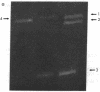
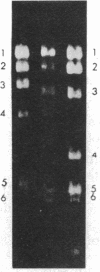
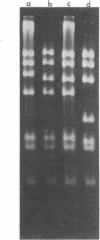
Abstract
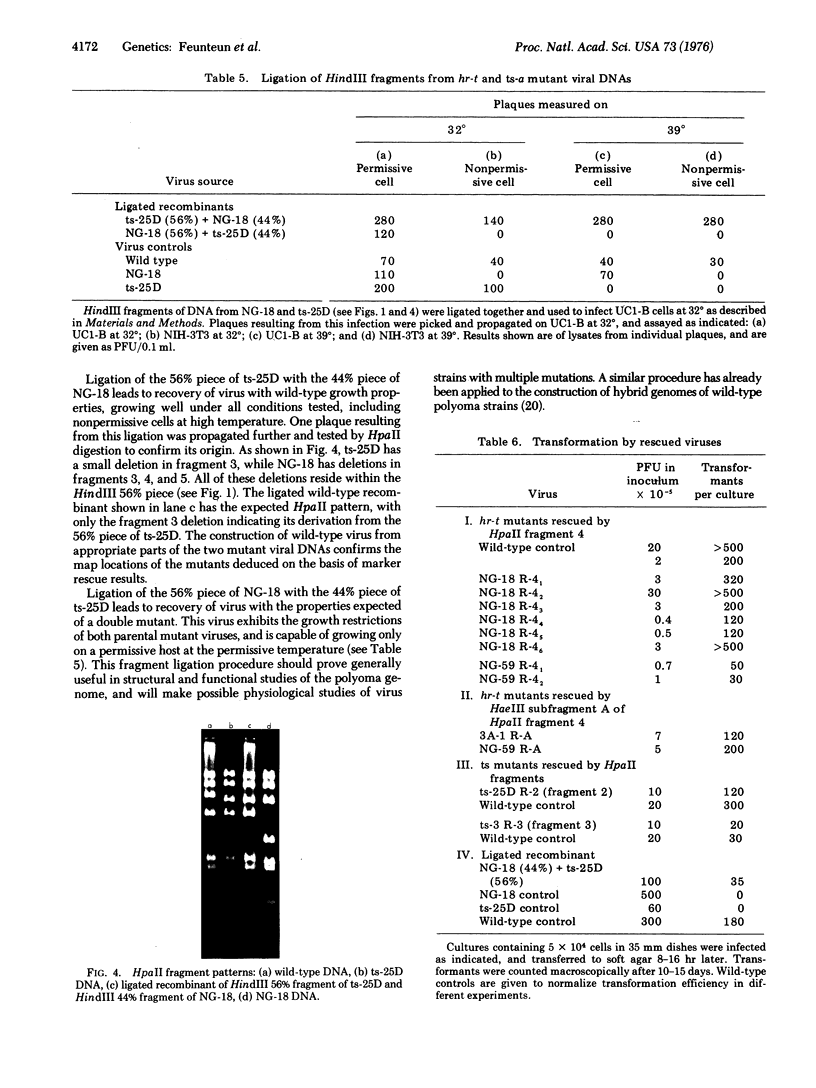
Polyoma virus mutants of four functionally distinct groups have been mapped by the marker rescue technique using restriction enzyme fragments of wild-type viral DNA. Nontransforming host-range mutants map in the proximal part of the early region of the viral genome. The same DNA fragment that restores a normal host range also restores normal transforming ability to these mutants. ts-25D, a temperature-sensitive (ts)-a class mutant, maps in the distal part of the early region. ts-3 and ts-1260 map in the proximal and distal parts of the late region, respectively.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barzilai R. SV40 DNA: quantitative conversion of closed circular to open circular form by an ethidium bromide-restricted endonuclease. J Mol Biol. 1973 Mar 15;74(4):739–742. doi: 10.1016/0022-2836(73)90062-4. [DOI] [PubMed] [Google Scholar]
- Benjamin T. L. Host range mutants of polyoma virus. Proc Natl Acad Sci U S A. 1970 Sep;67(1):394–399. doi: 10.1073/pnas.67.1.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Mayorca G., Callender J., Marin G., Giordano R. Temperature-sensitive mutants of polyoma virus. Virology. 1969 May;38(1):126–133. doi: 10.1016/0042-6822(69)90134-2. [DOI] [PubMed] [Google Scholar]
- Echkart W. Genetics of DNA tumor viruses. Annu Rev Genet. 1974;8:301–317. doi: 10.1146/annurev.ge.08.120174.001505. [DOI] [PubMed] [Google Scholar]
- Eckhart W. Complementation and transformation by temperature-sensitive mutants of polyoma virus. Virology. 1969 May;38(1):120–125. doi: 10.1016/0042-6822(69)90133-0. [DOI] [PubMed] [Google Scholar]
- Eckhart W., Dulbecco R. Properties of the ts3 mutant of polyoma virus during lytic infection. Virology. 1974 Aug;60(2):359–369. doi: 10.1016/0042-6822(74)90331-6. [DOI] [PubMed] [Google Scholar]
- Goldman E., Benjamin T. L. Analysis of host range of nontransforming polyoma virus mutants. Virology. 1975 Aug;66(2):372–384. doi: 10.1016/0042-6822(75)90210-x. [DOI] [PubMed] [Google Scholar]
- Hamer D. H., Thomas C. A., Jr Molecular cloning of DNA fragments produced by restriction endonucleases Sa1I and BamI. Proc Natl Acad Sci U S A. 1976 May;73(5):1537–1541. doi: 10.1073/pnas.73.5.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Hutchison C. A., 3rd, Edgell M. H. Genetic assay for small fragments of bacteriophage phi X174 deoxyribonucleic acid. J Virol. 1971 Aug;8(2):181–189. doi: 10.1128/jvi.8.2.181-189.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamen R., Lindstrom D. M., Shure H., Old R. W. Virus-specific RNA in cells productively infected or transformed by polyoma virus. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 1):187–198. doi: 10.1101/sqb.1974.039.01.025. [DOI] [PubMed] [Google Scholar]
- Lai C. J., Nathans D. A map of temperature-sensitive mutants of simian virus 40. Virology. 1975 Jul;66(1):70–81. doi: 10.1016/0042-6822(75)90179-8. [DOI] [PubMed] [Google Scholar]
- Lai C. J., Nathans D. Mapping temperature-sensitive mutants of simian virus 40: rescue of mutants by fragments of viral DNA. Virology. 1974 Aug;60(2):466–475. doi: 10.1016/0042-6822(74)90340-7. [DOI] [PubMed] [Google Scholar]
- MACPHERSON I., MONTAGNIER L. AGAR SUSPENSION CULTURE FOR THE SELECTIVE ASSAY OF CELLS TRANSFORMED BY POLYOMA VIRUS. Virology. 1964 Jun;23:291–294. doi: 10.1016/0042-6822(64)90301-0. [DOI] [PubMed] [Google Scholar]
- Mantel N., Boyer H. W., Goodman H. M. Mapping simian virus 40 mutants by construction of partial heterozygotes. J Virol. 1975 Sep;16(3):754–757. doi: 10.1128/jvi.16.3.754-757.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller L. K., Fried M. Construction of infectious polyoma hybrid genomes in vitro. Nature. 1976 Feb 19;259(5544):598–601. doi: 10.1038/259598a0. [DOI] [PubMed] [Google Scholar]
- Miller L. K., Fried M. Construction of the genetic map of the polyoma genome. J Virol. 1976 Jun;18(3):824–832. doi: 10.1128/jvi.18.3.824-832.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxman M. N., Takemoto K. K., Eckhart W. Polyoma T antigen synthesis by temperature-sensitive mutants of polyoma virus. Virology. 1972 Sep;49(3):675–682. doi: 10.1016/0042-6822(72)90524-7. [DOI] [PubMed] [Google Scholar]
- Paulin D., Cuzin F. Polyoma virus T antigen. I. Synthesis of modified heat-labile T angiten in cells transformed with the ts-a mutant. J Virol. 1975 Feb;15(2):393–397. doi: 10.1128/jvi.15.2.393-397.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffhausen B. S., Benjamin T. L. Deficiency in histone acetylation in nontransforming host range mutants of polyoma virus. Proc Natl Acad Sci U S A. 1976 Apr;73(4):1092–1096. doi: 10.1073/pnas.73.4.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. A., Sugden B., Sambrook J. Detection of two restriction endonuclease activities in Haemophilus parainfluenzae using analytical agarose--ethidium bromide electrophoresis. Biochemistry. 1973 Jul 31;12(16):3055–3063. doi: 10.1021/bi00740a018. [DOI] [PubMed] [Google Scholar]
- Shenk T. E., Carbon J., Berg P. Construction and analysis of viable deletion mutants of simian virus 40. J Virol. 1976 May;18(2):664–671. doi: 10.1128/jvi.18.2.664-671.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]