Strains pathogenic to mammals share phylogenetic and phenotypic features with strains approved for mosquito control.
Keywords: Oomycetes, Oomycota, fungal-like, Lagenidium, Lagenidium giganteum, Pythium insidiosum, pythiosis, lagenidiosis, biological control, fungi
Abstract
Infections of mammals by species in the phylum Oomycota taxonomically and molecularly similar to known Lagenidium giganteum strains have increased. During 2013–2014, we conducted a phylogenetic study of 21 mammalian Lagenidium isolates; we found that 11 cannot be differentiated from L. giganteum strains that the US Environmental Protection Agency approved for biological control of mosquitoes; these strains were later unregistered and are no longer available. L. giganteum strains pathogenic to mammals formed a strongly supported clade with the biological control isolates, and both types experimentally infected mosquito larvae. However, the strains from mammals grew well at 25°C and 37°C, whereas the biological control strains developed normally at 25°C but poorly at higher temperatures. The emergence of heat-tolerant strains of L. giganteum pathogenic to lower animals and humans is of environmental and public health concern.
During the 20th century, Pythium insidiosum was the only fungus-like species from the phylum Oomycota known to cause life-threatening infections in mammals and birds (1,2). In 2003, a novel group of pathogenic Oomycota was isolated in the southeastern United States from dogs with invasive cutaneous infections resembling pythiosis, but the strains had morphologic and molecular affinities with the genus Lagenidium (3,4). Subsequently, other strains similar to previously studied Lagenidium species were found to be associated with infections in dogs, cats, and humans (5–7). The disease caused by these 2 Oomycota species has been reported mainly in apparently healthy hosts; thus, these species’ evolutionary traits need to be investigated (1–7). Although detailed descriptions of the clinical, pathologic, and serologic features of Lagenidium spp. pathogenic to mammals have been published (3,4), their taxonomic relationship to Lagenidium strains approved for biological control and the capacity of their zoospores to infect mosquitoes have yet to be investigated (5,6).
The genus Lagenidium, introduced by Schenk in 1857 (8), comprises numerous saprotrophic species (9), as well as several members that are pathogenic to algae, phytoplankton, pollen, barnacles, blue crabs, mosquito larvae, shrimp, and mammals (3–5). Couch (10) isolated L. giganteum from Daphnia larvae in Virginia, USA. Later, Couch and Romney (11) confirmed that L. giganteum infects and kills mosquito larvae as it reproduces sexually. Their work stimulated interest in L. giganteum as a biological control agent of mosquitoes (11,12). In 1995, the US Environmental Protection Agency registered L. giganteum under the trade name Laginex as a biocontrol agent (13) but later deregistered it at the request of the manufacturer (http://www.gpo.gov/fdsys/pkg/FR-2011-09-28/pdf/2011-24832.pdf). L. giganteum strains used for testing and commercial development were deposited within the American Type Culture Collection (ATCC), and the ATCC strains and other not-well-characterized strains were released in nature for field studies (12–16). In this study, conducted during 2013–2014, we investigated 21 mammalian Lagenidium strains and compared them with L. giganteum isolates used for mosquito control.
Materials and Methods
Living Cultures
A complete list of the strains and their accession numbers is shown in the Table. In brief, we studied the following isolates: L. giganteum (ATCC 36492 = ATCC 52675 used as biological control and ATCC 48336 isolated from mosquito larvae); L. humanum (ATCC 76726 isolated from the skin of dead humans); L. giganteum pathogenic to mammals (culture collection at the Biomedical Laboratory Diagnostics, Michigan State University (MTLA): MTLA 01 = ATCCMYA-4933 type strain, MTLA-03, MTLA-04 = ATCCMYA-4934 (from a US cat), MTLA-05 = ATCCMYA-4935, MTLA-10, MTLA-12, MTLA-14, MTLA-15, MTLA-16, MTLA-17, and MTLA-18 from dogs); L. ajelloi (MTLA-06 = ATCCMYA-4936 type strain, MTLA-07 = ATCCMYA-4937, MTLA-19, MTLA-20, MTLA-21, MTLA-22, MTLA-23, isolated from dogs); L. vilelae (MTLA-24 = DNA extracted from fixed feline tissue and MTLA-25 cat strain); and L. albertoi (MTLA13 = ATCCMYA-4932 type strain).
Table. Strains, accession numbers, acronyms, optimal temperatures, and affected hosts on the studied Lagenidium isolates studied, 2011–2012.
| Taxonomic name | Strain identification† | Acronym | Primer |
Optimal temperature, °C | Host | ||||
|---|---|---|---|---|---|---|---|---|---|
| Internal transcriber spacer | CDC42 | COXII | HSP90 | TUB | |||||
| L. giganteum | NPI01 | Lgm1 | EF016915 | 0 | 0 | 0 | 0 | 25 | Mosquito larvae |
| L. giganteum | ATCC 48336 | Lgm2 | JQ745259 | 0 | 0 | 0 | 0 | 25 | Mosquito larvae |
| L. giganteum | ATCC 36492 | Lgm3 | AY151183 | JX985758 | AF086697 | JX999093 | JX999124 | 25 | Mosquito larvae |
| Lagenidium sp. | CBS 127533 | Ls1 | HQ395647 | 0 | 0 | 0 | 0 | ND | Nematodes |
| Lagenidium sp. A | CBS 126881 | Lspa1 | HQ111460 | 0 | 0 | 0 | 0 | ND | Nematodes |
| Lagenidium sp .A | CBS 126885 | Lspa2 | HQ111452 | 0 | 0 | 0 | 0 | ND | Nematodes |
| Lagenidium sp .A | CBS 126878 | Lspa3 | HQ111464 | 0 | 0 | 0 | 0 | ND | Nematodes |
| Lagenidium sp. A | CBS 126880 | Lspa4 | HQ111459 | 0 | 0 | 0 | 0 | ND | Nematodes |
| Lagenidium sp .B | CBS 126882 | Lspb1 | HQ111461 | 0 | 0 | 0 | 0 | ND | Nematodes |
| Lagenidium sp. B | CBS 126879 | Lspb2 | HQ111463 | 0 | 0 | 0 | 0 | ND | Nematodes |
| Lagenidium sp. B | CBS 126884 | Lspb3 | HQ111450 | 0 | 0 | 0 | 0 | ND | Nematodes |
| Lagenidium sp. B | CBS 126883 | Lspb4 | HQ111462 | 0 | 0 | 0 | 0 | ND | Nematodes |
| L. callinectes | NJM-0531 | Lc1 | AB285488 | 0 | 0 | 0 | 0 | 25 | Crustacea |
| L. callinectes | NJM-8989 | Lc2 | AB285487 | 0 | 0 | 0 | 0 | 25 | Crustacea |
| L. callinectes | ATCC 24973 | Lc3 | AB285486 | 0 | 0 | 0 | 0 | 25 | Crustacea |
| L. vilelae | CBS 127042 | Lv1 | HQ111455 | 0 | 0 | 0 | 0 | ND | Nematodes |
| Lagenidium sp. C | CBS 127284 | Lspc1 | HQ111472 | 0 | 0 | 0 | 0 | ND | Nematodes |
| Lagenidium sp. C | CBS 127285 | Lspc2 | HQ111470 | 0 | 0 | 0 | 0 | ND | Nematodes |
| Lagenidium sp. C | CBS 127283 | Lscd1 | HQ111471 | 0 | 0 | 0 | 0 | ND | Nematodes |
| Lagenidium sp. C | CBS 127276 | Lspc3 | HQ111451 | 0 | 0 | 0 | 0 | ND | Nematodes |
| Lagenidium sp. C | CBS 127281 | Lspd2 | HQ111473 | 0 | 0 | 0 | 0 | ND | Nematodes |
| Lagenidium sp. D | CBS 127277 | Lspd3 | HQ111454 | 0 | 0 | 0 | 0 | ND | Nematodes |
| L. caudatum | CBS 58485 | Lca1 | HQ643136 | 0 | AF290309 | 0 | 0 | 25 | Nematodes |
| L. myophilum | NJM-8403 | Lm1 | AB285497 | 0 | AF290311 | 0 | 0 | 20 | Shrimp |
| L. humanum | ATCC 76726 | Lh1 | JX970867 | JX985757 | AF290310 | JX999094 | JX999125 | 25 | Saprotrophic |
| L. giganteum mammals | ATCCMYA-4933 | Lgp1 | JF919611 | JF919613 | JX970853 | JX999097 | JX999127 | 37°C fast | Dog |
| L. giganteum mammals | MTLA-03 | Lgp2 | JX999099 | JF919612 | JX970852 | JX999099 | JX999128 | 37 fast | Dog |
| L. giganteum mammals | ATCCMYA-4934 | Lgp3 | JX999096 | JF919613 | JX970854 | JX999096 | JX999126 | 37 slow | Cat |
| L. giganteum mammals | ATCCMYA-4935 | Lgp4 | JX999098 | JF919614 | JX970855 | JX999098 | JX999129 | 37 fast | Dog |
| L. giganteum mammals | MTLA-10 | Lgp5 | KJ506111 | KJ506076 | KJ506083 | KJ506097 | KJ506111 | 37 fast | Dog |
| L. giganteum mammals | MTLA-12 | Lgp6 | KJ506112 | KJ506078 | KJ506084 | KJ506098 | KJ506112 | 37 fast | Dog |
| L. giganteum mammals | MTLA-14 | Lgp7 | KJ506113 | KJ506077 | KJ506085 | KJ506099 | KJ506113 | 37 fast | Dog |
| L. giganteum mammals | MTLA-15 | Lgp8 | KJ506116 | KJ506081 | KJ506087 | KJ506103 | KJ506116 | 37 fast | Dog |
| L. giganteum mammals | MTLA-16 | Lgp9 | KJ506115 | KJ506080 | KJ506086 | KJ506102 | KJ506115 | 37 fast | Dog |
| L. giganteum mammals | MTLA-17 | Lgp10 | KJ506117 | KJ506082 | KJ506088 | KJ506101 | KJ506117 | 37 fast | Dog |
| L. giganteum mammals | MTLA-18 | Lgp11 | KJ506114 | JX970841 | KJ506089 | KJ506100 | KJ506114 | 37 fast | Dog |
| L. ajelloi | ATCCMYA-4936 | La1 | JX970885 | JX970885 | JX970841 | JX999112 | JX999143 | 37 slow | Dog |
| L. ajelloi | ATCCMYA-4937 | La2 | JX970884 | JX970884 | JX970842 | JX999113 | JX999144 | 37 slow | Dog |
| L. ajelloi | MTLA-19 | La3 | KJ506134 | KJ506070 | KJ506092 | KJ506106 | KJ506120 | 37 slow | Dog |
| L. ajelloi | MTLA-20 | La4 | KJ506135 | KJ506071 | KJ506094 | KJ506109 | KJ506124 | 37 slow | Dog |
| L. ajelloi | MTLA-21 | La5 | KJ506137 | KJ506073 | KJ506095 | KJ506108 | KJ506122 | 37 slow | Dog |
| L. ajelloi | MTLA-22 | La6 | KJ506138 | KJ506069 | KJ506096 | KJ506107 | KJ506123 | 37 slow | Dog |
| L. ajelloi | MTLA-23 | La7 | KJ506136 | KJ506072 | KJ506093 | KJ506110 | KJ506121 | 37 slow | Dog |
| L. albertoi | ATCCMYA-4932 | Lal8 | JX970870 | JX970838 | JX970838 | JX999115 | JX999142 | 37 slow | Human |
| L. vilelae | MTLA-24 | Lv2 | KJ506133 | KJ506075 | KJ506091 | KJ506105 | KJ506119 | 37 slow | Cat |
| L. vilelae | MTLA-25-Tissue | Lv3 | KJ506132 | KJ506090 | KJ506090 | KJ506104 | KJ506118 | ND | Cat |
| Pythium insidiosum | MTPI-36 | Pi1 | AY486144 | 0 | 0 | 0 | 0 | 37 | Mammals |
| P. grandisporangium | ATCC 28295 | Pg1_c | AY151182 | 0 | 0 | 0 | 0 | 25 | Saprotrophic |
| P. aphanidermatum | STR-135 | Pa1_a | AY151180 | 0 | 0 | 0 | 0 | 25 | Plants |
| P. diliense | STR-66 | Pd1_a | AY151181 | 0 | 0 | 0 | 0 | 25 | Plants |
| P. flevoense | STR-W041 | Pf1_b | EU240168 | 0 | 0 | 0 | 0 | 25 | Plants/fish |
| P. diclinum | STR-P7824 | Pdi1_b | EF153675 | 0 | 0 | 0 | 0 | 25 | Plants |
| P. capillosum | CBS 222–94 | Pc1_b | AY598635 | 0 | 0 | 0 | 0 | 25 | Saprotrophic |
| P. sylvaticum | CBS 453.67 | Ps1_f | AY598657 | 0 | 0 | 0 | 0 | 25 | Plants |
| P. pleroticum | CBS 776.81 | Pp1_e | AY598642 | 0 | 0 | 0 | 0 | 25 | Plants |
| P. iwayamai | CBS 156.64 | Piw1_g | AY598648 | 0 | 0 | 0 | 0 | 25 | Plants |
| P. undulatum | CBS 157.69 | Pu1_j | AY598708 | 0 | 0 | 0 | 0 | 25 | Plants |
| P. ultimum | CBS 398.51 | Pul1_i | AY598642 | 0 | 0 | 0 | 0 | 25 | Plants |
| P. nunn | CBS 808.96 | Pn1_j | AY598709 | 0 | 0 | 0 | 0 | 25 | Fungi |
*ATCC, American Type Culture Collection; CBS, Centraalbureau voor Schimmelcultures; CDC42, cell division cycle 42; COXII, cytochrome oxidase II; HSP90, heat shock protein 90; ND, no data; TUB, tubulin; 0, no DNA sequences available. †MTLA and MTPI strains Biomedical Laboratory Diagnostics, Michigan State University culture collection.
Media and Culture Conditions
The isolates were grown on brain-heart infusion (BHI) (DIFCO, Detroit, MI, USA), corn meal agar (CMA) (BBL, Sparks, MD, USA), 2% Sabouraud dextrose agar (SDA), 2% Sabouraud dextrose broth, in triplicate. Cardinal temperatures of growth (25°C, 30°C, and 37°C measured during 3 days’ incubation) and their relation to the production of sexual and asexual structures were evaluated on the above media. Development of the different stages of vesicle and zoospore formation, cleavage, and release were assessed on colonized grass leaves in water cultures containing Ca++ and Mg++. Briefly, the evaluated strains (Table) were subcultured on SDA plates at 37°C for 24 h. After incubation, 5 × 5–mm diameter blocks were cut from the advancing edges of the culture and placed on top of a 2% water agar plates. Sterile 4 × 10–mm grass blades were laid on top of each block and incubated at 37°C for 24 h (L. giganteum = MTLA-01 from mammals). The grass blades were then collected and placed in a beaker that contained 50 mL of sporulation solution (made of 2 solutions: mix no. 1 contained NH42HPO4 [66.04 g], KH2PO4 [68.05 g], and K2HPO4 [87.09 g] in 500 mL H2O; and mix no. 2 contained CaCl2.2H2O [18.38 g] and MgCl2.6H2O [25.42 g] in 250 mL H2O). The sporulation solution was obtained by mixing 0.5 mL of solution no. 1 plus 0.1 mL of solution no. 2 in 1.0 L of distilled water. The beaker with the 50-mL of sporulation solution plus the parasitized grass blades was incubated at 37°C, and the development of sporangia and zoospores microscopically was evaluated every 30 min for the following 6 h (or longer when needed).
Morphologic Description of L. giganteum from Mammals
We evaluated the morphologic features of L. giganteum strains after subculture on BHI, CMA, and SDA at different intervals during 5 days’ incubation at 37°C or at room temperature (25°C). Briefly, 4 × 4–mm agar blocks were cut from the above media and mixed with 1 drop of lactophenol cotton blue (phenol 20 mL, lactic acid 20 mL, glycerol 40 mL, and distilled water 20 mL). The morphologic features of their hyphal and other structures developed in these media, including oogonia, were microscopically investigated. We evaluated the development of sporangia and zoospores in sporulation medium (see above). After incubation at the induction temperatures, the beaker containing 50 mL of sporulation medium plus the parasitized grass blades was inspected on an inverted microscope. If vesicles and zoospores developed, we removedthe grass blade with >10 vesicles, using tweezers, and placed it on a glass slide. Immediately, 5 μL of Merthiolate (0.02%) was added to the grass blade to stop the movement of the zoospores and facilitate measurements. Alternatively, grass blades containing vesicles with zoospores were also transferred to plates holding Culex pipiens larvae.
Experimental Mosquito Infection
The strains of L. giganteum from mammals (MTLA01, MTLA03, MTLA04, MTLA05, and MTLA10) (Table) were used to evaluate their capability to infect mosquito larvae of C. pipiens. Instar 3 C. pipiens larvae were collected and placed in 6-well plates (1 plate per experiment; Corning Inc., Corning, NY, USA) containing 5 mL of water and fed every other day with fish food flakes (TetraMin, Lancaster, PA, USA) until the end of the experiment. We transferred fresh grass blades, with >10 vesicles per blade containing numerous zoospores, to the plates holding 5 mosquito larvae per well (total of 30 larvae per plate). The plates were incubated at 25°C and observed daily by using an inverted microscope for the next 3 weeks. The criteria to select putative infected mosquito larvae were based on their swimming capabilities. We closely inspected slower swimmers for filamentous structures outside and between the larval segments and inside their bodies. Every trial included its positive (L. giganteum biological control ATCC 36492 = ATCC 52675) and negative controls (larvae without exposure to L. giganteum species), and the experiment was repeated twice (total 60 C. pipens larvae).
DNA Extraction and Molecular Procedures
The 20 strains investigated (Table) were inoculated into 250-mL flasks containing 100 mL of Sabouraud dextrose broth and incubated for 72 h at 37°C on a shaker rotating at 150 rpm. In addition, 1 formalin-fixed tissue sample from a cat was also used to extract total genomic DNA following the company protocols (QIAGEN, Valencia, CA, USA). After incubation, the cultures were killed with Merthiolate (0.02%, wt/vol) and then filtrated to separate the hyphal cell mass. The hyphal cell mass was then transferred to a mortar and ground in the presence of liquid nitrogen. DNA from the disrupted hyphae was treated with sodium dodecyl sulfate and proteinase K and then incubated at 60°C for 1 h, and genomic DNA of the hydrae was extracted with phenol, chloroform, isoamyl alcohol (Sigma, St. Louis, MO. USA). PCR was conducted by hot start amplification using the following primers: the universal primers for internal transcribed spacers (ITS): ITS1 = TCCGTAGGTGAACCTGCGG and ITS4 = TCCTCCGCTTATTGATATGC; cytochrome oxidase II (COXII): COX.LagF = 5′-CCACAAATTTCACTACATTGA-3′ and COX.LagR = 5′-TAGGATTTCAAGATCCTGC-3′; heat shock protein 90 (HSP90): HSP90.LagF = 5′-CAACCTBGGHACSATYGCCAAG-3′ and HSP90.LagR = 5′-ACRAAMGACARGTAYTCVGGCA-3′; cell division cycle 42 (CDC42): CDC42.LagF = 5′-GTSCCVACYGTVTTYGANAAYTA-3′ and CDC42.LagR = 5′-GCWSWGCAYTCVASRTAYTT-3′; Tubulin: TUB.LagF = 5′-GGTGGTGGTACCGGTTC-3′ and TUB.LagR = 5′-GACACACGCTTGAACATC-3′. In addition, the following primers were constructed to PCR amplify some of the L. ajelloi and L. vilelae strains: CDC42.Lag2F = 5′-GTGCCGACYGTGTTYGABAAC-3′ and CDC42.Lag2R = 5′-CTSTTCKGTTGTRATBGG-3′; and HSP90.Lag2F = 5′-GAGGCCTTCGTGGAAGCG-3′ and HSP90.Lag2R = 5′-GTTGTTCATTTTCTTGCGGG-3′. The PCR temperature-cycling parameters were as follows: 10 min at 95°C and 1 min for subsequent cycles, annealing for 1 min at 60°C, and elongation at 72°C for 2 min. The parameter was repeated for 40 cycles, followed by a final elongation of 2 min at 72°C for 7 min. The amplicons were ligated into pCR 2.1-TOPO vector (Invitrogen, Carlsbad, CA. USA), purified, and then sequenced by using BigDye Terminator chemistry in an ABI Prim 310 genetic analyzer (Perkin-Elmer, Foster City, CA. USA).
Phylogenetic Analysis
Genomic DNA sequences of the studied strains’ complete ITS and the partial gene sequences of the selected 4 exons (CDC42, COXII, HSP90, and TUB) were aligned with other Lagenidium and Pythium DNA sequences available at the National Center for Biotechnology Information by using ClustalW version 1.81 (http://www.clustal.org)with default settings and their alignments visually inspected. Pythium spp. groups a–c, e–g, and j (15) (Table) were used as outgroups. Phylogenetic and molecular evolutionary analyses were conducted by using MEGA6 (http://www.megasoftware.net). The aligned sequences were exported for parsimony analysis by using a heuristic search with tree bisection reconnection branch swapping (MEGA6) and distant analysis by neighbor-joining (MEGA6). We coded large insertions as 1 event by excluding all but 1 nt per insertion. The generated gaps were treated as missing data. Neighbor-joining analysis used either uncorrected distances or maximum-likelihood estimates of distance with a time-reversible model (6ST), and empirical base frequencies with no rate variation among sites, or a shape parameter of 0.5 γ-distribution with 4 rate categories. Branch support was estimated as percentage of parsimony tree (1,000 resampling, heuristic, branch swapping) or neighbor-joining trees (1,000 resampling, maximum-likelihood distances) within each branch. Concatenated DNA sequences of the 5 loci investigated were used in Bayesian analysis. We conducted the test in MrBayes version 3.2.1 ×64 (http://mrbayes.sourceforge.net) using the general time reversible + I + γ model, with 2 chains (1 heated), 2 runs, sampling every 100th generation for 1 × 106 generations, and exclusion of the first 2.5 × 105 samples (the burn-in) before analysis. Support for branches was estimated as the percentage of parsimony tree (1,000 resampling, heuristic, nni branch swapping) or neighbor-joining tree (1,000 resampling, maximum-likelihood distances) containing the branch, as well as by determining the Bayesian probability estimated as the percentage of Bayesian trees possessing the branch after discarding the burn-in sample.
Results
Phylogenetic Analysis
Using Pythium spp. groups a–c, e–g, and j (Table) as outgroups, we sorted the concatenated DNA sequences of 4 protein-coding genes and 1 ribosomal region in Bayesian and maximum-likelihood phylogenetic analyses 21 mammalian pathogenic Legenidium strains into 4 strongly supported clades in the combined analysis (Figure 1). In the largest of these clades, 11 of the 21 strains (Table) recovered from infected mammals form a monophyletic clade that includes other L. giganteum strains previously used to develop biological control agents of mosquitoes (Figure 1). Also included in this clade is an L. giganteum strain recovered from nematodes in Taiwan (Lsp1). The remaining 10 mammal-infecting Lagenidium strains were resolved with strong support into 3 independent clades that are basal to L. giganteum and identified in this study as L. ajelloi, L. albertoi, and L. vilelae (Figures 1, 2). These new species will be fully described elsewhere. Other clades of Lagenidium species, as yet unnamed, are also apparent in the phylogeny (Lspa to Ld; Figures 1, 2).
Figure 1.
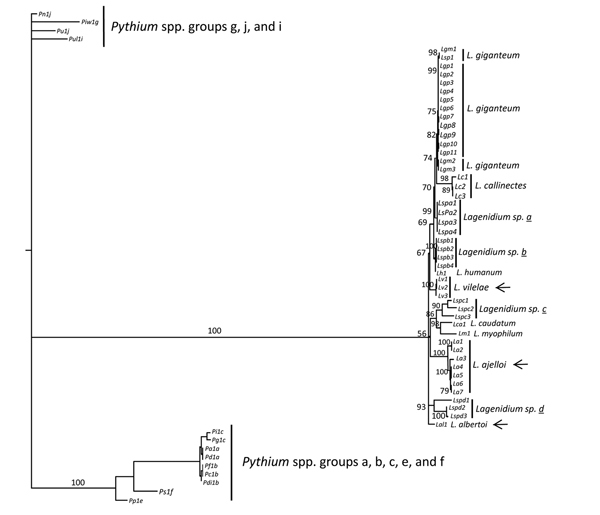
Bayesian phylogenetic analysis of concatenated 4 partial coding gene sequences (cell division cycle 42, cytochrome oxidase II, heat shock protein 90, and tubulin) and the complete internal transcribed spacers 1, 2, and 5.8S of Lagenidium DNA sequences. Thirteen Pythium species DNA sequences were included as the outgroup (groups a–c, e–g, j, and I [16 ]; Table). Support on key branches is the Bayesian probability for that branch followed by the percentage of 1,000 bootstrap resampled datasets containing the branch in neighbor-joining analyses of maximum-likelihood distances followed by the percentage of 1,000 bootstrap resampled datasets containing the branch in parsimony analyses using heuristic searches. In this analysis, the DNA sequences of L. giganteum mosquito control (Lg 1–3) and a Lagenidium sp. recovered from a nematode in Taiwan (Ls1, Lsp1 = HQ395647) clustered with L. giganteum from mammals (Lg 1–10). The pathogen of crab L. callinectes (Lc) was the sister group to the cluster. Three Lagenidium mammalian pathogenic novel species (L. ajelloi = La, L. albertoi = Lal, and L. vilelae = Lv) were placed in 3 distinctive strongly supported clades (arrows). The accession numbers, the abbreviations used to identify each species, and the Lagenidium and Pythium spp. DNA sequences are shown in the Table. ATCC, American Type Culture Collection; CBS, Centraalbureau voor Schimmelcultures.
Figure 2.
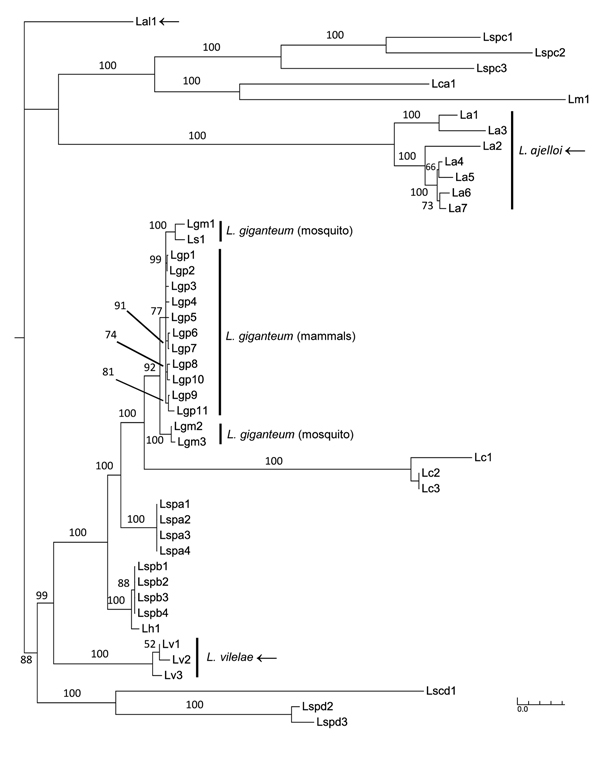
The Bayesian tree was constructed by concatenated aligned of Lagenidium spp. DNA sequences as in Figure 1 without outgroup to highlight the position of L. giganteum in the tree. L. giganteum from mammals (Lgp 1–11), L. giganteum mosquito control (Lgm 1–3 and Ls1 = HQ395647), and the novel species L. ajelloi = La, L. albertoi = Lal, and L. vilelae = Lv were placed in 4 strongly supported clades (arrows). Scale bar indicates nucleotide substitutions per site.
Heat-tolerance and Mosquito Larva Infection Experiments
The 11 L. giganteum strains from mammalian hosts grew faster at 37°C and 30°C than at 25°C, whereas L. giganteum biological control strains (ATCC 36492 = ATCC 52675 and ATCC 48336) grew well at 25°C and poorly at 30°C and almost failed to develop at 37°C in growth experiments conducted in CMA and SDA media (Figure 3). The biological control strains barely developed at 37°C but survived, as shown by their subsequent growth, after being transferred to 25°C.
Figure 3.
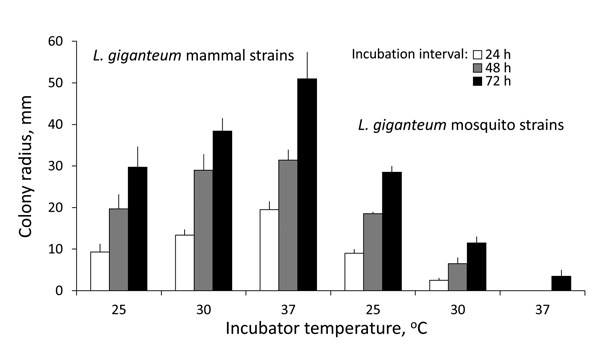
Cardinal temperatures of Lagenidium giganteum types in culture. Growth (mean colony radius and SEM, mm) of L. giganteum mammalian and mosquito strains at 3 temperatures at 24-, 48-, and 72-hour intervals postinoculation onto 2% Sabouraud dextrose agar. Repeated measures analysis of variance showed highly significant differences between strains (F1,33 = 165.0, p<0.0001) and a highly significant interaction of strain and incubation temperature across time intervals (F2,33 = 45.9, p<0.0001).
We tested the ability of the mammalian L. giganteum zoospores to infect mosquito larvae in 5 L. giganteum mammal strains (MTLA01, 03, 04, 05, and 10; Table) and 2 L. giganteum biocontrol strains (ATCC 48336 = Lgm2, ATCC 36492 = Lgm3; Table). The emerging mammalian pathogenic strains exhibited mosquito-infection capabilities similar to those of the strains approved by the US Environmental Protection Agency for mosquito control (Figure 4). At least 1 larvae per well was found infected. Infected larvae exhibited abnormal swimming behavior and died within 2 days (Figure 4). Of the 60 C. pipiens larvae (2 trials, 30 larvae per experiment), 12 were found infected at the end of both experiments (instar 3 or instar 4). Uninfected larvae continued their normal life cycle. Postmortem examination of infected larvae showed extensive infections with numerous hyphae spreading internally and emerging between segments (Figure 4, panels A–E). Under our experimental conditions, L. giganteum ATCC 36492 infected 18 of the 60 C. pipens larvae. In unexposed controls all larvae survived, larvae swam normally, and we found no evidence of infection. The identity of mammalian L. giganteum strains experimentally infecting mosquito larvae was confirmed by culture, DNA extraction, sequencing, and phylogenetic analysis at the conclusion of each experiment.
Figure 4.

Lagenidium giganteum from mammal experimental infection using Culex pipiens mosquito larvae. A) Composite of 2 photographs showing an instar 3 C. pipiens larvae infected with 1 of the 5 tested strains of L. giganteum recovered from dogs with lagenidiosis (MTLA01, type strain). Note the mycelioid structures emerging from the infected larvae (arrows). B, C) Enlargements of the 2 white boxes in (A) showing details of the mycelioid structures emerging between the segments of the larvae. D, E) Aggressiveness of the invading mycelioid structures (arrows) within the body of C. pipiens larvae.
Morphologic Characteristics of L. giganteum in Culture
Submerged colonies of both L. giganteum types (biological control and mammalian strains) are colorless or yellow with few aerial mycelia that present irregular radiating and undulating patterns on any of 3 media (CMA, BHI, or 2% SDA) (Figure 5, panels A, D). Typically, the elongated mycelium comprises ovoides or spherical 30- to 40-μm diameter segments forming chains and connected by broad to slender isthmuses; hyphae also form long irregular segments of >400 μm (Figure 5, panels B, C, E, F). We did not find oogonia in the evaluated strains. In liquid sporulation medium, hyphal segments develop 1–2 discharge tubes that produce terminal vesicles containing reniform zoospores 9–10 μm wide × 10–12 μm long (Figure 5, panel G). Zoospores were released in both mammal and mosquito L. giganteum strains after the discharge vesicle was disrupted (Figure 5, panel H), which then swam for 10–20 minutes before encysting. Germ tubes developed from encysted zoospores within 10–20 minutes after encystment.
Figure 5.
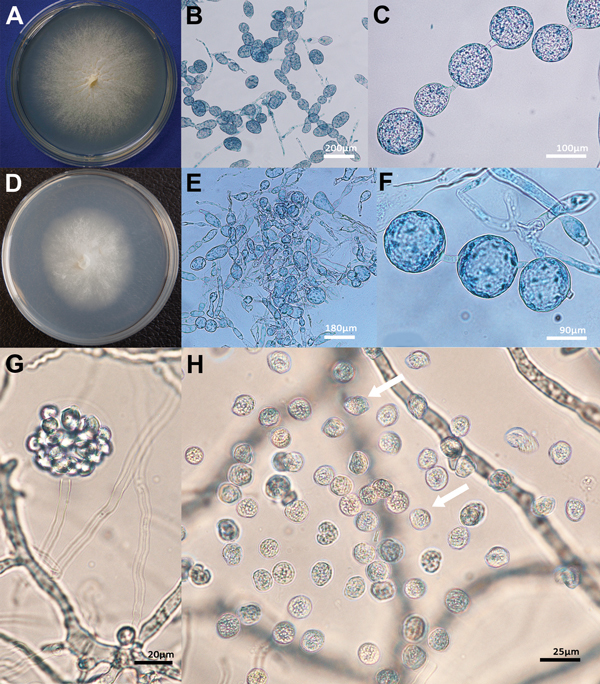
Morphologic features of isolates of Lagenidium giganteum mosquito control agent and L. giganteum mold from mammals. Panel A shows henotypic features in culture of the mammalian pathogen (ATCCMYA-4933, type strain) and panel D shows the biological control (ATCC 36492). The development of spherical and ovoid 40- to 170-μm swelling segments (panels B, C, E, F) was the main feature of both mammalian and biocontrol strains. Panel G shows A tubular body developed from an unseen segmented sporangium form a vesicle enclosing numerous zoospores in a mammalian L. giganteum strain. The kidney-shaped zoospores before release (G) and after release (H, arrows) agree with those in the original description of L. giganteum by Couch (10). ATCC, American Type Culture Collection.
Discussion
We detected a novel group of mammalian pathogenic L. giganteum strains unique for their capacity to experimentally infect invertebrate hosts and cause life-threatening infections in healthy lower animals and humans (5,7). Placement of both heat-sensitive (affecting mosquito larvae) and heat-tolerant (affecting mammals) L. giganteum strains in a strongly supported monophyletic clade in Bayesian analysis corroborates the taxonomic and phenotypic attributes shared by both types in this and other studies (3–5,10–15). In addition to L. giganteum, 3 other clades harbor mammal isolates: L. ajelloi, L. albertoi, and L. vilelae (Figures 1, 2). Strains in the remaining clades are from invertebrates, whether from nematodes (the 4 unnamed species, L. spa, L. spb, L. spc, and L. spd [Table]), crab eggs (L. callinectes), or shrimp (L. myophilum) (17). Lagenidium strains in the 3 clades closest to L. giganteum infect invertebrates, as can all isolates of L. giganteum, suggesting that the mammal-infecting L. giganteum isolates recently evolved the ability to infect warm-blooded hosts. However, the capacity to infect mammals is not unique to L. giganteum, and knowing that each of the 3 other mammal-infecting Lagenidium clades has closely related relatives that infect invertebrates, it seems likely that the trait of infecting mammals has arisen several times, independently, in the genus Lagenidium. From its name, 1 species, L. humanum, might be expected to be among the mammal parasites, but it has been isolated only from the dead skin of humans or snakes and cannot grow at 37°C (9). Regarding the trait of heat tolerance, species that are isolated from mammals, L. ajelloi, L. albertoi, L. giganteum, and L. vilelae also can grow at 37°C, but judging from cultures recovered from lower animals, some strains associated with invertebrates also might lack the heat-tolerance trait. Lagenidium strain Centraalbureau voor Schimmelcultures (CBS) 127042 recovered from nematodes and phylogenetically linked to L. vilelae may be a good example of this unusual feature (Figure 1).
Emergence of mammalian pathogens often is accompanied by host switching in a zoonotic context from 1 vertebrate species to another (18,19), but range extension from invertebrates to vertebrates as hypothesized here is rare. Waterfield et al. (20) outlined 3 scenarios in which bacteria pathogenic to humans might have evolved from pathogens associated with invertebrates, but the time for the process was thousands of years, whereas in our second scenario, we describe a process that might have happened within a decade. The 2 phenotypes of heat-tolerance and mammal pathogenicity appear to have repeatedly evolved in the genus Lagenidium, and several recent studies have shown that members of Oomycota can acquire pathogenicity genes by horizontal gene transfer (21–23). Before their release as agents of biocontrol, L. giganteum (ATCC 52675 = ATCC 36492) (12) and other strains proposed as biocontrol agents of insect were shown not to infect other arthropods and mammals (14,15). However, experimental infection of mammals by the other Oomycota pathogen, P. insidosum, is difficult to achieve, e.g., only rabbits have been successfully, experimentally infected by this pathogen of cats, cattle, dogs, horses, and humans (2). This experience indicates that the safety of Lagenidium species as pathogens of hosts other than mosquito might be difficult to assess because of a general difficulty over experimental infection of mammals by Oomycota species.
The evolutionary relationships among L. giganteum strains should be more thoroughly examined by using population genomics, which could lead to discovery of the basis of adaptation to specific hosts, as has been shown for other adaptive phenotypes in other filamentous microbial eukaryotes (24). The resent transcriptome analysis of a strain of L. giganteum recovered in nature from mosquito larvae further strengthens the relevance of our findings (25). The use of varieties or subspecies to name these 2 populations of L. giganteum was avoided until more comprehensive genomic data shed light on their true evolutionary relationships. Future genomic studies also could discriminate between the scenarios of gaining novel traits in specific ecologic niches or revealing types with hidden capabilities, both of which are of environmental and public health concern when the traits include putative pathogenicity of humans.
Acknowledgments
We thank the many veterinary practitioners and physicians who provided clinical samples from putative cases of lagenidiosis. We also thank R. Morris for comments on the manuscript.
This study was supported by the Department of Microbiology and Molecular Genetics, Michigan State University, and by grants R37AI21884, National Institute of Allergy and Infectious Diseases to E.D.W. and National Science Foundation DEB-125752 to J.W.T.
Biography
Dr. Vilela is a physician with a PhD in medical microbiology who works with unusual infectious fungal and fungal-like microbes at Biomedical Laboratory Diagnostics, Michigan State University. Her research interests include Lacazia loboi, Lagenidium spp., Pythium insidiosum, and Rhinosporidium seeberi.
Footnotes
Suggested citation for this article: Vilela R, Taylor JW, Walker ED, Mendoza L. Lagenidium giganteum pathogenicity in mammals. Emerg Infect Dis [Internet]. 2015 Feb [date cited]. http://dx.doi.org/10.3201/eid2102.141091
These authors contributed equally to this article.
References
- 1.De Cock AW, Mendoza L, Padhye AA, Ajello L, Kaufman L. Pythium insidiosum sp. nov., the etiologic agent of pythiosis. J Clin Microbiol. 1987;25:344–9 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gaastra W, Lipman LJ, De Cock AW, Exel TK, Pegge RB, Scheurwater J, et al. Pythium insidiosum: an overview. Vet Microbiol. 2010;146:1–16. 10.1016/j.vetmic.2010.07.019 [DOI] [PubMed] [Google Scholar]
- 3.Grooters AM. Pythiosis, lagenidiosis, and zygomycosis in small animals. Vet Clin North Am Small Anim Pract. 2003;33:695–720. 10.1016/S0195-5616(03)00034-2 [DOI] [PubMed] [Google Scholar]
- 4.Grooters AM, Hodgin EC, Bauer RW, Detrisac CJ, Znajda NR, Thomas RC. Clinicopathologic findings associated with Lagenidium sp. infection in 6 dogs: initial description of an emerging oomycosis. J Vet Intern Med. 2003;17:637–46. 10.1111/j.1939-1676.2003.tb02494.x [DOI] [PubMed] [Google Scholar]
- 5.Mendoza L, Vilela R. The mammalian pathogenic oomycetes. Curr Fungal Infect Rep. 2013;7:198–208.
- 6.Reinprayoon U, Permpalung N, Kasetsuwan N, Plongla R, Mendoza L, Chindamporn A. Lagenidium sp. ocular infection mimicking ocular pythiosis. J Clin Microbiol. 2013;51:2778–80 and. 10.1128/JCM.00783-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grooters AM, Proia LA, Sutton D, Hodgin EC. Characterization of a previously undescribed Lagenidium pathogen associated with soft tissue infection: initial description of a new human oomycosis [abstract]. Focus on Fungal Infections 14. 2004. Mar 24–26; New Orleans, Louisiana, USA. p. 174. [Google Scholar]
- 8.Schenk A. Algologische mittheilungen. Verhandl Phys Med Gessell Würzburg. 1859;9:12–31. [Google Scholar]
- 9.Karling JS. Predominantly holocarpic and eucarpic simple biflagellate phycomycetes. 2nd ed. Vaduz (Germany): J. Cramer; 1981. p. 89–154. [Google Scholar]
- 10.Couch JN. A new saprophytic species of Lagenidium, with notes and other forms. Mycologia. 1935;27:376–87. 10.2307/3754166 [DOI] [Google Scholar]
- 11.Couch JN, Roney SV. Sexual reproduction in Lagenidium giganteum. Mycologia. 1973;65:250–2. 10.2307/3757815 [DOI] [Google Scholar]
- 12.Kerwin JL, Washino RK. Field evaluation of Lagenidium giganteum (Oomycetes: Lagenidiales) and description of a natural epizootic involving a new isolate of the fungus. J Med Entomol. 1988;25:452–60 . [DOI] [PubMed] [Google Scholar]
- 13.Teng H-J, Lu L-C, Wu Y-L, Fang J-G. Evaluation of various control agents against mosquito larvae in rice paddies in Taiwan. J Vector Ecol. 2005;30:126–32 . [PubMed] [Google Scholar]
- 14.Kerwin JL, Washino RK. Ground and aerial application of the sexual stage of Lagenidium giganteum for control of mosquitoes with rice culture in the central valley of California. J Am Mosq Control Assoc. 1987;3:59–64 . [PubMed] [Google Scholar]
- 15.Kerwin JL, Dritz DA, Washino RK. Confirmation of the safety of Lagenidium giganteum (Oomycetes: Lagenidiales) to mammals. J Econ Entomol. 1990;83:374–6 . [DOI] [PubMed] [Google Scholar]
- 16.Lévesque CA, De Cock AW. Molecular phylogeny and taxonomy of the genus Pythium. Mycol Res. 2004;108:1363–83. 10.1017/S0953756204001431 [DOI] [PubMed] [Google Scholar]
- 17.Rogers-Talbert R. The fungus Lagenidium callinectes Couch (1942) on eggs of the blue crab in Chesapeake Bay. Biol Bull. 1948;95:214–28. 10.2307/1538026 [DOI] [PubMed] [Google Scholar]
- 18.Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, et al. Global trends in emerging infectious diseases. Nature. 2008;451:990–3. 10.1038/nature06536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morens DM, Fauci AS. Emerging infectious diseases: threats to human health and global stability. PLoS Pathog. 2013;9:e1003467. 10.1371/journal.ppat.1003467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waterfield NR, Wren BW, French-Constant RH. Invertebrates as a source of emerging human pathogens. Nat Rev Microbiol. 2004;2:833–41. 10.1038/nrmicro1008 [DOI] [PubMed] [Google Scholar]
- 21.Jiang RHY, Tyler BM. Mechanisms and evolution of virulence in oomycetes. Annu Rev Phytopathol. 2012;50:295–318. 10.1146/annurev-phyto-081211-172912 [DOI] [PubMed] [Google Scholar]
- 22.Seidl MF, Van den Ackerveken G, Govers F, Snel B. Reconstruction of oomycetes genome evolution identities differences in evolutionary trajectories leading to present-day large gene families. Genome Biol Evol. 2012;4:199–211. 10.1093/gbe/evs003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richards TA, Soanes DM, Jones MDM, Vasieva O, Leonard G, Paszkiewicz K, et al. Horizontal gene transfer facilitated the evolution of plant parasitic mechanisms in the oomycetes. Proc Natl Acad Sci U S A. 2011;108:15258–63. 10.1073/pnas.1105100108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ellison CE, Hall C, Kowbel D, Welch J, Brem RB, Glass NL, et al. Population genomics and local adaptation in wild isolates of a model microbial eukaryote. Proc Natl Acad Sci U S A. 2011;108:2831–6. 10.1073/pnas.1014971108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quiroz Velasquez PF, Abiff SK, Fins KC, Conway QB, Salazar NC, Delgado AP, et al. Transcriptome analysis of the entomopathogenic oomycete Lagenidium giganteum reveals putative virulence factors. Appl Environ Microbiol. 2014;80:6427–36. 10.1128/AEM.02060-14 [DOI] [PMC free article] [PubMed] [Google Scholar]


