M400272200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stephan JS, Yeh YY, Ramachandran V, Deminoff SJ, Herman PK. The tor and pka signaling pathways independently target the Atg1/Atg13 protein kinase complex to control autophagy. Proc Natl Acad Sci U S A. 2009;106:17049–17054. doi: 10.1073/pnas.0903316106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Natarajan K, Meyer MR, Jackson BM, Slade D, Roberts C, Hinnebusch AG, Marton MJ. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol Cell Biol. 2001;21:4347–4368. doi: 10.1128/MCB.21.13.4347-4368.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tallóczy Z, Jiang W, Virgin HW, IV, Leib DA, Scheuner D, Kaufman RJ, Eskelinen EL, Levine B. Regulation of starvation- and virus-induced autophagy by the eIF2α kinase signaling pathway. Proc Natl Acad Sci U S A. 2002;99:190–195. doi: 10.1073/pnas.012485299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan J, Kuroyanagi H, Kuroiwa A, Matsuda Y, Tokumitsu H, Tomoda T, Shirasawa T, Muramatsu M. Identification of mouse Ulk1, a novel protein kinase structurally related to C. elegans unc-51. Biochem Biophys Res Commun. 1998;246:222–227. doi: 10.1006/bbrc.1998.8546. [DOI] [PubMed] [Google Scholar]
- 24.Yan J, Kuroyanagi H, Tomemori T, Okazaki N, Asato K, Matsuda Y, Suzuki Y, Ohshima Y, Mitani S, Masuho Y, Shirasawa T, Muramatsu M. Mouse Ulk2, a novel member of the unc-51-like protein kinases: Unique features of functional domains. Oncogene. 1999;18:5850–5859. doi: 10.1038/sj.onc.1202988. [DOI] [PubMed] [Google Scholar]
- 25.Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan JL, Mizushima N. FIP200, a ulk-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, Kim DH. ULK-ATG13-FIP200 complexes mediate MTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mercer CA, Kaliappan A, Dennis PB. A novel, human Atg13 binding protein, Atg101, interacts with Ulk1 and is essential for macroautophagy. Autophagy. 2009;5:649–662. doi: 10.4161/auto.5.5.8249. [DOI] [PubMed] [Google Scholar]
- 28.Ganley IG, Lam du H, Wang J, Ding X, Chen S, Jiang X. ULK1·ATG13·FIP200 complex mediates mtor signaling and is essential for autophagy. J Biol Chem. 2009;284:12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, Guan JL, Oshiro N, Mizushima N. Nutrient-dependent mtorc1 association with the ULK1-ATG13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chan EY, Longatti A, McKnight NC, Tooze SA. Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domains using an Atg13-independent mechanism. Mol Cell Biol. 2009;29:157–171. doi: 10.1128/MCB.01082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim J, Kundu M, Viollet B, Guan K-L. AMPK and MTOR regulate autophagy through direct phosphorylation of ULK1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Inoki K, Li Y, Xu T, Guan K-L. Rheb GTPase is a direct target of TSC2 gap activity and regulates mTOR signaling. Genes Dev. 2003;17:1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inoki K, Zhu T, Guan K-L. Tsc2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 34.Loffler AS, Alers S, Dieterle AM, Keppeler H, Franz-Wachtel M, Kundu M, Campbell DG, Wesselborg S, Alessi DR, Stork B. Ulk1-mediated phosphorylation of AMPK constitutes a negative regulatory feedback loop. Autophagy. 2011;7:696–706. doi: 10.4161/auto.7.7.15451. [DOI] [PubMed] [Google Scholar]
- 35.Alessi DR, James SR, Downes CP, Holmes AB, Gaffney PR, Reese CB, Cohen P. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Bα. Curr Biol. 1997;7:261–269. doi: 10.1016/s0960-9822(06)00122-9. [DOI] [PubMed] [Google Scholar]
- 36.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the Rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 37.Manning BD, Tee AR, Logsdon MN, Blenis J, Cantley LC. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/Akt pathway. Mol Cell. 2002;10:151–162. doi: 10.1016/s1097-2765(02)00568-3. [DOI] [PubMed] [Google Scholar]
- 38.Petiot A, Ogier-Denis E, Blommaart EF, Meijer AJ, Codogno P. Distinct classes of phosphatidylinositol 3'-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J Biol Chem. 2000;275:992–998. doi: 10.1074/jbc.275.2.992. [DOI] [PubMed] [Google Scholar]
- 39.Arico S, Petiot A, Bauvy C, Dubbelhuis PF, Meijer AJ, Codogno P, Ogier-Denis E. The tumor suppressor pten positively regulates macroautophagy by inhibiting the phosphatidylinositol 3-kinase/protein kinase B pathway. J Biol Chem. 2001;276:35243–35246. doi: 10.1074/jbc.C100319200. [DOI] [PubMed] [Google Scholar]
- 40.Kametaka S, Okano T, Ohsumi M, Ohsumi Y. Apg14p and apg6/vps30p form a protein complex essential for autophagy in the yeast, Saccharomyces cerevisiae. J Biol Chem. 1998;273:22284–22291. doi: 10.1074/jbc.273.35.22284. [DOI] [PubMed] [Google Scholar]
- 41.Kihara A, Noda T, Ishihara N, Ohsumi Y. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol. 2001;152:519–530. doi: 10.1083/jcb.152.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Obara K, Sekito T, Ohsumi Y. Assortment of phosphatidylinositol 3-kinase complexes--Atg14p directs association of complex i to the pre-autophagosomal structure in Saccharomyces cerevisiae. Mol Biol Cell. 2006;17:1527–1539. doi: 10.1091/mbc.E05-09-0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Furuya N, Yu J, Byfield M, Pattingre S, Levine B. The evolutionarily conserved domain of Beclin 1 is required for Vps34 binding, autophagy and tumor suppressor function. Autophagy. 2005;1:46–52. doi: 10.4161/auto.1.1.1542. [DOI] [PubMed] [Google Scholar]
- 44.Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, Maejima I, Shirahama-Noda K, Ichimura T, Isobe T, Akira S, Noda T, Yoshimori T. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11:385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- 46.Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, Chait BT, Heintz N, Yue Z. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11:468–476. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, Corazzari M, Fuoco C, Ucar A, Schwartz P, Gruss P, Piacentini M, Chowdhury K, Cecconi F. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447:1121–1125. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- 48.Sun Q, Fan W, Chen K, Ding X, Chen S, Zhong Q. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class iii phosphatidylinositol 3-kinase. Proc Natl Acad Sci U S A. 2008;105:19211–19216. doi: 10.1073/pnas.0810452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matsunaga K, Morita E, Saitoh T, Akira S, Ktistakis NT, Izumi T, Noda T, Yoshimori T. Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14L. J Cell Biol. 2010;190:511–521. doi: 10.1083/jcb.200911141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y, Liang C, Jung JU, Cheng JQ, Mule JJ, Pledger WJ, Wang HG. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9:1142–1151. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 52.Zalckvar E, Berissi H, Mizrachy L, Idelchuk Y, Koren I, Eisenstein M, Sabanay H, Pinkas-Kramarski R, Kimchi A. Dap-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Rep. 2009;10:285–292. doi: 10.1038/embor.2008.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, Kim H, Neufeld TP, Dillin A, Guan K-L. ULK1 induces autophagy by phosphorylating Beclin-1 and activating Vps34 lipid kinase. Nat Cell Biol. 2013;15:741–750. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim J, Kim YC, Fang C, Russell RC, Kim JH, Fan W, Liu R, Zhong Q, Guan K-L. Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell. 2013;152:290–303. doi: 10.1016/j.cell.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, Lippincott-Schwartz J. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol. 2009;11:1433–1437. doi: 10.1038/ncb1991. [DOI] [PubMed] [Google Scholar]
- 57.Young ARJ, Chan EYW, Hu XW, KöUhl R, Crawshaw SG, High S, Hailey DW, Lippincott-Schwartz J, Tooze SA. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci. 2006;119:3888–3900. doi: 10.1242/jcs.03172. [DOI] [PubMed] [Google Scholar]
- 58.Yla-Anttila P, Vihinen H, Jokitalo E, Eskelinen E-L. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy. 2009;5:1180–1185. doi: 10.4161/auto.5.8.10274. [DOI] [PubMed] [Google Scholar]
- 59.Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Polson HE, de Lartigue J, Rigden DJ, Reedijk M, Urbé S, Clague MJ, Tooze SA. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy. 2010;6:506–522. doi: 10.4161/auto.6.4.11863. [DOI] [PubMed] [Google Scholar]
- 61.Karanasios E, Stapleton E, Manifava M, Kaizuka T, Mizushima N, Walker SA, Ktistakis NT. Dynamic association of the ULK1 complex with omegasomes during autophagy induction. J Cell Sci. 2013;126:5224–5238. doi: 10.1242/jcs.132415. [DOI] [PubMed] [Google Scholar]
- 62.Tooze SA, Yoshimori T. The origin of the autophagosomal membrane. Nat Cell Biol. 2010;12:831–835. doi: 10.1038/ncb0910-831. [DOI] [PubMed] [Google Scholar]
- 63.Hamasaki M, Furuta N, Matsuda A, Nezu A, Yamamoto A, Fujita N, Oomori H, Noda T, Haraguchi T, Hiraoka Y, Amano A, Yoshimori T. Autophagosomes form at er-mitochondria contact sites. Nature. 2013;495:389–393. doi: 10.1038/nature11910. [DOI] [PubMed] [Google Scholar]
- 64.He C, Baba M, Cao Y, Klionsky DJ. Self-interaction is critical for Atg9 transport and function at the phagophore assembly site during autophagy. Mol Biol Cell. 2008;19:5506–5516. doi: 10.1091/mbc.E08-05-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Noda T, Kim J, Huang W-P, Baba M, Tokunaga C, Ohsumi Y, Klionsky DJ. Apg9p/cvt7p is an integral membrane protein required for transport vesicle formation in the cvt and autophagy pathways. J Cell Biol. 2000;148:465–480. doi: 10.1083/jcb.148.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mari M, Griffith J, Rieter E, Krishnappa L, Klionsky DJ, Reggiori F. An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J Cell Biol. 2010;190:1005–1022. doi: 10.1083/jcb.200912089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yamamoto H, Kakuta S, Watanabe TM, Kitamura A, Sekito T, Kondo-Kakuta C, Ichikawa R, Kinjo M, Ohsumi Y. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J Cell Biol. 2012;198:219–233. doi: 10.1083/jcb.201202061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Reggiori F, Shintani T, Nair U, Klionsky DJ. Atg9 cycles between mitochondria and the pre-autophagosomal structure in yeasts. Autophagy. 2005;1:101–109. doi: 10.4161/auto.1.2.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.He C, Song H, Yorimitsu T, Monastyrska I, Yen W-L, Legakis JE, Klionsky DJ. Recruitment of Atg9 to the preautophagosomal structure by atg11 is essential for selective autophagy in budding yeast. J Cell Biol. 2006;175:925–935. doi: 10.1083/jcb.200606084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yen W-L, Legakis JE, Nair U, Klionsky DJ. Atg27 is required for autophagy-dependent cycling of atg9. Mol Biol Cell. 2007;18:581–593. doi: 10.1091/mbc.E06-07-0612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Orsi A, Razi M, Dooley HC, Robinson D, Weston AE, Collinson LM, Tooze SA. Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, are required for autophagy. Mol Biol Cell. 2012;23:1860–1873. doi: 10.1091/mbc.E11-09-0746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Webber JL, Tooze SA. Coordinated regulation of autophagy by p38α MAPK through mAtg9 and p38IP. EMBO J. 2010;29:27–40. doi: 10.1038/emboj.2009.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim J, Dalton VM, Eggerton KP, Scott SV, Klionsky DJ. Apg7p/cvt2p is required for the cytoplasm-to-vacuole targeting, macroautophagy, and peroxisome degradation pathways. Mol Biol Cell. 1999;10:1337–1351. doi: 10.1091/mbc.10.5.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tanida I, Mizushima N, Kiyooka M, Ohsumi M, Ueno T, Ohsumi Y, Kominami E. Apg7p/Cvt2p: A novel protein-activating enzyme essential for autophagy. Mol Biol Cell. 1999;10:1367–1379. doi: 10.1091/mbc.10.5.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shintani T, Mizushima N, Ogawa Y, Matsuura A, Noda T, Ohsumi Y. Apg10p, a novel protein-conjugating enzyme essential for autophagy in yeast. EMBO J. 1999;18:5234–5241. doi: 10.1093/emboj/18.19.5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y. A protein conjugation system essential for autophagy. Nature. 1998;395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- 77.Mizushima N, Sugita H, Yoshimori T, Ohsumi Y. A new protein conjugation system in human. The counterpart of the yeast Apg12p conjugation system essential for autophagy. J Biol Chem. 1998;273:33889–33892. doi: 10.1074/jbc.273.51.33889. [DOI] [PubMed] [Google Scholar]
- 78.Kuma A, Mizushima N, Ishihara N, Ohsumi Y. Formation of the approximately 350-kDa Apg12-Apg5·Apg16 multimeric complex, mediated by Apg16 oligomerization, is essential for autophagy in yeast. J Biol Chem. 2002;277:18619–18625. doi: 10.1074/jbc.M111889200. [DOI] [PubMed] [Google Scholar]
- 79.Mizushima N, Kuma A, Kobayashi Y, Yamamoto A, Matsubae M, Takao T, Natsume T, Ohsumi Y, Yoshimori T. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J Cell Sci. 2003;116:1679–1688. doi: 10.1242/jcs.00381. [DOI] [PubMed] [Google Scholar]
- 80.Mizushima N, Noda T, Ohsumi Y. Apg16p is required for the function of the Apg12p-Apg5p conjugate in the yeast autophagy pathway. EMBO J. 1999;18:3888–3896. doi: 10.1093/emboj/18.14.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Geng J, Klionsky DJ. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. EMBO Rep. 2008;9:859–864. doi: 10.1038/embor.2008.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ohsumi Y. Molecular dissection of autophagy: Two ubiquitin-like systems. Nat Rev Mol Cell Biol. 2001;2:211–216. doi: 10.1038/35056522. [DOI] [PubMed] [Google Scholar]
- 83.Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, Yoshimori T, Ohsumi M, Takao T, Noda T, Ohsumi Y. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol. 2000;151:263–276. doi: 10.1083/jcb.151.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tanida I, Ueno T, Kominami E. Human light chain 3/MAP1LC3B is cleaved at its carboxyl-terminal Met121 to expose Gly120 for lipidation and targeting to autophagosomal membranes. J Biol Chem. 2004;279:47704–47710. doi: 10.1074/jbc.M407016200. [DOI] [PubMed] [Google Scholar]
- 85.Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, Noda T, Ohsumi Y. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 86.Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell. 2008;19:2092–2100. doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T, Inagaki F, Ohsumi Y. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem. 2007;282:37298–37302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- 88.Romanov J, Walczak M, Ibiricu I, Schuchner S, Ogris E, Kraft C, Martens S. Mechanism and functions of membrane binding by the Atg5-Atg12/Atg16 complex during autophagosome formation. EMBO J. 2012;31:4304–4317. doi: 10.1038/emboj.2012.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Noda NN, Fujioka Y, Hanada T, Ohsumi Y, Inagaki F. Structure of the Atg12-Atg5 conjugate reveals a platform for stimulating Atg8-PE conjugation. EMBO Rep. 2013;14:206–211. doi: 10.1038/embor.2012.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci. 2004;117:2805–2812. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- 93.Tanida I, Komatsu M, Ueno T, Kominami E. GATE-16 and GABARAP are authentic modifiers mediated by Apg7 and Apg3. Biochem Biophys Res Commun. 2003;300:637–644. doi: 10.1016/s0006-291x(02)02907-8. [DOI] [PubMed] [Google Scholar]
- 94.Tanida I, Tanida-Miyake E, Komatsu M, Ueno T, Kominami E. Human Apg3p/Aut1p homologue is an authentic E2 enzyme for multiple substrates, GATE-16, GABARAP, and MAP-LC3, and facilitates the conjugation of hApg12p to hApg5p. J Biol Chem. 2002;277:13739–13744. doi: 10.1074/jbc.M200385200. [DOI] [PubMed] [Google Scholar]
- 95.Weidberg H, Shvets E, Shpilka T, Shimron F, Shinder V, Elazar Z. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 2010;29:1792–1802. doi: 10.1038/emboj.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Klionsky DJ. The molecular machinery of autophagy: unanswered questions. J. Cell Sci. 2005;118:7–18. doi: 10.1242/jcs.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Harding TM, Morano KA, Scott SV, Klionsky DJ. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J. Cell Biol. 1995;13:591–602. doi: 10.1083/jcb.131.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Darsow T, Rieder SE, Emr SD. A multispecificity syntaxin homologue, Vam3p, essential for autophagic and biosynthetic protein transport to the vacuole. J. Cell Biol. 1997;138:517–529. doi: 10.1083/jcb.138.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sato TK, Darsow T, Emr SD. Vam7p, a SNAP-25-like molecule, and Vam3p, a syntaxin homolog, function together in yeast vacuolar protein trafficking. Mol. Cell. Biol. 1998;18:5308–5319. doi: 10.1128/mcb.18.9.5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mollard GF, Von, Stevens TH. The Saccharomyces cerevisiae v-SNARE Vti1p is required for multiple membrane transport pathways to the vacuole. Mol. Biol. Cell. 1999;10:1719–1732. doi: 10.1091/mbc.10.6.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang C-W, Stromhaug PE, Kauffman EJ, Weisman LS, Klionsky DJ. Yeast homotypic vacuole fusion requires the Ccz1-Mon1 complex during the tethering/docking stage. J. Cell Biol. 2003;163:973–85. doi: 10.1083/jcb.200308071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang C-W, Stromhaug PE, Shima J, Klionsky DJ. The Ccz1-Mon1 protein complex is required for the late step of multiple vacuole delivery pathways. J. Biol. Chem. 2002;277:47917–27. doi: 10.1074/jbc.M208191200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jäger S, Bucci C, Tanida I, Ueno T, Kominami E, Saftig P, Eskelinen E-L. Role for Rab7 in maturation of late autophagic vacuoles. J. Cell Sci. 2004;117:4837–48. doi: 10.1242/jcs.01370. [DOI] [PubMed] [Google Scholar]
- 104.Sanchez-Wandelmer J, Reggiori F. Amphisomes: out of the autophagosome shadow. EMBO J. 2013;2013;32:3116–8. doi: 10.1038/emboj.2013.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yang Z, Huang J, Geng J, Nair U, Klionsky DJ. Atg22 recycles amino acids to link the degradative and recycling functions of autophagy. Mol. Biol. Cell. 2006;17:5094–5104. doi: 10.1091/mbc.E06-06-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.World Health Organization . Global health risks: Mortality and burden of disease attributable to selected major risks. WHO Press; 2009. [Google Scholar]
- 107.Nemchenko A, Chiong M, Turer A, Lavandero S, Hill JA. Autophagy as a therapeutic target in cardiovascular disease. J Mol Cell Cardiol. 2011;51:584–593. doi: 10.1016/j.yjmcc.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang ZV, Rothermel BA, Hill JA. Autophagy in hypertensive heart disease. J Biol Chem. 2010;285:8509–8514. doi: 10.1074/jbc.R109.025023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rothermel BA, Hill JA. Autophagy in load-induced heart disease. Circ Res. 2008;103:1363–1369. doi: 10.1161/CIRCRESAHA.108.186551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rifki OF, Hill JA. Cardiac autophagy: Good with the bad. J Cardiovasc Pharmacol. 2012;60:248–252. doi: 10.1097/FJC.0b013e3182646cb1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hill JA, Olson EN. Cardiac plasticity. N Engl J Med. 2008;358:1370–1380. doi: 10.1056/NEJMra072139. [DOI] [PubMed] [Google Scholar]
- 112.Chiong M, Wang ZV, Pedrozo Z, Cao DJ, Troncoso R, Ibacache M, Criollo A, Nemchenko A, Hill JA, Lavandero S. Cardiomyocyte death: Mechanisms and translational implications. Cell Death Dis. 2011;2:e244. doi: 10.1038/cddis.2011.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lee E, Koo Y, Ng A, Wei Y, Luby-Phelps K, Juraszek A, Xavier RJ, Cleaver O, Levine B, Amatruda JF. Autophagy is essential for cardiac morphogenesis during vertebrate development. Autophagy. 2014;10:572–587. doi: 10.4161/auto.27649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chobanian AV, Bakris GL, Black HR, Cushman WC, Green LA, Izzo JL, Jr., Jones DW, Materson BJ, Oparil S, Wright JT, Jr., Roccella EJ, National HIgh Blood Pressure Education Program Coordinating Committee Seventh report of the joint national committee on prevention detection, evaluation, and treatment of high blood pressure. Hypertension. 2003;42:1206–1252. doi: 10.1161/01.HYP.0000107251.49515.c2. [DOI] [PubMed] [Google Scholar]
- 115.Ocaranza MP, Michea L, Chiong M, Lagos CF, Lavandero S, Jalil JE. Recent insights and therapeutic perspectives of angiotensin-(1-9) in the cardiovascular system. Clin Sci (Lond) 2014;127:549–557. doi: 10.1042/CS20130449. [DOI] [PubMed] [Google Scholar]
- 116.Nakai A, Yamaguchi O, Takeda T, Higuchi Y, Hikoso S, Taniike M, Omiya S, Mizote I, Matsumura Y, Asahi M, Nishida K, Hori M, Mizushima N, Otsu K. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med. 2007;13:619–624. doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- 117.Porrello ER, D'Amore A, Curl CL, Allen AM, Harrap SB, Thomas WG, Delbridge LM. Angiotensin II type 2 receptor antagonizes angiotensin ii type 1 receptor-mediated cardiomyocyte autophagy. Hypertension. 2009;53:1032–1040. doi: 10.1161/HYPERTENSIONAHA.108.128488. [DOI] [PubMed] [Google Scholar]
- 118.Steckelings UM, Unger T. Angiotensin receptors and autophagy: Live and let die. Hypertension. 2009;53:898–899. doi: 10.1161/HYPERTENSIONAHA.109.131425. [DOI] [PubMed] [Google Scholar]
- 119.Kovsan J, Bluher M, Tarnovscki T, Kloting N, Kirshtein B, Madar L, Shai I, Golan R, Harman-Boehm I, Schon MR, Greenberg AS, Elazar Z, Bashan N, Rudich A. Altered autophagy in human adipose tissues in obesity. J Clin Endocrinol Metab. 2011;96:E268–277. doi: 10.1210/jc.2010-1681. [DOI] [PubMed] [Google Scholar]
- 120.Xu X, Hua Y, Nair S, Zhang Y, Ren J. Akt2 knockout preserves cardiac function in high-fat diet-induced obesity by rescuing cardiac autophagosome maturation. J Mol Cell Biol. 2013;5:61–63. doi: 10.1093/jmcb/mjs055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ebato C, Uchida T, Arakawa M, Komatsu M, Ueno T, Komiya K, Azuma K, Hirose T, Tanaka K, Kominami E, Kawamori R, Fujitani Y, Watada H. Autophagy is important in islet homeostasis and compensatory increase of beta cell mass in response to high-fat diet. Cell Metab. 2008;8:325–332. doi: 10.1016/j.cmet.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 122.Jung HS, Chung KW, Won Kim J, Kim J, Komatsu M, Tanaka K, Nguyen YH, Kang TM, Yoon KH, Kim JW, Jeong YT, Han MS, Lee MK, Kim KW, Shin J, Lee MS. Loss of autophagy diminishes pancreatic beta cell mass and function with resultant hyperglycemia. Cell Metab. 2008;8:318–324. doi: 10.1016/j.cmet.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 123.Lim YM, Lim H, Hur KY, Quan W, Lee HY, Cheon H, Ryu D, Koo SH, Kim HL, Kim J, Komatsu M, Lee MS. Systemic autophagy insufficiency compromises adaptation to metabolic stress and facilitates progression from obesity to diabetes. Nat Commun. 2014;5:4934. doi: 10.1038/ncomms5934. [DOI] [PubMed] [Google Scholar]
- 124.Quan W, Hur KY, Lim Y, Oh SH, Lee JC, Kim KH, Kim GH, Kim SW, Kim HL, Lee MK, Kim KW, Kim J, Komatsu M, Lee MS. Autophagy deficiency in beta cells leads to compromised unfolded protein response and progression from obesity to diabetes in mice. Diabetologia. 2012;55:392–403. doi: 10.1007/s00125-011-2350-y. [DOI] [PubMed] [Google Scholar]
- 125.Fujitani Y, Ebato C, Uchida T, Kawamori R, Watada H. Beta-cell autophagy: A novel mechanism regulating beta-cell function and mass: Lessons from beta-cell-specific atg7-deficient mice. Islets. 2009;1:151–153. doi: 10.4161/isl.1.2.9057. [DOI] [PubMed] [Google Scholar]
- 126.Marsh BJ, Soden C, Alarcon C, Wicksteed BL, Yaekura K, Costin AJ, Morgan GP, Rhodes CJ. Regulated autophagy controls hormone content in secretory-deficient pancreatic endocrine beta-cells. Mol Endocrinol. 2007;21:2255–2269. doi: 10.1210/me.2007-0077. [DOI] [PubMed] [Google Scholar]
- 127.Sciarretta S, Zhai P, Shao D, Maejima Y, Robbins J, Volpe M, Condorelli G, Sadoshima J. Rheb is a critical regulator of autophagy during myocardial ischemia: Pathophysiological implications in obesity and metabolic syndrome. Circulation. 2012;125:1134–1146. doi: 10.1161/CIRCULATIONAHA.111.078212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mellor KM, Bell JR, Young MJ, Ritchie RH, Delbridge LM. Myocardial autophagy activation and suppressed survival signaling is associated with insulin resistance in fructose-fed mice. J Mol Cell Cardiol. 2011;50:1035–1043. doi: 10.1016/j.yjmcc.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 129.Masini M, Bugliani M, Lupi R, del Guerra S, Boggi U, Filipponi F, Marselli L, Masiello P, Marchetti P. Autophagy in human type 2 diabetes pancreatic beta cells. Diabetologia. 2009;52:1083–1086. doi: 10.1007/s00125-009-1347-2. [DOI] [PubMed] [Google Scholar]
- 130.Marambio P, Toro B, Sanhueza C, Troncoso R, Parra V, Verdejo H, Garcia L, Quiroga C, Munafo D, Diaz-Elizondo J, Bravo R, Gonzalez MJ, Diaz-Araya G, Pedrozo Z, Chiong M, Colombo MI, Lavandero S. Glucose deprivation causes oxidative stress and stimulates aggresome formation and autophagy in cultured cardiac myocytes. Biochim Biophys Acta. 2010;1802:509–518. doi: 10.1016/j.bbadis.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 131.Maruyama R, Goto K, Takemura G, Ono K, Nagao K, Horie T, Tsujimoto A, Kanamori H, Miyata S, Ushikoshi H, Nagashima K, Minatoguchi S, Fujiwara T, Fujiwara H. Morphological and biochemical characterization of basal and starvation-induced autophagy in isolated adult rat cardiomyocytes. Am J Physiol Heart Circ Physiol. 2008;295:H1599–1607. doi: 10.1152/ajpheart.91449.2007. [DOI] [PubMed] [Google Scholar]
- 132.Kanamori H, Takemura G, Maruyama R, Goto K, Tsujimoto A, Ogino A, Li L, Kawamura I, Takeyama T, Kawaguchi T, Nagashima K, Fujiwara T, Fujiwara H, Seishima M, Minatoguchi S. Functional significance and morphological characterization of starvation-induced autophagy in the adult heart. Am J Pathol. 2009;174:1705–1714. doi: 10.2353/ajpath.2009.080875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Salabei JK, Hill BG. Implications of autophagy for vascular smooth muscle cell function and plasticity. Free Radic Biol Med. 2013;65:693–703. doi: 10.1016/j.freeradbiomed.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.De Meyer GR, Martinet W. Autophagy in the cardiovascular system. Biochim Biophys Acta. 2009;1793:1485–1495. doi: 10.1016/j.bbamcr.2008.12.011. [DOI] [PubMed] [Google Scholar]
- 136.Cao DJ, Hill JA. Titrating autophagy in cardiac plasticity. Autophagy. 2011;7:1078–1079. doi: 10.4161/auto.7.9.16176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lavandero S, Troncoso R, Rothermel BA, Martinet W, Sadoshima J, Hill JA. Cardiovascular autophagy: Concepts, controversies, and perspectives. Autophagy. 2013;9:1455–1466. doi: 10.4161/auto.25969. [DOI] [PubMed] [Google Scholar]
- 138.Wang ZV, Ferdous A, Hill JA. Cardiomyocyte autophagy: Metabolic profit and loss. Heart Fail Rev. 2013;18:585–594. doi: 10.1007/s10741-012-9350-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ryter SW, Lee SJ, Smith A, Choi AM. Autophagy in vascular disease. Proc Am Thorac Soc. 2010;7:40–47. doi: 10.1513/pats.200909-100JS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhang H, Bosch-Marce M, Shimoda LA, Tan YS, Baek JH, Wesley JB, Gonzalez FJ, Semenza GL. Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J Biol Chem. 2008;283:10892–10903. doi: 10.1074/jbc.M800102200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 141.Matsui Y, Takagi H, Qu X, Abdellatif M, Sakoda H, Asano T, Levine B, Sadoshima J. Distinct roles of autophagy in the heart during ischemia and reperfusion: Roles of amp-activated protein kinase and beclin 1 in mediating autophagy. Circ Res. 2007;100:914–922. doi: 10.1161/01.RES.0000261924.76669.36. [DOI] [PubMed] [Google Scholar]
- 142.Troncoso R, Vicencio JM, Parra V, Nemchenko A, Kawashima Y, Del Campo A, Toro B, Battiprolu PK, Aranguiz P, Chiong M, Yakar S, Gillette TG, Hill JA, Abel ED, Leroith D, Lavandero S. Energy-preserving effects of igf-1 antagonize starvation-induced cardiac autophagy. Cardiovasc Res. 2012;93:320–329. doi: 10.1093/cvr/cvr321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Valentim L, Laurence KM, Townsend PA, Carroll CJ, Soond S, Scarabelli TM, Knight RA, Latchman DS, Stephanou A. Urocortin inhibits beclin1-mediated autophagic cell death in cardiac myocytes exposed to ischaemia/reperfusion injury. J Mol Cell Cardiol. 2006;40:846–852. doi: 10.1016/j.yjmcc.2006.03.428. [DOI] [PubMed] [Google Scholar]
- 144.Yan L, Vatner DE, Kim SJ, Ge H, Masurekar M, Massover WH, Yang G, Matsui Y, Sadoshima J, Vatner SF. Autophagy in chronically ischemic myocardium. Proc Natl Acad Sci U S A. 2005;102:13807–13812. doi: 10.1073/pnas.0506843102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Huang C, Liu W, Perry CN, Yitzhaki S, Lee Y, Yuan H, Tsukada YT, Hamacher-Brady A, Mentzer RM, Jr., Gottlieb RA. Autophagy and protein kinase c are required for cardioprotection by sulfaphenazole. Am J Physiol Heart Circ Physiol. 2010;298:H570–579. doi: 10.1152/ajpheart.00716.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Decker RS, Wildenthal K. Lysosomal alterations in hypoxic and reoxygenated hearts. I. Ultrastructural and cytochemical changes. Am J Pathol. 1980;98:425–444. [PMC free article] [PubMed] [Google Scholar]
- 147.Gurusamy N, Lekli I, Gorbunov NV, Gherghiceanu M, Popescu LM, Das DK. Cardioprotection by adaptation to ischaemia augments autophagy in association with bag-1 protein. J Cell Mol Med. 2009;13:373–387. doi: 10.1111/j.1582-4934.2008.00495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Huang C, Yitzhaki S, Perry CN, Liu W, Giricz Z, Mentzer RM, Jr., Gottlieb RA. Autophagy induced by ischemic preconditioning is essential for cardioprotection. J Cardiovasc Transl Res. 2010;3:365–373. doi: 10.1007/s12265-010-9189-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ma X, Liu H, Foyil SR, Godar RJ, Weinheimer CJ, Hill JA, Diwan A. Impaired autophagosome clearance contributes to cardiomyocyte death in ischemia/reperfusion injury. Circulation. 2012;125:3170–3181. doi: 10.1161/CIRCULATIONAHA.111.041814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ma X, Liu H, Foyil SR, Godar RJ, Weinheimer CJ, Diwan A. Autophagy is impaired in cardiac ischemia-reperfusion injury. Autophagy. 2012;8:1394–1396. doi: 10.4161/auto.21036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hein S, Arnon E, Kostin S, Schonburg M, Elsasser A, Polyakova V, Bauer EP, Klovekorn WP, Schaper J. Progression from compensated hypertrophy to failure in the pressure-overloaded human heart: Structural deterioration and compensatory mechanisms. Circulation. 2003;107:984–991. doi: 10.1161/01.cir.0000051865.66123.b7. [DOI] [PubMed] [Google Scholar]
- 152.Zhu H, Tannous P, Johnstone JL, Kong Y, Shelton JM, Richardson JA, Le V, Levine B, Rothermel BA, Hill JA. Cardiac autophagy is a maladaptive response to hemodynamic stress. J Clin Invest. 2007;117:1782–1793. doi: 10.1172/JCI27523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Cao DJ, Wang ZV, Battiprolu PK, Jiang N, Morales CR, Kong Y, Rothermel BA, Gillette TG, Hill JA. Histone deacetylase (hdac) inhibitors attenuate cardiac hypertrophy by suppressing autophagy. Proc Natl Acad Sci U S A. 2011;108:4123–4128. doi: 10.1073/pnas.1015081108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kostin S, Pool L, Elsasser A, Hein S, Drexler HC, Arnon E, Hayakawa Y, Zimmermann R, Bauer E, Klovekorn WP, Schaper J. Myocytes die by multiple mechanisms in failing human hearts. Circ Res. 2003;92:715–724. doi: 10.1161/01.RES.0000067471.95890.5C. [DOI] [PubMed] [Google Scholar]
- 155.Saijo M, Takemura G, Koda M, Okada H, Miyata S, Ohno Y, Kawasaki M, Tsuchiya K, Nishigaki K, Minatoguchi S, Goto K, Fujiwara H. Cardiomyopathy with prominent autophagic degeneration, accompanied by an elevated plasma brain natriuretic peptide level despite the lack of overt heart failure. Intern Med. 2004;43:700–703. doi: 10.2169/internalmedicine.43.700. [DOI] [PubMed] [Google Scholar]