Abstract
The density and duration of pneumococcal carriage are considered to affect the likelihood of transmission and invasive disease. Because of its importance in both spreading and causing disease, carriage has been suggested as an endpoint in future vaccine studies. Culture is the current gold standard for detection, but may not be sensitive enough to detect changes at low density. Healthy adult volunteers received an intranasal inoculation of Streptococcus pneumoniae serotype 6B. Pneumococcal density in nasal washes collected at six time-points post-inoculation was determined by culture and quantitative PCR (qPCR). Natural pneumococcal carriers detected at initial screening were followed in parallel. In 331 nasal washes from 79 volunteers, the sensitivity and specificity of pneumococcal detection by qPCR, as compared with culture, were 92.3% and 75.9%. The estimation of pneumococcal density by culture and qPCR was highly correlated (rs = 0.73, p <0.0001), although qPCR had a lower detection limit. Pneumococcal density fluctuated within a carriage episode, and occasionally fell below the detection limit of both methods. The duration of carriage episodes was underestimated when only one method was used. Similar fluctuations in density were observed in natural carriers. Pneumococcal carriage is a dynamic event. Culture and qPCR are complementary for surveying the density and duration of pneumococcal carriage episodes.
Keywords: Carriage, culture, density, qPCR, Streptococcus pneumoniae
Introduction
Colonization of the nasopharynx by Streptococcus pneumoniae (pneumococcus) is common, with carriage rates in children aged <5 years of 40–90%, and stabilizing at approximately 10% in adults [1–3]. Understanding colonization is important, as it is implicated in both person-to-person transmission and spread within the body, potentially leading to meningitis, bacteraemia, or pneumonia. To what extent the density and duration of pneumococcal carriage affect the risk of transmission and invasive disease is being increasingly scrutinized [4,5].
The WHO currently recommends nasopharyngeal swabs to detect pneumococcal carriage in children [6]. In adults, the best sampling method is less clear. We have previously suggested that nasal wash (NW) is better than nasopharyngeal swabs for detecting potential nasopharyngeal pathogens, and it provides the opportunity to examine the immune response [7]. Recently, trans-oral swabs have been suggested to be superior to trans-nasal swabs for detecting pneumococcal carriage in adults [8]. Detection of live pneumococci by culture is the current gold standard [6]. However, culture and molecular methods can be combined to increase carriage detection, including detection of multiple serotypes [9]. This can be achieved by including an enrichment step prior to culture and molecular analysis, or by combining data from both culture and molecular methods, which allows density to be determined for each method separately [10].
Understanding carriage is also important in the context of vaccination. Pneumococcal conjugate vaccines have resulted in a decrease in the rates of invasive disease and carriage of vaccine-serotype pneumococci [11,12]. However, whereas carriage of vaccine-type pneumococci has declined, carriage of non-vaccine-type pneumococci has increased [13,14]. The long-term impact of vaccination can be measured by using carriage data, which can provide an estimate of post-vaccine changes in invasive disease incidence [15]. Colonization has been suggested as an endpoint because it is relatively easy to measure and is more common than disease, and therefore requires a much smaller sample size in a vaccine trial [16–18]. However, to identify small changes in the density or duration of colonization, sensitive methods to accurately detect these changes will be important in estimating vaccine effects.
In this study, we used an experimental human pneumococcal carriage model to longitudinally follow carriage episodes, and compared detection of carriage by culture and quantitative PCR (qPCR). We focused on carriage density and duration in experimental carriers, but also compared both parameters in natural carriers, who were followed in parallel.
Materials and Methods
Recruitment and ethics
Healthy volunteers were enrolled, with informed consent, in an experimental human pneumococcal carriage trial [19]. All participants were non-smoking adults aged 18–60 years who had no close contact with at-risk individuals, including young children and the elderly. Ethical approval was obtained from the National Health Service Research Ethics Committee (11/NW/0592), and the study was sponsored by the Royal Liverpool and Broadgreen University Hospitals Trust.
Experimental nasopharyngeal challenge
Pneumococcal stock preparation and inoculation were performed as previously described [19]. Briefly, a serotype 6B clinical isolate (BHN418) [20] was grown to an optical density at 600 nm of 0.2–0.3 in Vegitone broth (Oxoid, Basingstoke, UK), and stored in 1-mL aliquots containing 10% glycerol at −80°C. Serotype confirmation was performed by latex agglutination (Statens Serum Institute, Copenhagen, Denmark), and bacterial stock purity and penicillin sensitivity were confirmed by an independent reference laboratory (Public Health England, London, UK). Eighty-four volunteers received a pneumococcal inoculation of between 10 000 and 320 000 CFUs per naris.
Quantification of pneumococci by culture
NW samples were collected before inoculation and at days 2, 7, 14, 21, 28 and 35 post-inoculation, and were processed as previously described [19]. If pneumococci were detected at the pre-inoculation screening, the volunteer was not challenged, but returned for a NW at the same time-points as those in whom pneumococci were detected. Plates were inspected after 24 h of incubation at 37°C in 5% CO2. α-haemolytic, draughtsman-like colonies were Gram-stained and subcultured for optochin sensitivity and bile solubility. Pneumococcal serotype was confirmed by latex agglutination.
Carriage density by culture was determined as previously described, with minor modifications [19]. Briefly, the NW bacterial pellet was resuspended in 100 μL of skimmed-milk tryptone glucose glycerol (STGG) medium, and the total volume of the suspension was determined. The suspension was then serially diluted on blood agar. On the next day, CFUs/μL were determined, and multiplied by the suspension volume. This value was then divided by the amount of NW returned by the volunteer to obtain CFUs/mL of NW.
Bacterial DNA extraction
Directly after NW collection, 2 mL was added to 4 mL of RNAprotect Bacteria Reagent (Qiagen, Manchester, UK). After 5 min of incubation at room temperature, the sample was transferred to the laboratory on ice and stored at −80°C. The thawed suspension was pelleted in a 2-mL tube by three centrifugation steps at 19 090 g for 20 min at 4°C. The pellet was resuspended in 0.3 mL of lysis buffer with protease (Agowa Mag mini DNA extraction kit; LGC Genomics, Berlin, Germany), 50 mg of sterilized zirconia/silica beads (diameter of 0.1 mm; Biospec Products, Bartlesville, OK, USA), and 0.3 mL of phenol (Phenol BioUltra; Sigma-Aldrich, Zwijndrecht, The Netherlands). The sample was mechanically disrupted by bead beating in a TissueLyser LT (Qiagen, Venlo, The Netherlands) twice at 50 Hz for 2 min. After centrifugation, the aqueous phase was transferred to a sterile 1.5-mL tube. Binding buffer was added at twice the volume of the aqueous phase plus 10 μL of magnetic beads, after which the sample was incubated in a mixing machine for 30 min at room temperature. The magnetic beads were washed with 200 μL of both wash buffer 1 and wash buffer 2, and eluted with 63 μL of elution buffer, according to the manufacturer's instructions.
Quantification of pneumococcal DNA by qPCR
Carriage density by qPCR was determined by partial amplification of the lytA gene. The primer and probe sequences were as follows: forward primer, 5′-ACGCAATCTAGCAGATGAAGCA-3′; reverse primer, 5′-TCGTGCGTTTTAATTCCAGCT-3′; probe, 5′-(FAM)-GCCGAAAACGCTTGATACAGGGAG-(BHQ1)-3′ [21]. The 20-μL PCR mix consisted of 1 × TaqMan Universal PCR Master Mix (Life Technologies, Bleiswijk, The Netherlands), 0.1 μM each primer, 0.1 μM probe, and 1 μL of the extracted DNA. Thermal cycling was performed in an ABI 7500 Fast Real-Time PCR System (Life Technologies) under the following cycling conditions: 2 min at 50°C, 10 min at 95°C, and 40 cycles of 15 s at 95°C and 1 min at 60°C. A standard curve of a ten-fold dilution series of genomic DNA extracted from S. pneumoniae (TIGR4, ATCC BAA-334) was used. The genomic DNA was extracted with the Qiagen Genomic-tip 20/G Kit (Qiagen), and quantified with a spectrophotometer (Nanodrop ND-1000; Thermo Fisher Scientific, Landsmeer, The Netherlands). The conversion from weight of pneumococcal DNA to number of S. pneumoniae DNA copies was based on the weight of one genome copy of TIGR4 calculated as the genome length in base pairs times the weight of a DNA base pair (650 Da). The lower limit of detection was set at 40 cycles. A NW was considered to be positive if both duplicates yielded a qPCR signal below 40 cycles.
Statistical analysis
Differences in proportions of samples positive for carriage were statistically tested with a chi-square test (Fisher's exact test if there were more than ten cases in a cell). Quantitative data were compared by the use of Spearman's rank correlation coefficient. Graphical and statistical analyses were performed with GraphPad prism version 5.0 (GraphPad Software, La Jolla, CA, USA). All p-values were two-tailed, and a p-value of ≤0.05 was considered to be significant.
Results
Comparison of culture and qPCR for the detection of pneumococci
Three hundred and thirty-one NW samples collected from 79 volunteers were tested by microbiological culture and qPCR for the presence of S. pneumoniae. The proportion of samples positive for carriage by qPCR was significantly higher than the proportion of samples positive for carriage by culture (42.6% vs. 27.5%, p <0.0001) (Table 1). The sensitivity of detection by qPCR was 92.3% (84/91) as compared with culture, and the specificity was 75.9% (183/241). In 2.1% of samples, pneumococci were detected by culture but not qPCR, and in 17.2% of samples pneumococci were detected only by qPCR.
TABLE 1.
Comparison of bacterial culture and quantitative PCR (qPCR) for the detection of Streptococcus pneumoniae
| Culture-positive (%) | Culture-negative (%) | Total (%) | |
|---|---|---|---|
| qPCR-positive (%) | 84 (25.4) | 57 (17.2) | 141 (42.6) |
| qPCR-negative (%) | 7 (2.1) | 183 (55.3) | 190 (57.4) |
| Total | 91 (27.5) | 241 (72.8) | 331 (100) |
The proportion of carriage-positive volunteers was also determined over time (Fig. 1). Across all time-points, the number of volunteers positive for carriage by qPCR was higher than the number positive for carriage by culture, but this was only significant at 2 days post-challenge (p 0.009).
FIG 1.
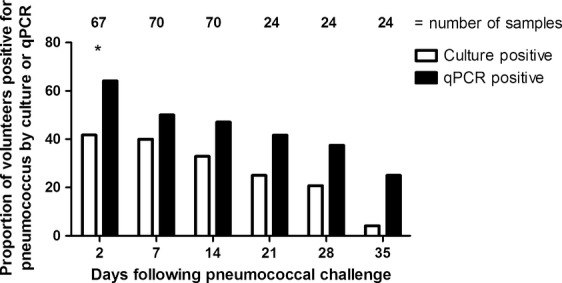
Proportion of carriage-positive volunteers detected by bacterial culture or quantitative PCR (qPCR) over time. For detection by culture, nasal wash samples were plated on blood agar with gentamicin. For qPCR detection, nasal wash was added to RNAprotect and frozen until DNA extraction. A volunteer was considered to be positive by culture (white bar) if Gram-positive, α-haemolytic, optochin-sensitive colonies were detected after 48 h. Serotype was confirmed by latex agglutination. The target gene for qPCR was lytA. Samples with a Ct value of <40 were considered to be qPCR-positive (black bar). The number of samples analysed is indicated above each time-point. *Statistical significance (p ≤0.05)
Correlation of density detected by culture and by qPCR
The pneumococcal density correlation between culture and qPCR was determined for 147 samples. One sample was excluded from analysis because the number of colonies on the culture plate was too high for accurate determination of CFUs/mL. Quantification of pneumococci by culture and by qPCR were positively correlated (rs = 0.73, p <0.0001) (Fig. 2). The detection limit for qPCR was the number of copies still detectable after 40 cycles, or 101 DNA copies/mL. The limit of detection for culture was set at the lowest density detected, or 0.2 CFUs/mL.
FIG 2.
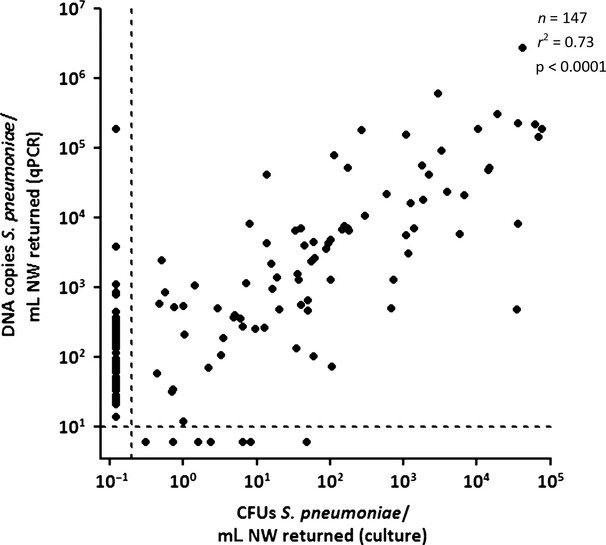
Correlation between bacterial culture and qPCR in quantifying Streptococcus pneumoniae in nasal wash (NW) samples. Quantification of pneumococci by culture and by qPCR are positively correlated. The Spearman rank correlation coefficient for samples positive by both culture and qPCR is 0.73. Dotted lines represent lower limits of detection. A p-value of ≤0.05 was considered to be significant.
When pneumococcal detection by culture was stratified by qPCR density, the difference between the two methods became apparent. If the qPCR density was >103 copies/mL, 94.8% (55/58) of samples were both culture-positive and qPCR-positive (Table 2). However, at densities ranging between 101 and 102, only 34.55% (29/84) of qPCR-positive samples were also positive by culture (p <0.0001). Below the qPCR limit of detection, 7.7% (7/91) of samples were still positive by culture. In all but one of these samples, the culture density was lower than 10 CFUs/mL.
TABLE 2.
Detection of pneumococci in nasal wash by bacterial culture and quantitative PCR (qPCR), categorized according to qPCR density
| Density by qPCR (copies/mL) | No. culture-positive/no. qPCR-positive (%) |
|---|---|
| <10 | 7/0 |
| 101 | 6/37 (16.2) |
| 102 | 23/47 (48.9) |
| 103 | 30/32 (93.8) |
| 104 | 14/14 (100) |
| 105 | 11/12 (91.7) |
| Total | 91/142 (64.1) |
Fluctuations in carriage density are accurately detected when both culture and qPCR are used for detection
Culture and qPCR were used to examine pneumococcal detection over time in four experimentally colonized and three naturally colonized volunteers. The fluctuations in density during a carriage episode were similar for both experimentally and naturally colonized volunteers (Fig. 3). As shown in Fig. 3a, volunteer 1 was culture-negative but qPCR-positive at two consecutive time-points (days 7 and 14). In contrast, volunteer 2 was culture-positive but qPCR-negative on day 28. In both cases, the carriage status varied according to the method. This variation was also seen in natural carriers (Fig. 3c). Both methods were beneficial for determining the length of a carriage episode. Volunteers 3 and 4 (Fig. 3b) had at least two samples positive by both culture and qPCR, but then at least three consecutive samples negative by culture but positive by qPCR. This pattern was also seen in natural carriers of serotype 33 (Fig. 3c).
FIG 3.
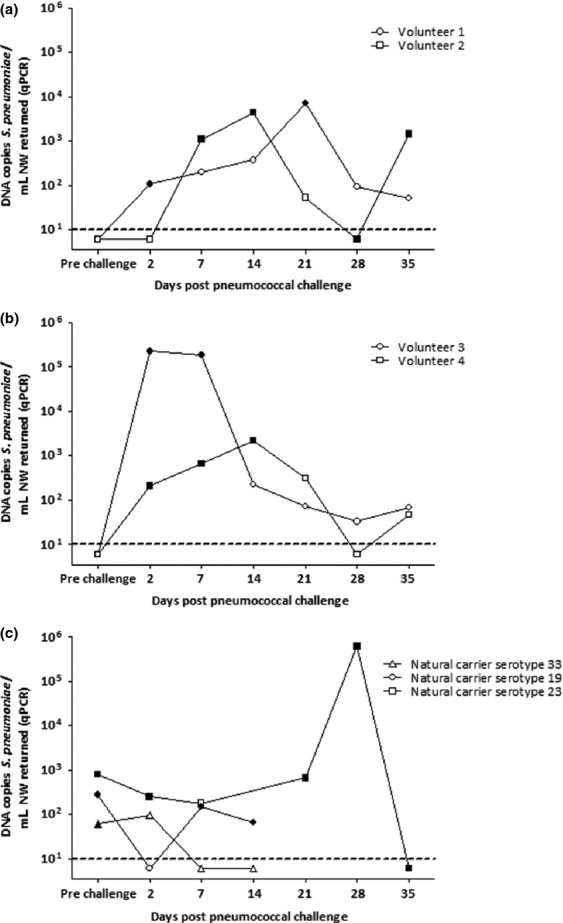
Culture and quantitative PCR (qPCR) are complementary for following a carriage episode. (a, b) At low density, carriage results can vary by method, which can impact on (a) the number of carriage episodes and (b) the duration of carriage episodes. (c) Natural carriage episodes mimic the variation seen in experimental carriage. Filled shapes: carriage-positive by culture. Open shapes: carriage-negative by culture. Dotted lines represent lower limits of detection. NW, nasal wash.
Discussion
Longitudinal NW samples from adults who were intranasally challenged with pneumococci were assessed for carriage density by culture and qPCR. We detected a higher pneumococcal carriage rate with qPCR than with culture but pneumococcal densities measured with the two methods were positively correlated. The culture positivity rate decreased in parallel with the decrease in density measured by qPCR. Culture and qPCR were complementary in assessing the number and length of carriage episodes in both experimental and natural carriage.
The strength of this study is the determination of density by both culture and qPCR during a controlled carriage episode. We observed a high correlation between the two methods in calculating density. Absolute numbers were 1–2 log10 lower by culture, which is in line with observations previously reported by Albrich et al. [4]. The difference may be attributable to the inability of qPCR to distinguish between live and dead bacteria—an important distinction in low-density samples. Lower culture density may also be a result of the pneumococcal tendency to grow in pairs and chains. More than one pneumococcus can form a single colony, but qPCR will detect each individual bacterium.
Detection of pneumococcal carriage by culture is the gold standard [6] and Isolation of the pneumococcus allows further tests to be performed to determine serotype and antibiotic sensitivity. Culture is also more feasible in resource-limited settings. Non-culture methods are increasingly being used to detect pneumococci. Several studies have focused on gene targets by using qPCR, with lytA being the most widely used gene, and have shown that a qPCR-based method is more sensitive than culture [21,22]. There have been concerns over the specificity of lytA and the possibility that it may not discriminate between S. pneumoniae and viridans streptococci; however, our specificity (75.9%) was similar to that reported by Carvalho et al. [21] in cerebrospinal fluid samples (70%). To increase sensitivity, some studies have included an enrichment step, but this cannot be used when quantification is an endpoint [8,10].
A lower detection rate by culture than by qPCR could be explained by the potential for subclinical—or low–density—carriage. Turner et al. [9] showed that detection of multiple serotypes in nasopharyngeal samples is significantly underestimated when using standard WHO protocol, which is able to detect a predominant serotype but is not efficient at detecting low-abundance serotypes. This is linked with serotype replacement, where it is unclear whether the increase in non-vaccine-type pneumococci is attributable to the elimination of vaccine-type pneumococci, or whether the replacement occurs because non-vaccine types are carried at low density and are simply ‘unmasked’ when vaccine types are inhibited or removed [11,13]. Low-density carriage could explain the differences between culture and qPCR seen here; at a density of <102, the low abundance is missed by culture but picked up by qPCR.
It has been shown in children that, at a density of <105 CFUs/mL, significantly fewer carriers are detected by culture, not only for pneumococcus but also for Staphylococcus aureus and Haemophilus influenzae [22]. However, as we have shown here and discussed previously, we are able to detect carriage by culture at <10 copies/mL, and suggest that, when a fresh NW sample is used, culture and qPCR are equally sensitive up to 102 CFUs/mL [23].
The parallel decreases in culture positivity rates and pneumococcal densities measured by qPCR suggest that qPCR is more suitable for detecting low levels of carriage. However, qPCR is not informative regarding the viability of pneumococci in the nasopharynx. As seen in Fig. 3b, continuous qPCR detection when culture results are negative may represent prolonged low-density carriage, living cells that are in a culture-unfavourable metabolic state, or remaining pneumococcal debris. Also, intermittent positive culture results could be misconstrued as separate carriage episodes when they actually represent a single carriage episode as supported by qPCR detection (Fig. 3a). With application of both methods, the chance of missing pneumococcal carriage may be reduced. In our controlled environment, the most advantageous approach for the longitudinal study of a carriage episode is to use both methods.
Inconsistencies between culture and qPCR detection could also be explained by decreasing success rates at the threshold of detection. Culture densities were low (<10 CFUs/mL) in six of seven qPCR-negative samples, and, in 54 of 57 culture-negative samples, qPCR densities were <103 copies/mL. The higher density inconsistencies could be attributable to experimental error; culture-positive/qPCR-negative samples could be the result of faulty DNA extraction or the exclusion of qPCR signals of <10 copies/mL (>40 cycles). Four of the seven exclusively culture-positive samples were analysed again with a different lytA primer set, but remained negative for pneumococcal DNA [24]. We chose a threshold of 40 cycles, which was used in the development of the lytA qPCR assay. We observed that the success rate of detection with the standard curve was still 100% at 40 cycles, and that the standard curve still followed a straight line at 40 cycles. In addition, almost all samples detected between 35 and 40 cycles were positive in both duplicates. These observations imply that the lower limit of detection of our method had not yet been reached at 40 cycles. This cut-off is two cycles higher than that used in the study with children, possibly explaining the differences in detection limits between the two studies [21,22].
Our study surveyed adult pneumococcal carriage in consecutive NW samples. However, this sampling method is less suited for those who are unable to actively participate in sample collection (e.g. young children). Further studies will need to determine whether our observations hold true for different sampling methods and populations.
Carriage is a dynamic event, and we have shown that an experimental human carriage model mimics a natural carriage episode. The data from our study indicate that culture and qPCR are highly complementary for studying pneumococcal carriage episodes, especially when the carriage density is low. This is important information for future vaccine studies in which carriage density or duration may be an endpoint.
Acknowledgments
The authors thank B. Henriques-Normark (Karolinska Institutet) for donating the 6B pneumococcal strain, the clinical research team: Angela D. Wright, Andrea M. Collins, Carole A. Hancock, and David Shaw, and all of the volunteers who participated in the study. The study was co-sponsored by the Royal Liverpool and Broadgreen University Hospitals NHS Trust and the Liverpool School of Tropical Medicine. This work was supported by the Bill and Melinda Gates Foundation (OPP1035281 Grand Challenge Exploration award to S. B. Gordon), the National Institutes for Health Research Comprehensive Local Research Network, and the EU-FP7 (ENIAC Joint Undertaking—CAJAL4EU). This work was presented at the Society for General Microbiology Annual Conference, April 2014, in Liverpool, UK as a poster.
Transparency Declaration
All authors declare that no conflicts of interest.
References
- 1.Adetifa IM, Antonio M, Okoromah CA, et al. Pre-vaccination nasopharyngeal pneumococcal carriage in a Nigerian population: epidemiology and population biology. PLoS One. 2012;7:e30548. doi: 10.1371/journal.pone.0030548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hussain M, Melegaro A, Pebody RG, et al. A longitudinal household study of Streptococcus pneumoniae nasopharyngeal carriage in a UK setting. Epidemiol Infect. 2005;133:891–898. doi: 10.1017/S0950268805004012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Regev-Yochay G, Raz M, Dagan R, et al. Nasopharyngeal carriage of Streptococcus pneumoniae by adults and children in community and family settings. Clin Infect Dis. 2004;38:632–639. doi: 10.1086/381547. [DOI] [PubMed] [Google Scholar]
- 4.Albrich WC, Madhi SA, Adrian PV, et al. Use of a rapid test of pneumococcal colonization density to diagnose pneumococcal pneumonia. Clin Infect Dis. 2012;54:601–609. doi: 10.1093/cid/cir859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stralin K, Herrmann B, Abdeldaim G, Olcen P, Holmberg H, Molling P. Comparison of sputum and nasopharyngeal aspirate samples and of the PCR gene targets lyta and spn9802 for quantitative PCR for rapid detection of pneumococcal pneumonia. J Clin Microbiol. 2014;52:83–89. doi: 10.1128/JCM.01742-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Satzke C, Turner P, Virolainen-Julkunen A, et al. Standard method for detecting upper respiratory carriage of Streptococcus pneumoniae: updated recommendations from the World Health Organization pneumococcal carriage working group. Vaccine. 2013;32:165–179. doi: 10.1016/j.vaccine.2013.08.062. [DOI] [PubMed] [Google Scholar]
- 7.Gritzfeld JF, Roberts P, Roche L, El Batrawy S, Gordon SB. Comparison between nasopharyngeal swab and nasal wash, using culture and PCR, in the detection of potential respiratory pathogens. BMC Res Notes. 2011;4:122. doi: 10.1186/1756-0500-4-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trzcinski K, Bogaert D, Wyllie A, et al. Superiority of trans-oral over trans-nasal sampling in detecting Streptococcus pneumoniae colonization in adults. PLoS One. 2013;8:e60520. doi: 10.1371/journal.pone.0060520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turner P, Hinds J, Turner C, et al. Improved detection of nasopharyngeal cocolonization by multiple pneumococcal serotypes by use of latex agglutination or molecular serotyping by microarray. J Clin Microbiol. 2011;49:1784–1789. doi: 10.1128/JCM.00157-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.da Gloria Carvalho M, Pimenta FC, Jackson D, et al. Revisiting pneumococcal carriage by use of broth enrichment and PCR techniques for enhanced detection of carriage and serotypes. J Clin Microbiol. 2010;48:1611–1618. doi: 10.1128/JCM.02243-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Brien KL, Millar EV, Zell ER, et al. Effect of pneumococcal conjugate vaccine on nasopharyngeal colonization among immunized and unimmunized children in a community-randomized trial. J Infect Dis. 2007;196:1211–1220. doi: 10.1086/521833. [DOI] [PubMed] [Google Scholar]
- 12.Mbelle N, Huebner RE, Wasas AD, Kimura A, Chang I, Klugman KP. Immunogenicity and impact on nasopharyngeal carriage of a nonavalent pneumococcal conjugate vaccine. J Infect Dis. 1999;180:1171–1176. doi: 10.1086/315009. [DOI] [PubMed] [Google Scholar]
- 13.Weinberger DM, Malley R, Lipsitch M. Serotype replacement in disease after pneumococcal vaccination. Lancet. 2011;378:1962–1973. doi: 10.1016/S0140-6736(10)62225-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bottomley C, Roca A, Hill PC, Greenwood B, Isham V. A mathematical model of serotype replacement in pneumococcal carriage following vaccination. J R Soc Interface. 2013;10:20130786. doi: 10.1098/rsif.2013.0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weinberger DM, Bruden DT, Grant LR, et al. Using pneumococcal carriage data to monitor postvaccination changes in invasive disease. Am J Epidemiol. 2013;178:1488–1495. doi: 10.1093/aje/kwt156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldblatt D, Ramakrishnan M, O'Brien K, Grp P-WM. Using the impact of pneumococcal vaccines on nasopharyngeal carriage to aid licensing and vaccine implementation; a Pneumocarr meeting report March 27–28, 2012, Geneva. Vaccine. 2013;32:146–152. doi: 10.1016/j.vaccine.2013.06.040. [DOI] [PubMed] [Google Scholar]
- 17.Auranen K, Rinta-Kokko H, Goldblatt D, et al. Colonisation endpoints in Streptococcus pneumoniae vaccine trials. Vaccine. 2013;32:153–158. doi: 10.1016/j.vaccine.2013.08.061. [DOI] [PubMed] [Google Scholar]
- 18.Auranen K, Rinta-Kokko H, Goldblatt D, et al. Design questions for Streptococcus pneumoniae vaccine trials with a colonisation endpoint. Vaccine. 2013;32:159–164. doi: 10.1016/j.vaccine.2013.06.105. [DOI] [PubMed] [Google Scholar]
- 19.Gritzfeld JF, Wright AD, Collins AM, et al. Experimental human pneumococcal carriage. J Vis Exp. 2013;72:50115. doi: 10.3791/50115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Browall S, Norman M, Tangrot J, et al. Intraclonal variations among Streptococcus pneumoniae isolates influence the likelihood of invasive disease in children. J Infect Dis. 2014;209:377–388. doi: 10.1093/infdis/jit481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carvalho Mda G, Tondella ML, McCaustland K, et al. Evaluation and improvement of real-time PCR assays targeting lyta, ply, and psaa genes for detection of pneumococcal DNA. J Clin Microbiol. 2007;45:2460–2466. doi: 10.1128/JCM.02498-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chien YW, Vidal JE, Grijalva CG, et al. Density interactions among Streptococcus pneumoniae Haemophilus influenzae and Staphylococcus aureus in the nasopharynx of young Peruvian children. Pediatr Infect Dis J. 2013;32:72–77. doi: 10.1097/INF.0b013e318270d850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gritzfeld JF, Gordon SB, Cremers A. Detection limits in pneumococcal carriage. Pediatr Infect Dis J. 2013;32:425–426. doi: 10.1097/INF.0b013e31827add5f. [DOI] [PubMed] [Google Scholar]
- 24.McAvin JC, Reilly PA, Roudabush RM, et al. Sensitive and specific method for rapid identification of Streptococcus pneumoniae using real-time fluorescence PCR. J Clin Microbiol. 2001;39:3446–3451. doi: 10.1128/JCM.39.10.3446-3451.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]


