Abstract
Ehrlichia sp. DNA was amplified from 4 Ehrlichia-seroreactive horses from Mérida, Nicaragua. Sequencing of 16S rDNA, sodB, and groEL genes indicated that the bacterium is most likely a novel Ehrlichia species. The tick vector and the potential for canine and human infection remain unknown.
Keywords: bacteria, tickborne pathogen, Anaplasmataceae, Ehrlichia, equine, horse, PCR, DNA sequencing, Central America, Nicaragua
Worldwide, ehrlichioses are considered emerging infectious diseases of animals and humans. Transmitted by ticks, ehrlichae are obligate intracellular, gram-negative bacteria that infect animals and humans (1). Recognized species include E. canis, E. chaffeensis, E. ewingii, E. muris, and E. ruminantium (1). Other species identified in North America are Panola Mountain (2) and the E. muris–like agent (3). Two recently identified new species are Ehrlichia sp. AvBat, isolated from Argas vespertilionis ticks in France (4), and E. mineirensis, isolated from hemolymph of Rhipicephalus microplus ticks in Brazil (5).
Although studies from Brazil and Oklahoma (USA) have documented reactivity to Ehrlichia spp. (6,7) in horse serum, no reports have documented isolation or PCR detection of Ehrlichia spp. infection in horses worldwide. In North America, cervids are reservoir hosts for E. chaffeensis, which after tick transmission causes monocytic ehrlichiosis in humans (1). Although equids are not known hosts for E. chaffeensis, bacterial DNA has been amplified from ticks (Dermacentor nitens and Amblyomma cajennense) collected from horses in Panama (8). Anaplasma phagocytophilum and Borrelia burgdorferi, causes of granulocytic anaplasmosis and borreliosis (Lyme disease), respectively, are transmitted by Ixodes scapularis and I. pacificus ticks in North America and infect cats, dogs, horses, and humans.
In Mérida, Nicaragua, the potential for infection of horses by tickborne pathogens is a concern because of the horses’ often poor body condition and heavy tick infestations. In 2013, to determine exposure of equids to >1 tickborne organism, visiting veterinary students collected blood samples from horses.
The Study
With approval from the Oregon State University Animal Care and Use Committee (Animal Care and Use Proposal no. 4329), blood samples were collected from 92 horses being evaluated for medical conditions (e.g., anorexia, weight loss, lameness, administration of endoparasiticides and ectoparasiticides) or before elective surgery (e.g., castration, wound repair) at the clinic in Mérida from August 28 through September 4, 2013. After jugular venipuncture, 6 mL of blood was collected into EDTA tubes.
Each whole blood sample was tested for antibodies against Anaplasma spp. (A. phagocytophilum and A. platys), B. burgdorferi sensu stricto, and Ehrlichia spp. (E.canis, E. chaffeensis, and E. ewingii) by using the ELISA-based assay SNAP 4DxPlus (IDEXX Laboratories, Inc., Westbrook, ME, USA) according to the manufacturer’s instructions (9). The assay does not use a host species–specific conjugate and can therefore be used in research to screen mammals other than dogs. According to assay results, 51 (55%) horse serum samples were Ehrlichia spp. seroreactive. One sample was B. burgdorferi seroreactive, whereas none were Anaplasma spp. seroreactive.
The 51 Ehrlichia spp.–reactive serum samples were subsequently stored at 28°C for up to 1 week during transport to the United States. For PCR testing, samples were shipped (US Department of Agriculture import permit no. 13846) to the Intracellular Pathogens Research Laboratory, Center for Comparative Medicine and Translational Research, North Carolina State University, College of Veterinary Medicine. DNA extraction from 200 μL of EDTA-anticoagulated whole blood was performed by using a QIAsymphony DNA Mini Kit (QIAGEN, Valencia, CA, USA; catalog no. 931236). Previously described PCRs were used to amplify a 420-bp fragment of the 16S rRNA gene, a 620-bp fragment of the GroEL gene, and a 300-bp fragment of the Ehrlichia spp. sodB gene (2,10,11). A larger, 600-bp, fragment of the sodB gene was amplified from 1 sample that was positive by PCR by using the following unpublished primers: sodbEhrl600-F 5′-ATGTTTACTTTACCTGAACTTCCATATC-3′ and sodbEhrl600-R 5′-ATCTTTGAGCTGCAAAATCCCAATT-3′. Positive (Anaplasma or Ehrlichia spp. plasmid DNA) and negative (RNase-free molecular-grade water and a DNA extraction control consisting of uninfected canine genomic DNA) controls were used for each assay. Amplified DNA was sequenced directly by GENEWIZ, Inc. (Research Triangle Park, NC, USA), and alignments were compared with those of GenBank sequences by using AlignX software (Vector NTI Advance version 11.5; Invitrogen, Carlsbad, CA, USA).
Of the 51 samples tested by 16S rDNA PCR, the rDNA amplicon sequences were identical for 4 (8%). Sequence comparisons of the amplified products with Ehrlichia spp., Anaplasma spp., and Neorickettsia risticii sequences in GenBank are summarized in Table. Identical GroEL and sodB DNA sequences were amplified from 3 of 4 horses.
Table. Base pair similarities for DNA sequences of novel Ehrlichia species obtained from horses in Nicaragua in 2013* .
| Bacteria | GenBank accession nos.† | Gene, no. positive/no. tested (%) |
||
|---|---|---|---|---|
| 16S rRNA | GroEl | SodB | ||
| E. ruminantium | CR925678 | 361/374 (96.5) | 534/590 (90.5) | 508/599 (84.8) |
| E. canis | CP000107 | 356/374 (95.2) | 540/590 (91.5) | 496/599 (82.8) |
| E. chaffeensis | CP000236 | 360/374 (96.2) | 529/590 (89.7) | 491/599 (82.0) |
| E. ewingii | NR_044747, AF195273, KC778986 | 352/374 (94.1) | 530/590 (89.8) | 241/306 (78.8) |
| A. marginale | CP006847 | 339/374 (90.6) | 417/590 (70.8) | 348/599 (58.1) |
| A. phagocytophilum | CP006618 | 348/374 (93.0) | 439/590 (74.4) | 367/599 (61.2) |
| N. risticii | CP001431 | 306/374 (81.8) | 395/590 (66.9) | 358/599 (59.8) |
*Sequences for partial 16S rRNA, GroEl, and SodB genes are compared with GenBank database sequences from Ehrlichia ruminantium, E. canis, E. chaffeensis, E. ewingii, Anaplasma marginale, A. phagocytophilum, and Neorickettsia risticii. †GenBank sequence accession numbers for 16S rRNA, GroEl, and SodB for Ehrlichia sp. from horses in Nicaragua are KJ434178, KJ434179, and KJ434180, respectively.
Conclusions
According to serologic, PCR amplification, and DNA sequencing results, tick-infested horses in Mérida, Nicaragua, might be infected with a potentially novel Ehrlichia species. Initial serologic screening with the rapid ELISA indicated that exposure to >1 Ehrlichia species is common among horses in Nicaragua (55%). For dogs, SNAP 4DxPlus results can be positive after exposure to E. canis, E. chaffeensis, E. ewingii, and potentially E. muris and Panola Mountain ehrlichiae (2,3,6). Before this study, equine exposure to A. phagocytophilum (previous designation E. equi) in Nicaragua was considered more likely because equine exposure has occurred in Guatemala; this rickettsial organism is pathogenic for cats, dogs, horses, and humans (12). However, in North America, A. phagocytophilum is transmitted by I. scapularis and I. pacificus ticks, which have not been reported in Nicaragua. In future studies of horses in Nicaragua, ticks will be collected for identification. In Guatemala, R. microplus and A. cajennense ticks were the predominant species found on cattle, whereas D. nitens and A. cajennense ticks were most commonly found on horses (12). Of note, in Guatemala, tick infestation levels were substantially higher and body condition scores lower for horses than for cattle. Also, cattle were exposed to an agent with serologic cross-reactivity and close genetic relatedness to E. ruminantium.
The partial 16S rDNA sequences obtained from these horses most likely represent a novel species of Ehrlichia. This conclusion is further supported by sequence analysis of 2 protein-coding genes, sodB and groEL. Partial sequences from sodB and groEL genes demonstrated similarity to Ehrlichia spp. sequences found in GenBank, but they were not 100% identical to any sequences deposited to date. When 16S rDNA for rickettsiae are compared, Fournier et al. recommend that gene homology for organisms of identical species and genus be 99.8% and 98.1%, respectively (13). Although identical species are typically defined as being >99% identical with a reference sequence, the percentage identity needed to define a separate species is debated, ranging from 97% to 99.5% (14). Calculation of values can be based on alignment methods, reference databases, and number of basepairs in the sequence. Fournier et al. recommend that for novel species identification and rickettsiae classification, protein-coding genes should be used, specifically the 4 protein-coding genes gltA, ompA, ompB, and gene D in addition to the 16S rRNA gene. To date, 16S rDNA sequences or whole-genome sequencing have been used to classify Ehrlichia species and strains. Collectively, and as depicted in the Figure (in which phylogenetic alignment trees for all 3 genes tested in this study were constructed by using reference sequences from representative members of the genera Anaplasma, Ehrlichia, and Neorickettsia), the genetic findings in this study support infection of horses in Nicaragua with a novel Ehrlichia species. To confirm this possibility, future efforts will focus on cell culture isolation of the Ehrlichia organism from horses in Nicaragua.
Figure.
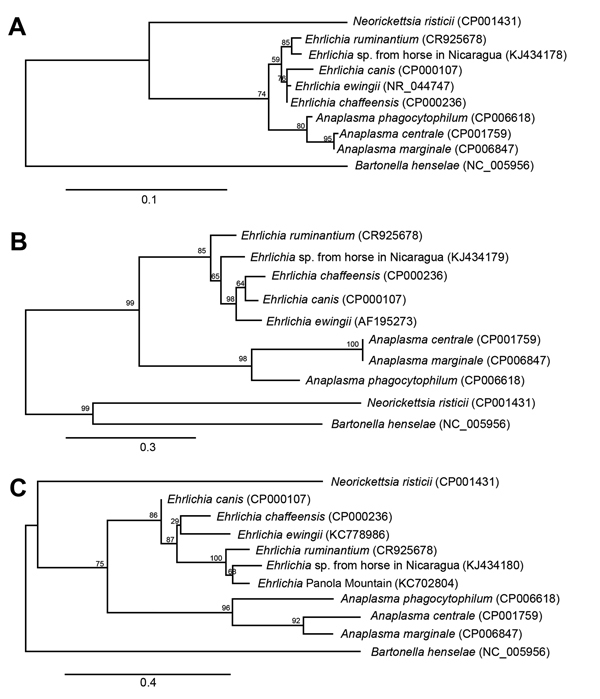
Phylogenetic trees of Ehrlichia sp. from horses in Nicaragua and selected bacterial species (GenBank accession numbers for reference sequences in parenthesis) based on partial sequences from genes coding for 16SrRNA (A), GroEL (B), and SodB (C). Sequences were aligned by using MUSCLE version 3.7 (http://www.ebi.ac.uk/Tools/msa/muscle/), and alignments were refined by using Gblocks version 0.91b (http://www.idtdna.com/gblocks.com). Phylogenetic trees were constructed by using PhyML version 3.0 aLRT (http://code.google.com/p/phyml/) under the HKY85 model, and the resulting trees were rendered by using TreeDyn version 198.3 (http://www.treedyn.org/). Scale bars indicate number of substitutions per site, and the numbers in the branches represent percentage support of the node.
Vectorborne pathogens can infect any host species bitten by infected ticks. At least 4 Ehrlichia species have been implicated as being pathogenic for canids and humans (1–3). Thus, future studies should also determine whether dogs, other animals, and humans in Nicaragua are exposed to and infected with this potentially novel Ehrlichia species.
Acknowledgments
We thank the Department of Biomedical Sciences, Oregon State University College of Veterinary Medicine, for funding this study; the veterinary students who participated in the 2013 Nicaragua trip and helped collect the blood samples used for this study; and Dana Riggins for technical assistance in processing samples for PCR.
This research was supported in part by the State of North Carolina and donations to the North Carolina State University College of Medicine Foundation for Vector-Borne Disease Research. B.A.Q. receives funding from IDEXX Laboratories Inc.
Biography
Ms. O’Nion is a student in the veterinary medical curriculum at Oregon State University. Her research interests involve an international One Health approach to vectorborne infectious diseases of human and veterinary medical importance.
Footnotes
Suggested citation for this article: O’Nion VL, Montilla HJ, Qurollo BA, Maggi RG, Hegarty BC, Tornquist J, et al. Potentially novel Ehrlichia species in horses, Nicaragua. Emerg Infect Dis. 2015 Feb [date cited]. http://dx.doi.org/10.3201/eid2102.140290
References
- 1.Dumler JS, Walker DH. Tick-borne ehrlichioses. Lancet Infect Dis. 2001;1:21–8. 10.1016/S1473-3099(09)70296-811871406 [DOI] [Google Scholar]
- 2.Qurollo BA, Davenport AC, Sherbert BM, Grindem CB, Birkenheuer AJ, Breitschwerdt EB. Infection with Panola Mountain Ehrlichia sp. in a dog with atypical lymphocytosis and clonal T-cell expansion. J Vet Intern Med. 2013;27:1251–5. 10.1111/jvim.12148 [DOI] [PubMed] [Google Scholar]
- 3.Hegarty BC, Maggi RG, Koskinen P, Beall MJ, Eberts M, Chandrashekar R, et al. Ehrlichia muris infection in a dog from Minnesota. J Vet Intern Med. 2012;26:1217–20. 10.1111/j.1939-1676.2012.00968.x [DOI] [PubMed] [Google Scholar]
- 4.Socolovschi C, Kernif T, Raoult D, Parola P. Borrelia, Rickettsia, and Ehrlichia species in bat ticks, France, 2010. Emerg Infect Dis. 2012;18:1966–75 and. 10.3201/eid1812.111237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cabezas-Cruz A, Vancová M, Zweygarth E, Ribeiro MF, Grubhoffer L, Passos LM. Ultrastructure of Ehrlichia mineirensis, a new member of the Ehrlichia genus. Vet Microbiol. 2013;167:455–8. 10.1016/j.vetmic.2013.08.001 [DOI] [PubMed] [Google Scholar]
- 6.Vieira RF, Vieira TS, Nascimento Ddo A, Martins TF, Krawczak FS, Labruna MB, et al. Serological survey of Ehrlichia species in dogs, horses and humans: zoonotic scenery in a rural settlement from southern Brazil. Rev Inst Med Trop Sao Paulo. 2013;55:335–40. 10.1590/S0036-46652013000500007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duell JR, Carmichael R, Herrin BH, Holbrook TC, Talley J, Little SE. Prevalence and species of ticks on horses in central Oklahoma. J Med Entomol. 2013;50:1330–3. 10.1603/ME13117 [DOI] [PubMed] [Google Scholar]
- 8.Bermudez SE, Eremeeva ME, Karpathy SE, Samudio F, Zambrano ML, Zaldivar Y, et al. Detection and identification of rickettsial agents in ticks from domestic mammals in Eastern Panama. J Med Entomol. 2009;46:856–61. 10.1603/033.046.0417 [DOI] [PubMed] [Google Scholar]
- 9.Chandrashekar R, Daniluk D, Moffitt S, Lorentzen L, Williams J. Serologic diagnosis of equine borreliosis: evaluation of an in-clinic enzyme-linked immunosorbent assay (SNAP 4DX). Intern J Appl Res Vet Med. 2008;3:145–50. [Google Scholar]
- 10.Eddlestone SM, Diniz PPVP, Neer TM. Gaunt SD, Corstvet R, Cho D, et al. Doxycycline clearance of experimentally induced chronic Ehrlichia canis infection in dogs. J Vet Intern Med. 2007;21:1237–42. [DOI] [PubMed]
- 11.Barber RM, Li Q, Diniz PPVP. Evaluation of brain tissue or cerebrospinal fluid with broadly reactive polymerase chain reaction for Ehrlichia, Anaplasma, spotted fever group Rickettsia, Bartonella, and Borrelia species in canine neurological diseases (109 cases). J Vet Intern Med. 2010;24:372–8. 10.1111/j.1939-1676.2009.0466.x [DOI] [PubMed] [Google Scholar]
- 12.Teglas M, Matern E, Lein S, Foley P, Mahan SM, Foley J. Ticks and tick-borne disease in Guatemalan cattle and horses. Vet Parasitol. 2005;131:119–27. 10.1016/j.vetpar.2005.04.033 [DOI] [PubMed] [Google Scholar]
- 13.Fournier PE, Dumler JS, Greub G, Zhang J, Wu Y, Raoult D. Gene sequence-based criteria for identification of new Rickettsia, isolates and description of Rickettsia heilongjiangensis sp. nov. J Clin Microbiol. 2003;41:5456–65. 10.1128/JCM.41.12.5456-5465.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schlaberg R, Simmon KE, Fisher MA. A systematic approach for discovering novel, clinically relevant bacteria. Emerg Infect Dis. 2012;18:422–30. 10.3201/eid1803.111481 [DOI] [PMC free article] [PubMed] [Google Scholar]


