Summary
Phosphonates (C-PO32−) have application as antibiotics, herbicides and detergents. In some environments, these molecules represent the predominant source for phosphorus, and several microbes have evolved dedicated enzymatic machineries for phosphonate degradation. For example, most common naturally occurring phosphonates can be catabolized to either phosphonoacetaldehyde or phosphonoacetate, which can then be hydrolyzed to generate inorganic phosphate and acetaldehyde or acetate, respectively. The phosphonoacetaldehyde oxidase gene (phnY) links these two hydrolytic processes and provides a previously unknown catabolic mechanism for phosphonoacetate production in the microbial metabolome. Here, we present biochemical characterization of PhnY and high-resolution crystal structures of the apo state, as well as complexes with substrate, cofactor and product. Kinetic analysis of active site mutants demonstrates how a highly conserved aldehyde dehydrogenase active site has been modified in nature to generate activity with a phosphonate substrate.
INTRODUCTION
Phosphonates are ubiquitous molecules characterized by the presence of an inert carbon-phosphorus (C-PO32−) bond in contrast to a labile oxygen-phosphorus (O-PO32−) bond found in phosphoric esters. Phosphonates are nearly isosteric with the corresponding phosphoric esters and preserve the tetrahedral geometry and net charge distribution around the phosphorus atom, but are recalcitrant towards chemical and enzymatic hydrolytic reactions that can cleave the more labile O-PO32− bond. Thus, phosphonates can act as competitive inhibitors of biochemical processes that involve phosphoryl group transfer reactions (Metcalf and van der Donk, 2009). Nature has utilized this chemical mimicry for the development of several natural product phosphonates that have potent antibiotic, herbicidal, and other bio-inhibitory activities.
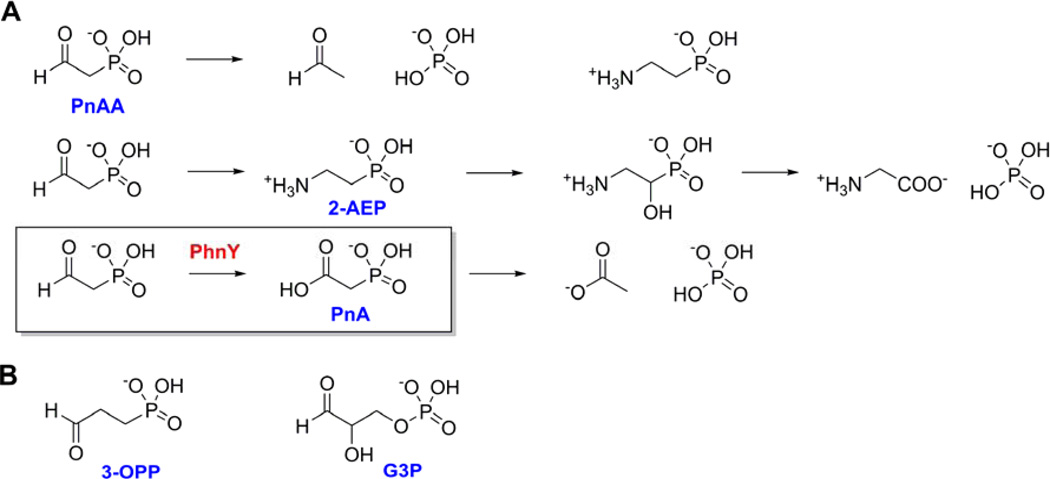
The stability of the C-PO32− bond renders phosphonate molecules as reservoirs for carbon and phosphorus in various microbial niches. In order to take advantage of these resources in nutrient scarce environments, nature has evolved pathways for the catabolism of reduced phosphorus compounds for their subsequent utilization as carbon and phosphate sources (McGrath et al., 2013; Nair and van der Donk, 2011; Peck and van der Donk, 2013). The genetic basis of utilization of phosphite by marine Prochlorococcus (Martinez et al., 2012) and other bacteria has been determined (Costas et al., 2001). Furthermore, the biosynthesis of methylphosphonate by the marine bacterium Nitrosopumilus maritimus (Metcalf et al., 2012) is postulated to result in its utilization by the widely distributed C-P lyase pathway (Hove-Jensen et al., 2011, 2012; Kamat et al., 2011) leading to production of methane in aerobic oceanic ecosystems. The most widely occurring natural phosphonate, 2-aminoethyl phosphonate (2-AEP), is degraded by at least three other experimentally characterized processes (Figure 1A) in addition to the C-P lyase pathway. Phosphonoacetaldehyde (PnAA), produced by transamination of 2-AEP, is enzymatically hydrolyzed to yield acetaldehyde and inorganic phosphate (Pi) (Morais et al., 2000). Alternatively, 2-AEP can be hydroxylated and then oxidized to yield glycine and Pi (McSorley et al., 2012) (note that McSorley et al designate another enzyme as “PhnY” that is unrelated to the PhnY enzyme described in this study). Furthermore, we have previously demonstrated the conversion of 2-AEP-derived PnAA to phosphonoacetate (PnA) (Figure 1A, box), which is then hydrolyzed by a PnA hydrolase enzyme (PhnA) to yield acetate and Pi in the soil dwelling bacterium Sinorhizobium meliloti 1021 (Borisova et al., 2011). Though the PnA hydrolase activity had been characterized previously from many different ecological niches (White and Metcalf, 2007), the discovery of the NAD+ dependent PnAA dehydrogenase enzyme PhnY established PnA to be a biogenic phosphonate. NADP+-dependent PnAA dehydrogenation activity was also reported in the cell-extracts of the marine bacterium Roseovarius nubinhibens ISM (Cooley et al., 2011). Bioinformatic analysis demonstrates that 2-AEP degradation pathways involving PhnY homologs are present across many different bacterial species and ecological niches (Borisova et al., 2011; Cooley et al., 2011; Kim et al., 2011).
Figure 1. Phosphonate degradation pathways.
(A) Three enzymatic pathways for the breakdown of PnAA. Top: PnAA hydrolysis to acetaldehyde and Pi. Middle: PnAA-derived 2-AEP is hydroxylated and oxidized to yield glycine and Pi. Bottom: Oxidation of PnAA by PhnY (boxed) and subsequent hydrolysis by PhnA to yield acetate and Pi. (B) Chemical structures of 3-OPP and G3P.
PnAA is a common intermediate in various phosphonate biosynthetic pathways and is produced by the enzymatic decarboxylation of phosphonopyruvate (Zhang et al., 2003). PnAA is then transaminated to generate 2-AEP (Kim et al., 2002), reduced to generate 2-hydroxyethyl phosphonate in dehydrophos, phosphinothricin, and fosfomycin biosynthesis (Peck et al., 2012; Shao et al., 2008; Woodyer et al., 2007), or condensed with oxaloacetate to generate 2-keto-4-hydroxy-5-phosphonopentanoic acid in rhizocticin biosynthesis (Borisova et al., 2010). Oxidation of PnAA to PnA is probably not a biosynthetic reaction, as thus far PnA has not been observed or postulated to exist in any known phosphonate biosynthetic scheme. Hence, PhnY provides the only known biogenic source for PnA in the microbial metabolome, allowing utilization of a common biological phosphonate, PnAA, as a growth source.
Here, we present detailed biochemical and crystallographic characterization of the PhnY enzyme from S. meliloti 1021 (henceforth referred to as PhnY). Crystal structures of the enzyme and kinetic analysis of site-specific mutants establish that PhnY bears structural and mechanistic similarity to the aldehyde dehydrogenase superfamily of enzymes. We demonstrate that PhnY is broadly substrate tolerant, and can accept 3-oxopropyl phosphonate (3-OPP, or phosphonopropionaldehyde) and glyceraldehyde-3-phosphate (G3P) as substrates (Figure 1B). The crystal structure of PhnY in the presence of G3P leads to the postulate that PhnY does not discriminate between phosphonates and phosphate-esters, but rather that the physical dimension of the enzyme active site, comprised of various basic residue side chains, modulates substrate preference.
RESULTS
Overall structure of PhnY
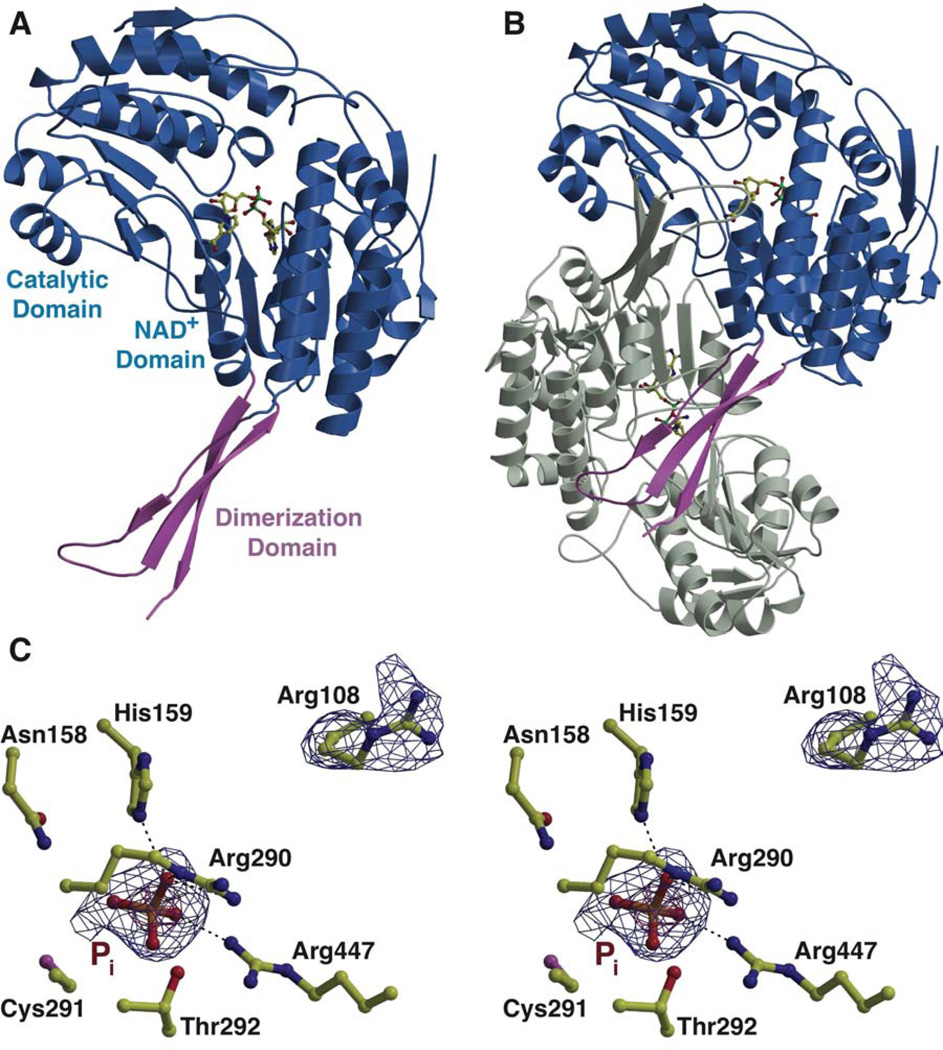
The structure of PhnY was determined in the apo state to 2.1 Å resolution. Crystallographic phases were determined by single wavelength anomalous diffraction (relevant data collection statistics are provided in Table 1). The crystallographic asymmetric unit consists of eight protein chains comprising four sets of biologically relevant homodimers (Figure 2A,B, see also Supplementary Figure S1). The arrangement of protein chains as sets of homodimers is consistent with the behavior of PhnY as a dimer in solution, as judged by size exclusion chromatography.
Table 1.
Data collection, phasing and refinement statistics.
| PhnY-apo | PhnY-PnAA | PhnY-PnAA- NAD+ |
PhnY-PnA- NAD+ |
PhnY-G3P-NAD+ | |
|---|---|---|---|---|---|
| Data collection | |||||
| a, b, c (Å), β(°) | 95.72, 172.79, 142.58, 107.57 | 93.35, 172.88, 138.97, 106.73 | 94.74, 172.13, 140.05, 107.75 | 95.16, 172.77, 142.60, 107.28 | 94.45, 172.70, 139.77, 106.90 |
| Wavelength (Å) | 0.97856 | 0.97872 | 0.97856 | 0.97856 | 0.97872 |
| Resolution (Å)1 | 50.00 – 2.10 (2.20 – 2.10) |
50.00 – 2.10 (2.20 – 2.10) |
40.00 – 2.00 (2.03–2.00) |
50.00 – 2.07 (2.11–2.07) |
40.00 – 2.25 (2.29–2.25) |
| Total reflections | 994,116 | 585,836 | 1,922,065 | 1,709,837 | 893,695 |
| Unique reflections | 246,726 | 236,736 | 286,921 | 250,280 | 198,176 |
| Rsym(%) | 12.1 (55.0) | 9.8 (53.6) | 12.7 (84.3) | 7.0 (51.6) | 10.9 (43.1) |
| I/σ(I) | 7.5 (1.6) | 9.4 (1.65) | 18.3 (2.4) | 18.8 (2.1) | 12.1 (2.0) |
| Completeness (%) | 96.1 (87.9) | 96.5 (90.3) | 99.9 (99.6) | 94.1 (74.9) | 97.3 (96.3) |
| Redundancy | 4.0 (2.4) | 2.5 (1.8) | 6.7 (6.2) | 6.8 (4.7) | 4.5 (3.7) |
| Refinement | |||||
| Resolution (Å) | 24.87 – 2.10 | 39.11 – 2.10 | 39.50 – 2.00 | 28.58 – 2.07 | 24.97 – 2.25 |
| No. reflections used | 234,253 | 224,891 | 272,514 | 237,960 | 188,026 |
| Rwork / Rfree2 | 19.8/23.7 | 21.0/25.2 | 18.2/23.0 | 18.1/21.7 | 20.4/24.9 |
| Number of atoms | |||||
| Protein | 29,144 | 29,008 | 29,008 | 29,163 | 29,008 |
| Ligand | 40 | 56 | 56 | 64 | 80 |
| Cofactor | - | - | 189 | 352 | 197 |
| Water | 2329 | 1916 | 2571 | 2861 | 1678 |
| B-factors | |||||
| Protein | 32.80 | 24.32 | 26.69 | 27.08 | 24.36 |
| Ligand | 35.21 | 21.84 | 25.81 | 37.96 | 33.94 |
| Cofactor | - | - | 32.28 | 32.66 | 36.62 |
| Water | 37.88 | 28.51 | 32.73 | 34.23 | 27.13 |
| R.m.s deviations | |||||
| Bond lengths (Å) | 0.009 | 0.016 | 0.008 | 0.012 | 0.014 |
| Bond angles (°) | 1.147 | 1.567 | 1.153 | 1.429 | 1.342 |
Highest resolution shell is shown in parenthesis.
R-factor = Σ(|Fobs|−k|Fcalc|)/Σ|Fobs| and R-free is the R value for a test set of reflections consisting of a random 5% of the diffraction data not used in refinement.
Figure 2. Overall three dimensional crystal structure of PhnY, see also Supplemental Figure S1.
Structure of the PhnY (A) monomer and (B) homodimer showing the relative positions of the three domains with one monomer colored grey, and the second monomer colored with the cofactor binding and catalytic domains in blue, and the oligomerization domain in pink. Cofactor NAD+ is shown in ball-and-stick representation with carbon atoms colored yellow. (C) Stereo view showing the active site features of PhnY in complex with Pi. The PhnY carbon atoms are shown in yellow ball-and-stick representation. Superimposed is a difference Fourier electron density map (contoured at 2.7σ over background in blue, and 6.0σ in red) calculated with coefficients |Fobs| − |Fcalc| and phases from the final refined model with the coordinates of the inorganic phosphate deleted prior to one round of refinement.
Each PhnY monomer shows architectural similarities to other members of the non-phosphorylating aldehyde dehydrogenase (ALDH) superfamily (Figure 2A). Residues 1–123, 147–256 and 461–475 together constitute the ‘cofactor-binding domain’ and adopt a classical α/β Rossmann fold. Residues 257–460 form the ‘catalytic domain’ with a distinct α/β fold comprised of seven central β strands flanked by α helices on either side. Residues 124–146, together with residues 476–486 constitute the three-membered antiparallel β sheet ‘oligomerization domain’ that provides the principal contacts for dimerization (Figure 2B). The loops at the interface of the cofactor-binding domain and the catalytic domain provide nearly all the residues implicated in substrate binding and catalysis (vide infra).
In the structure of the unliganded enzyme, electron density corresponding to a phosphate ion (presumably from the crystallization medium) is observed at the interface of the cofactor binding and catalytic domains. The phosphate oxygen atoms are coordinated by hydrogen bonds with the side chains of His159, Arg290, Thr292, and Arg447 (Figure 2C). The Arg108 side chain, which is in the vicinity of the phosphate-binding site, shows alternative conformations throughout the different molecules in the asymmetric unit. In one conformation, Arg108 points towards the phosphate binding-pocket with the guanidino group within interaction distance with the phosphate. In the alternate conformation, this residue points away from the active site and is stabilized by interactions with the side chain of Asp111. This basic, phosphate-binding pocket is strictly conserved among ALDHs that catalyze oxidation of phosphorylated substrates, but is missing in ALDHs that function on non-phosphorylated substrates (Supplementary Figure S1).
Cocrystal structure with the physiological phosphonate substrate PnAA
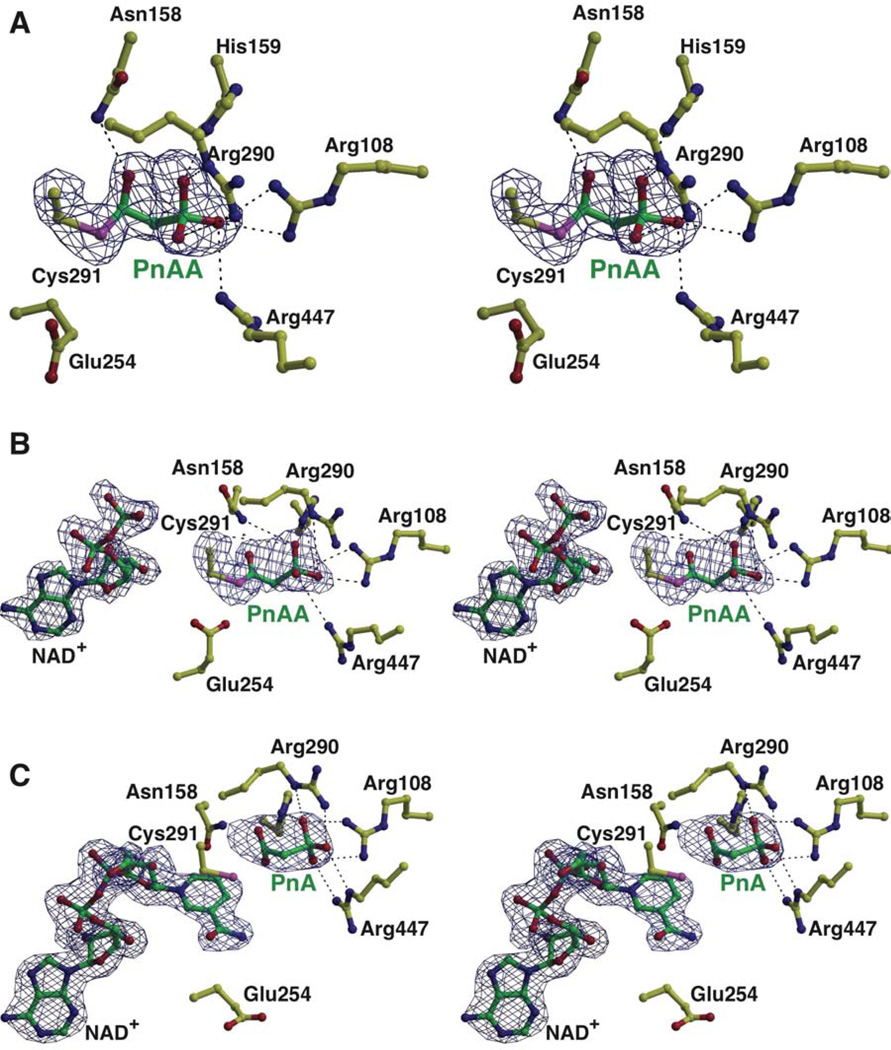
The 2.1 Å resolution cocrystal structure of PhnY in complex with PnAA shows unambiguous density for the ligand in the active site of all eight copies in the crystallographic asymmetric unit (Figure 3A). PnAA is bound at the interface of the cofactor-binding and catalytic domains. The side chain thiol of Cys291 undergoes a 120° rotation relative to the apo structure, which positions the Cys291 side chain for interactions with the substrate. The Cys291-Sγ atom in this conformation is 3.0 Å and 3.3 Å away from the backbone amide nitrogen atoms of Cys291 and Thr292, respectively. Similar interactions have been implicated in modulating the nucleophilicity of the Cys side chain for nucleophilic attack in related members of the ALDH superfamily (Cobessi et al., 2000).
Figure 3. PhnY active site in complex with substrate PnAA, product PnA and cofactor NAD+.
All atoms are shown in ball-and-stick representation with PhnY side chain carbon atoms colored yellow, and PnAA, PnA and NAD+ carbon atoms colored green. (A) See also Supplemental Figure S2. Stereo view showing the active site features of PhnY in the binary complex with the substrate PnAA. Superimposed is a difference Fourier electron density map (contoured at 2.5σ over background in blue) calculated as before. Stabilizing interactions of PnAA with PhnY are shown as black dashes. (B) See also Supplemental Figure S3. Stereo view of the PhnY active site in the ternary complex with substrate PnAA and cofactor NAD+. Superimposed is the difference electron density map contoured at 2.5σ over background in blue, calculated as before. Note that the nicotinamide and ribose rings for the cofactor have not been modeled. (C) See also Supplemental Figures S4, S5. Stereo view of the PhnY active site in the ternary complex with product PnA and cofactor NAD+. Superimposed is a difference Fourier electron density map (contoured at 2.5σ over background in blue,) calculated as before. Stabilizing interactions for NAD+ are described in Supplementary Figure S4. Note the presence of the nicotinamide ring and the ribose sugar.
Strong and continuous electron density can be observed between the Cys291-Sγ and the PnAA-Cβ atom (at a distance of 1.8 Å) indicative of a covalent bond between these two atoms. Unbiased difference Fourier electron density maps are indicative of tetrahedral geometry at the PnAA-Cβ atom that is covalently bound to the Cys291 sulfur atom (Figure 3A, see also Supplementary Figure S2). This tetrahedral geometry is presumably generated by nucleophilic attack of the Cys291-Sγ thiolate at the Re face of planar PnAA to generate a tetrahedral thiohemiacetal intermediate with an (R) stereochemical configuration. Consequently, we have modeled the substrate in the PhnY-PnAA binary complex as a thiohemiacetal instead of an aldehyde. To the best of our knowledge, this is the first instance of the observation of the thiohemiacetal intermediate in a member of the ALDH superfamily of enzymes.
The oxygen atom of the thiohemiacetal intermediate is positioned 3.1 Å away from the amide side chain of Asn158, and 3.2 Å away from the main chain amide nitrogen atom of Cys291 (Figure 3A). By analogy to the serine/cysteine protease reaction mechanism (Hedstrom, 2002), these two interactions constitute the ‘oxyanion hole’ that stabilizes the tetrahedral intermediate. Binding of PnAA orders the side chain of Arg108 in a solitary conformation pointing towards the active site, such that the guanidine moiety now also interacts with the phosphonate oxygen atoms.
Cocrystal structure of ternary complex with substrate PnAA and cofactor NAD+
In the 2.0 Å resolution PhnY-NAD+-PnAA ternary complex, unambiguous electron density can be observed for all atoms of PnAA. However electron density is absent for the nicotinamide ring and the adjoining ribose of the cofactor. Hence only the adenine diphosphate (ADP) moiety of the cofactor has been modeled in the final structure (Figure 3B, see also Supplementary Figure S3).
The substrate PnAA in the PhnY-NAD+-PnAA ternary complex is positioned analogously to the thiohemiacetal intermediate described previously, with all the interactions with the side chains of Asn158, His159, Arg108, Arg290, Thr292 and Arg447 conserved. However, the Fo − Fc difference electron density map is indicative of planar geometry around the PnAA-Cβ atom (Figure 3B). This planar geometry presumably arises due to the transfer of the hydride from the PnAA-Cβ to the cofactor, and the concomitant conversion of the tetrahedral thiohemiacetal intermediate to a sp2 hybridized planar thioester. The bond angles around the PnAA-Cβ have thus been refined to a planar geometry. Variability is observed in the positioning of the Glu254 side chain in the vicinity of the thioester intermediate. Based on sequence homology (Supplementary Figure S1), this residue is the catalytic base that primes a water molecule for nucleophilic attack on the thioester intermediate that is generated in the catalytic cycle for PhnY (vide infra). In some polypeptide chains, the Glu254 side chain points towards the thioester (Chains A, B and H), while it points away from the thioester in other chains (Chains C, D, E, F and G), without stabilizing interactions for the Glu254 side chain carboxylate in either of the two conformations. This lack of interactions for the Glu254 side chain is reflected in the significantly higher B-factors for the Glu254 side chain (51.1 Å2) as compared to the Glu254 main chain atoms (23.0 Å2). Incubation of a mutant of PhnY in which Glu254 was mutated to Ala resulted in suppressed deacylation activity allowing the detection of a covalent intermediate by electrospray mass spectrometry (Supplementary Figure S3). The resolution of the mass spectrum did not allow distinction between the thiohemiacetal or thioester, but the higher chemical stability of the latter suggests that the trapped intermediate corresponds to the thioester.
Structure of the ternary complex with NAD+ and PnA
The 2.1 Å resolution cocrystal structure of PhnY in complex with NAD+ and PnA shows clear and continuous density for the entire cofactor in all copies in the crystallographic asymmetric unit (Figure 3C, see also Supplementary Figures S4 and S5). The adenine ring of NAD+ is positioned in a hydrophobic pocket defined by the side chains of Pro214, Phe232, Val238, Leu241 and Ile242 (Supplementary Figure S4). The adenine ribose sugar hydroxyls are stabilized by interactions with Lys181. The 2’ hydroxyl makes additional contacts with Glu184, while the 3’ hydroxyl contacts the backbone carbonyl of Thr155. The Glu184 side chain likely dictates the specificity of PhnY for NAD+ as the cofactor (Borisova et al., 2011) as the additional phosphate group in NADP+ would clash with the Glu184 side chain. The pyrophosphate moiety of NAD+ makes hydrogen bonds with the main chain atoms of Ser235 and Phe157, and a water-mediated hydrogen bond with the Thr183 side chain. Hydrogen bonds with the side chain of Glu385 provide the only interaction for the nicotinamide ribose hydroxyl groups. The Cys291-Sγ is positioned at the Re-face of C4 of the nicotinamide ring. The Si face of the nicotinamide ring is buttressed by the side-chain of Thr233. The only interactions with the nicotinamide ring amide are two water-mediated hydrogen bonds with the side chain of Glu465 and the main chain amide nitrogen of Gly455. This relative lack of interactions manifests itself as higher B-factors for the adenine and nicotinamide rings of the cofactor, as compared to the rest of the molecule (Supplementary Figure S5). A comparative decrease in conformational ordering of the cofactor, starting from the adenine to the nicotinamide terminus of NAD+, is evident in the quality of the difference electron density maps (Figure 3C).
The product PnA binds analogously to the substrate PnAA within the active site (Figure 3C). However, no covalent bond exists between Cys291-Sγ and PnA-Cβ atoms, evident from the lack of continuous electron density. One of the PnA carboxylate oxygen atoms is within hydrogen bonding distance to the main chain amide nitrogen of Cys291 (3.0 Å) and the side chain imidazole Nε2 nitrogen atom of His159 (3.3 Å). The other PnA carboxylate oxygen atom is bereft of any interactions with the polypeptide.
Kinetic analysis of the wild type and site directed mutants of PhnY
Using the complex structures as guides, we generated site-specific PhnY mutants, and characterized their kinetic behavior. Kinetic characterization of the wild-type and variants enzymes were carried out as described previously (Borisova et al., 2011). For the wild type enzyme, the kinetic parameters determined in this study, kcat = 2.2 ± 0.1 s−1 and KM = 3.2 ± 0.7 µM for PnAA, are in good agreement with the previously published values (Table 2; Supplementary Figure S6) (Borisova et al., 2011).
Table 2.
Steady state kinetic parameters for various PhnY constructs and WT GAPDH. See also Supplemental Figure S6.
| KM,Pn (µM) | KM,NAD+ (µM) | kcat (s−1) | |
|---|---|---|---|
| PhnY constructs with PnAA | |||
| Wild type | 3.2 ± 0.7 | 58 ± 9 | 2.2 ± 0.1 |
| Arg108Ala | 9.7 ± 0.8 | 18 ± 2 | 0.051 ± 0.001 |
| Asn158Ala | 29 ± 9 | 40 ± 6 | 0.010 ± 0.001 |
| Glu254Ala | n.d.1 | n.d. | n.d. |
| Arg290Ala | 5.1 ± 0.6 | 75 ± 8 | 0.12 ± 0.003 |
| Cys291Ala | n.d. | n.d. | n.d. |
| Glu385Ala2 | 19 ± 3 | 370 ± 10 | 0.19 ± 0.03 |
| Arg447Ala | 150 ± 20 | 54 ± 1 | 0.076 ± 0.003 |
| PhnY constructs with G3P | |||
| Wild type | 97 ± 7 | 530 ± 50 | 0.098 ± 0.003 |
| Cys291Ala | n.d. | n.d. | n.d. |
| PhnY constructs with 3-OPP | |||
| Wild type | 3300 ± 100 | n.d. | 1.5 ± 0.1 |
| Cys291Ala | n.d. | n.d. | n.d. |
| GAPDH with various substrates | |||
| PnAA | n.d. | n.d. | n.d. |
| 3-OPP | 1500 ± 300 | 330 ± 20 | 0.038 ± 0.022 |
| G3P (racemic) | 25 ± 7 | 22 ± 1 | 22 ± 2 |
n.d.: Not determined. Low levels of detectable activity precluded kinetic parameter determination.
: This mutant displayed substrate inhibition (see Figure S6); the Ki,PnAA was 420 ± 70 µM.
No activity was observed when either Cys291 or Glu254 was replaced by Ala, confirming the vital roles each plays in catalysis. The Asn158Ala mutation resulted in a 200-fold decrease in kcat. Replacement of Arg108 by Ala led to a 40-fold decrease in kcat and a 3-fold increase in KM,PnAA, and the Arg290Ala mutation resulted in a similar 20-fold decrease in kcat with a slightly elevated KM,PnAA relative to wild type. The Arg447Ala mutation reduced the kcat 30-fold and increased the KM,PnAA 50-fold. Glu385 provides the only interaction with the nicotinamide ribose 2’ and 3’ hydroxyl groups; the Glu385Ala mutation resulted in a 10-fold reduction in kcat, with an approximately 6-fold higher KM,NAD+.
Kinetics and ternary complex structures of PhnY-NAD+ with non-physiological substrates-3-OPP and G3P
The active site of PhnY is highly similar to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) from Streptococcus mutans, a well-characterized enzyme of primary metabolism that catalyzes the conversion of G3P to 3-phosphoglycerate. A one-to-one mapping of active site side chains was observed, with the exception of GAPDH-Tyr155 being changed to His159 in PhnY. In order to explore whether this structural similarity between the two active sites translates to substrate promiscuity for PhnY, we explored the activity of PhnY with G3P, the physiological substrate of GAPDH. We additionally chemically synthesized a one carbon atom longer mimic of PnAA-3-OPP (Figure 1B). PhnY could indeed accept G3P and 3-OPP as substrates. Oxidation of 3-OPP by PhnY proceeded with nearly identical kcat, but a 1000-fold increase in KM compared to the KM for PnAA (Table 2). However, the oxidation of G3P proceeded with a 20-fold reduction in kcat for the enzyme and a 30-fold increase in the KM for G3P compared to KM,PnAA. No activity was observed for the PhnY Cys291Ala mutant enzyme using either 3-OPP or G3P.
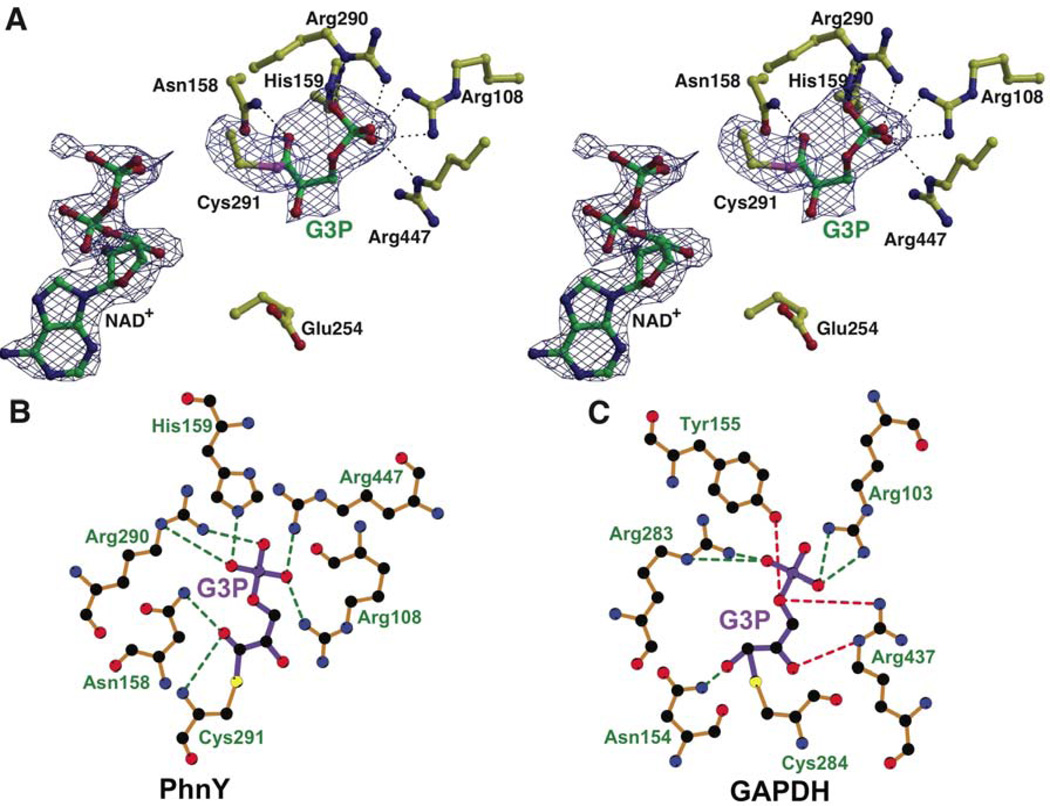
The crystal structure of the PhnY-G3P-NAD+ ternary complex, determined to a limiting resolution of 2.25 Å, reveals truncated density for the cofactor analogous to the PhnY-PnAA-NAD+ crystal structure described previously. The G3P-Cγ atom is covalently bonded to the Cys291-Sγ side chain. The geometry around the G3P-Cγ atom is indicative of planar sp2 hybridization (Figure 4A, see also Supplementary Figure S8). The thioester oxygen atom is 3.2 Å away from the side chain amide of Asn158, and 3.0 Å away from the main chain amide nitrogen of Cys291. In most PhnY monomers, the G3P-Cβ hydroxyl makes a water mediated hydrogen-bonding contact with the side chain of Glu465. No interactions are present for the G3P bridging oxygen of the phosphate ester. The side chain of Glu254 points away from the thioester intermediate in all polypeptides within the asymmetric unit.
Figure 4. Ternary complex of PhnY with G3P and truncated cofactor NAD+, see also Supplemental Figure S8.
(A) Stereo view of G3P and NAD+ binding to PhnY. All atoms are colored as before. Superimposed is the difference electron density map contoured at 2.5σ in blue, calculated as before. (B) Interactions are shown as green dashes for the binding of (B) G3P with PhnY, and (C) G3P with GAPDH. G3P bonds are colored purple, and enzyme residue bonds are colored brown. The interactions for the G3P phosphoryl oxygen atom and Cβ hydroxyl, which are absent in PhnY, are shown in red dashes. Note the different conformations of G3P in the PhnY active site and GAPDH active site. Figure was generated by LigPlot+ (Laskowski and Swindells, 2011) using refined coordinates of PhnY-G3P-NAD+ complex for panel B, and PDB 2ESD for panel C.
In the PhnY-G3P-NAD+ ternary complex, the side chains of Arg108, His159, Arg290 and Arg447 bind the phosphoryl oxygen atoms of G3P, in a manner analogous to PnAA (Figure 4A,B). These interactions are analogous to the interactions of G3P within the GAPDH active site, which are provided by the corresponding GAPDH residues—Arg103, Tyr155, Arg283 and Arg437 (Figure 4C). Direct interactions for the bridging phosphoryl ester oxygen atom of G3P, mediated by GAPDH residues Arg437 and Tyr155 (red dashed lines in Figure 4C), are absent in PhnY. However, there are no residues in the PhnY active site that would occlude phosphorylated substrates.
DISCUSSION
Prior work has led to the characterization of a novel pathway for phosphonate degradation in S. meliloti 1021, which utilizes PhnY to convert PnAA into PnA (Borisova et al., 2011). Oxidation of PnAA primes the resultant carboxylic acid for subsequent enzymatic hydrolysis by PhnA (Agarwal et al., 2011; Cooley et al., 2011; Kim et al., 2011). PhnY provides the first, and presently the only known biochemical basis for the synthesis of PnA in the microbial metabolome. The structural data presented here demonstrate that PhnY belongs to the family of CoA-independent non-phosphorylating aldehyde dehydrogenases. Structural comparisons with other ALDHs reveal how minor changes in a highly conserved ALDH active site can increase the repertoire for this class of enzymes to include phosphonate substrates, and expand upon the role of ALDHs in the biodegradation and bioremediation of inert molecules.
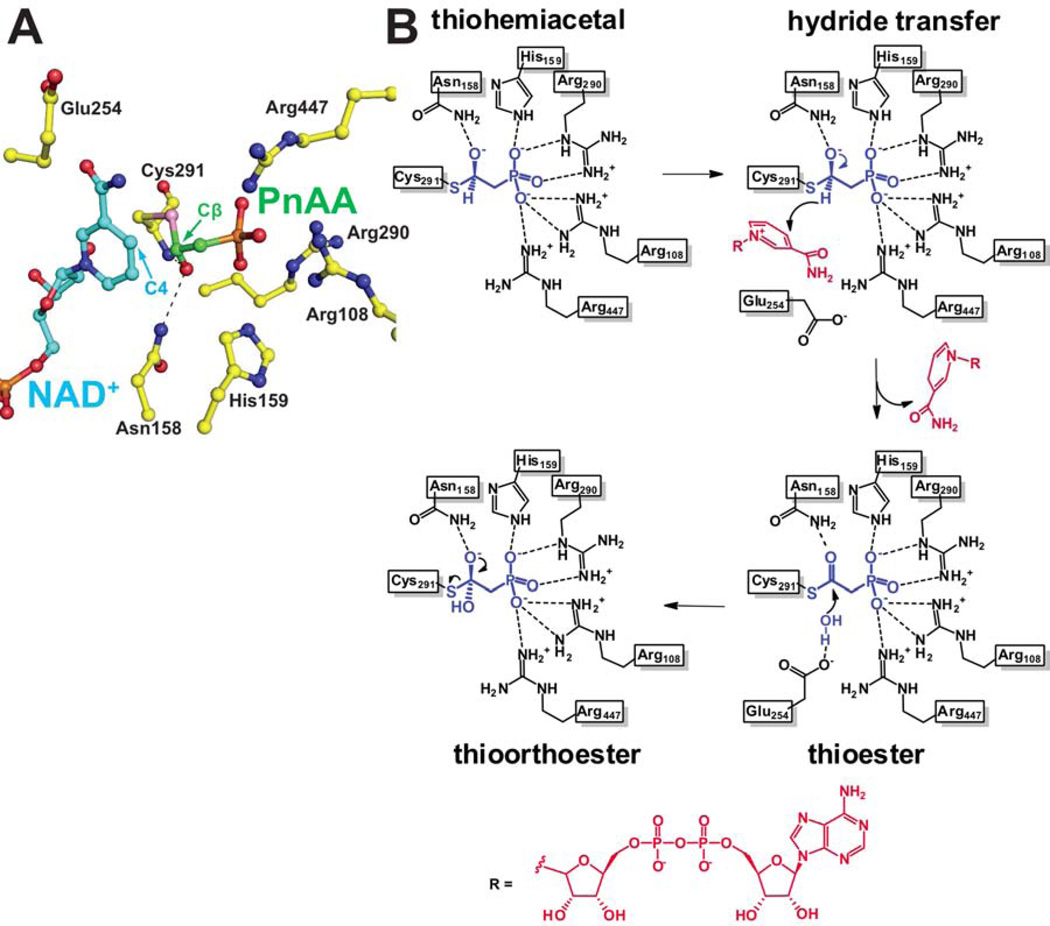
Model for productive hydride transfer conformation for the cofactor
Our efforts at obtaining a ternary complex of wild type PhnY with PnAA and NAD+ resulted in only partial electron density for the cofactor (Figure 3B), with density for the hydride acceptor nicotinamide ring and the adjoining ribose missing. Truncated density for the cofactor was also observed in the PhnY-G3P-NAD+ ternary complex crystals (Figure 4A). Soaking the PhnY-PnAA-NAD+ crystals with excess of NAD+ for up to 2 days also did not result in increased occupancy of the cofactor nicotinamide ring. However, a complete description of the cofactor was achieved in the PhnY-PnA-NAD+ ternary complex (Figure 3C).
In order to describe the hydride transfer conformation of the cofactor, we aligned the coordinates of the PhnY-PnAA binary complex, with those of the PhnY-PnA-NAD+ ternary complex. An analysis of the relative positioning of the C4 atom of the nicotinamide ring (the hydride acceptor) and the PnAA-Cβ atom (hydride donor) leads us to propose that this model represents the experimentally elusive hydride transfer conformation for PhnY (Figure 5A). The nicotinamide C4 and PnAA-Cβ atoms are positioned in close proximity with each other, with a distance of 2.1 Å between the two. The tetrahedral geometry of the thiohemiacetal intermediate in the PhnY-PnAA binary complex (Figure 3A, Supplementary Figure S2) would thus position the hydride for transfer to the C4 atom of the nicotinamide. It should be noted that in this model, the Glu254 side chain is occluded from the active site due to steric clash with the NAD+ nicotinamide ring (Figure 5A).
Figure 5. A model for productive hydride conformation of the cofactor and a proposed mechanistic scheme for conversion of PnAA to PnA by PhnY, see also Supplemental Figure S7.
(A) The coordinates for Cys291 and PnA were omitted from the PhnY-PnA-NAD+ ternary complex crystal structure and replaced with those of Cys291-PnAA thiohemiacetal from the PhnY-PnAA binary complex crystal structure. The cofactor is shown in ball-and-stick representation with carbon atoms colored blue, and carbon atoms for PnAA are colored green. The hydride donor PnAA-Cβ and the hydride acceptor nicotinamide-C4 atoms are labeled. Black dashes represent the ‘oxyanion hole’ stabilizing interactions for the thiohemiacetal intermediate. (B) Substrate binding interactions with the enzyme side chains are shown as black dashes. The reaction intermediates are shown in blue. The cofactor atoms are shown in red. The relative geometric positions of the hydride and hydroxyl of the thiohemiacetal intermediate, and the two hydroxyls in the thioorthoester intermediate are inferred from the crystal structures as shown in Figure 3A.
Proposed catalytic mechanism for PhnY involving a flexible cofactor
Three classes of ALDHs have been characterized, all of which share a common first acylation half reaction that includes hydride transfer to the cofactor. Variability is observed in the second deacylation half reaction, based on the deacylating nucleophile—hydroxyl anion, phosphate, or a coenzyme A (CoA) thiolate. Our understanding of the CoA independent non-phosphorylating ALDHs is based primarily on the extensive structural and mechanistic characterization of GAPDH (Cobessi et al., 2000; D'Ambrosio et al., 2006; Marchal and Branlant, 1999; Marchal et al., 2000). Based on the PhnY cocrystal structures and kinetic data presented in this study, and the structural similarity of the PhnY and GAPDH active sites, a mechanistic proposal for the conversion of PnAA to PnA by PhnY is shown in Figure 5B.
In the acylation half reaction, PnAA is covalently bound to the side chain of Cys291 forming a (R)-thiohemiacetal intermediate at the interface of the catalytic and cofactor binding domains. Residues Arg108, His159, Arg290, Thr292 and Arg447 coordinate phosphoryl oxygen atoms. The side chain of Asn158 and main chain amide nitrogen of Cys291 constitute the ‘oxyanion hole’ to stabilize the intermediate. The cofactor is aptly positioned for hydride transfer from the thiohemiacetal to the C4 carbon atom of the nicotinamide ring from the Re-face (Figure 5A), generating a thioester intermediate. The hydride transfer conformation of the cofactor occludes the side chain of Glu254 from approaching the active site, due to steric clash between the Si-face of the nicotinamide ring and the Glu254 side chain (Figure 5A, see also Supplementary Figure S7). Following hydride transfer, the nicotinamide departs from the active site, not necessarily involving release of the cofactor by the cofactor-binding domain. In the succeeding deacylation half reaction, the side chain of Glu254 is now positioned close to the thioester intermediate to deprotonate a water molecule for nucleophilic attack at the thioester. This attack leads to the formation of a thioorthoester intermediate, which is resolved by the departure of the Cys291 side chain and generation of the product PnA.
In the proposed reaction mechanism, the reduced cofactor is not released from the enzyme prior to deacylation, but rather the nicotinamide ring ‘flips-out’ from the enzyme active site to adopt alternate conformations, as has been proposed for the GAPDH enzyme (D'Ambrosio et al., 2006). This allows for the proper positioning of the catalytic base (Glu254 for PhnY), which is required for the deacylation half reaction to occur. At least four different conformations of the cofactor have been structurally described in the literature (Cobessi et al., 2000; D'Ambrosio et al., 2006; Langendorf et al., 2010; Perez-Miller and Hurley, 2003). A conserved feature of all these cofactor conformations is the relatively higher thermal (B) factors for the nicotinamide ring and the ribose sugar attached the nicotinamide ring, and lower B values for the ribose sugar bound to the adenine ring, and the pyrophosphate moiety, as also observed in this study (Supplementary Figure S5). This increase in B-values is also accompanied by a relative reduction in occupancy for the nicotinamide ring of the cofactor in these examples.
Relaxed substrate specificity of PhnY and evolution of enzyme function in the ALDH family
The active sites of PhnY and GAPDH are strikingly similar, apart from the replacement of GAPDH-Tyr155 by PhnY-His159. In order to determine whether this similarity in active sites would lead to substrate promiscuity within the ALDH superfamily, we sought to determine whether PhnY could use G3P as a substrate and whether GAPDH could accept phosphonate substrates. GAPDH oxidized 3-OPP with a large decrease in catalytic efficiency, and the rate of PnAA oxidation was below the detection limit (Table 2). In comparison, G3P was a better substrate for PhnY, albeit with a 20-fold reduction in kcat as compared to GAPDH (Marchal et al., 2000).
The observed reduction in the catalytic efficiency of G3P oxidation by PhnY as compared to GAPDH may be a result of unfavorable electrostatic interactions. In the PhnY-G3P-NAD+ ternary complex (Figure 4A,B), G3P is contorted so that the thiohemiacetal oxyanion is positioned only 2.9 Å away from one of the phosphoryl oxygen atoms. On the other hand for the GAPDH-G3P-NAD+ ternary complex (Figure 4C) (PDB: 2ESD) (D'Ambrosio et al., 2006), G3P is fully extended and the corresponding thiohemiacetal oxyanion does not have such destabilizing electrostatic repulsions from the phosphoryl oxygen atoms. Thus, the physical dimensions of the PhnY and GAPDH active sites seem to dictate substrate specificities. A larger GAPDH active site allows for G3P to be fully extended in the GAPDH active site while a smaller PhnY active site distorts G3P so that the thiohemiacetal intermediate is bound in an energetically less favorable conformation. In addition, the hydroxyl group in G3P makes contact with the side chain of Arg437 in GAPDH, whereas this contact is absent in the ternary complex of PhnY with G3P and cofactor.
A second postulate for the reduction in G3P oxidation activity by PhnY is provided by the orientation of the Glu254 side chain for the deacylation half reaction. In the PhnY-G3P-NAD+ ternary complex, the Glu254 side chain is occluded away from the thioester in all copies in the asymmetric unit. However, Glu254 shows variable positioning within the PhnY-PnAA-NAD+ ternary complex. Superimposing the two structures reveals that the productive deacylation orientation of Glu254 in the PhnY-G3P-NAD+ ternary complex is occluded by the G3P-Cβ hydroxyl due to a steric clash (Supplementary Figure S8). This in turn hinders the deacylation half reaction, which is rate limiting for GAPDH (Marchal et al., 2000).
In order to test these two postulates, we synthesized 3-OPP and determined the kinetic parameters for its oxidation by PhnY and GAPDH. In 3-OPP the aldehyde is separated from the phosphoryl moiety by one more bond than in PnAA, but one bond less than in G3P. PhnY could indeed accept 3-OPP as a substrate. As expected, the kcat for 3-OPP oxidation is comparable to that for PnAA as the rate limiting deacylation step is not hindered, in contrast to G3P (Marchal et al., 2000). However, the KM for 3-OPP oxidation is 1000-fold greater than that for PnAA (Table 2). This observation lends credence to the postulate that substrates larger than PnAA will be unfavorably accommodated within the constrained PhnY active site. This preference is not dependent on the substrate being a phosphate or a phosphonate. The lack of a hydroxyl group and a bridging oxygen on 3-OPP and the interactions they make with active site residues may explain the greatly reduced catalytic activity of GAPDH towards this substrate compared to G3P. These results are reflective of the diversity in catalytic activities that can be built upon the conserved ALDH superfamily structural core.
Significance
The discovery of the NAD+ dependent phosphonoacetaldehyde dehydrogenase enzyme PhnY established a biogenic origin for phosphonoacetate, an activated C-P bond-containing compound that is used as a phosphorus and carbon source in several microorganisms. The structural and biochemical studies of PhnY presented here provide insights into how the widely conserved aldehyde dehydrogenase active site can be adapted for phosphonate substrates. The cocrystal structure of PhnY with substrate phosphonoacetaldehyde reveals the first observation of a thiohemiacetal intermediate visualized in an aldehyde dehydrogenase superfamily enzyme.
MATERIALS AND METHODS
Chemicals
G3P was obtained from Sigma-Aldrich (G5251-25MG) and used without further purification. Media components were purchased from Thermo Fisher Scientific or VWR (West Chester, PA). DNA primers were synthesized by Integrated DNA Technologies Inc (Coralville, IA). Restriction enzymes were obtained from New England Biolabs (Ipswich, MA). DNA sequencing was performed by ACGT Inc (Wheeling, IL). Phosphonoacetaldehyde was freshly prepared before use as described earlier (Borisova et al., 2011).
Cloning, protein expression, purification, and crystallization
Cloning, expression, and purification of recombinant PhnY from E. coli has been described previously (Borisova et al., 2011). The mutant constructs Arg108Ala, Asn158Ala, His159Ala, Glu254Ala, Arg290Ala, Cys291Ala, Thr292Ala, Glu385Ala and Arg447Ala were generated by standard site-directed mutagenesis protocols and confirmed by plasmid DNA sequencing. Wild type and mutant enzymes were purified according to procedures described earlier (Borisova et al., 2011). Briefly, the plasmids were transformed into competent E. coli Rosetta2 strain, and recombinant protein expression was induced by addition of 0.2 mM of IPTG for 16 hours at 18° C. The cells were collected by centrifugation, and resuspended in lysis buffer A - 20 mM Tris (pH 8.0) 500 mM NaCl 10% glycerol. Cells were lysed by multiple passes through a C5 Avestin cell homogenizer, and the lysate by clarified by centrifugation. The lysate was loaded on a 5 mL Ni-NTA column equilibrated in lysis buffer A. The column was extensively washed with lysis buffer A supplemented with 30 mM imidazole, and eluted by a linear gradient to 200 mM imidazole. Pure protein fractions, as judged by SDS-PAGE, were pooled and dialyzed in 20 mM Tris (pH 8.9) 100 mM NaCl for 12 hours. The hexahistidine tag was removed by digestion with thrombin (1 unit/mg of protein) followed by ion exchange chromatography and size exclusion chromatography (Superdex 75 16/60, GE Healthcare) in 20 mM HEPES (pH 7.5) 100 mM KCl buffer. The protein was concentrated using Amicon centrifugal filters. This process routinely yielded 5 mg of highly pure protein per liter of culture.
The crystals of PhnY were grown at 15° C using hanging drop vapor-diffusion method by mixing 1 µL protein sample at 8 mg/mL concentration, with an equal volume of reservoir mother liquor. Apo-form crystals were obtained in mother liquor containing 22–25% PEG3350, 0.1 M sodium cacodylate (pH 6.5), 0.2 M sodium phosphate dibasic and 5–10% glycerol within two days and grew to their maximum size in one week. For crystallization of cofactor, substrate and product complex cocrystals, the protein was incubated with 5 mM final concentration of PnAA, PnA and NAD+, and the salt in mother liquor was substituted to 0.2 M ammonium chloride. Crystals were cryoprotected by brief soaking in mother liquor supplemented with 25% glycerol and vitrified in liquid nitrogen.
GAPDH was cloned from Streptococcus mutans Clarke ATCC® 25175 and expressed as an N-terminal His6-fusion protein from a pET15b vector in E. coli Rosetta2. Cells were grown to OD600 ~0.6 in LB, expression was induced with 0.3 mM IPTG at 18° C for 16 h, and cells were harvested by centrifugation. The cell pellet from 1 L of culture was resuspended in lysis buffer B (50 mM HEPES pH 7.5, 200 mM KCl, 10% glycerol, 5 mM DTT, 20 mM imidazole, 10 U DNAse, 25 mg lysozyme), lysed with a French press (15,000 psi), and the lysate was clarified by centrifugation. The clarified lysate was loaded onto a pre-equilibrated column with Ni-NTA resin. The column was washed with lysis buffer B, lysis buffer B with 50 mM imidazole, and lysis buffer B with 250 mM imidazole. After concentrating with an Amicon centrifuge filter, the protein buffer was exchanged to 50 mM HEPES pH 7.5, 200 mM KCl, 10% glycerol, 5 mM DTT using a NAP-25 filter. This process yielded ~60 mg GAPDH/L cell culture.
Phasing and structure determination
The structure of PhnY was solved by single wavelength anomalous dispersion (SAD) using data collected from a crystal of selenomethionine labeled protein. A four-fold redundant data set was collected from a single crystal of selenomethionine derivatized PhnY to a limiting resolution of 2.2 Ǻ (overall Rmerge = 11.7, I/σ(I) = 2 in the highest resolution shell) utilizing a Mar 300 CCD detector (LS-CAT, Sector 21 ID-D, Advanced Photon Source, Argonne, IL). Data were indexed and scaled using either the HKL-2000 package (Otwinowski et al., 2003) or XDS (Kabsch, 2010). Heavy atom sites were determined using HySS and refined using Phaser, as implemented in the Phenix suite of programs (Zwart et al., 2008), resulting in an initial Figure of Merit of 0.237. Initial phases were further improved through cycles of non-crystallographic symmetry averaging resulting in excellent quality experimental maps. Most of the main chain could be built using ARP/wARP (Perrakis et al., 1997). The remainder of the model was fitted using XtalView (McRee, 1999) or Coot (Emsley and Cowtan, 2004), and further improved by rounds of refinement with REFMAC (Murshudov et al., 1997; Murshudov et al., 1999) interspersed with rounds of manual building. Final refinement using geometric restraints and non crystallographic restraints was carried out using the Phenix package (Zwart et al., 2008). Cross-validation used 5% of the data in the calculation of the free R factor (Kleywegt and Brunger, 1996).
The cocrystal structures of PhnY in complex with PnAA, PnAA-NAD+, PnA-NAD+, and G3P-NAD+, were determined, to resolutions of 2.1 Å, 2.0 Å, 2.07 Å, and 2.25 Å, respectively, by molecular replacement using the coordinates of apo PhnY as a search probe. Each of the structures was refined and validated using the procedures detailed above. Cross-validation was routinely used throughout the course of model building and refinement using 5% of the data in the calculation of the free R factor. For each of the structures, the stereochemistry of the model was monitored throughout the course of refinement using PROCHECK (Laskowski et al., 1996). Relevant data collection and refinement statistics are provided in Table 1.
Synthesis of 3-OPP
Following a modified literature procedure (Rudinskas and Hullar, 1976), 2.0 g (7.5 mmol) of diethyl (3,3-diethoxypropyl)phosphonate was added to a three-necked flask equipped with a condenser under N2. TMSCl (3.8 mL, 30 mmol) was added, and the mixture was refluxed for 2 h. Although residual starting material was still present, refluxing for longer periods of time led to an increase in byproducts, which were difficult to remove. After 2 h, the mixture was cooled to 50° C, and 2 mL of H2O was added to hydrolyze the residual TMSCl and deprotect the silyl groups and unmask the aldehyde. The reaction was stirred for 30 min. H2O (5 mL) was added, and the solvent was removed using a rotary evaporator to afford a yellow oil. 1H, 31P, and [1H-31P] gHMBC NMR analysis and ESI-MS characterization confirmed the presence of both 3-OPP with a small amount of diethyl (3-oxopropyl)phosphonate as the sole byproduct. Spectral data for 3-OPP: NMR: 1H (500 MHz, D2O): δ 9.52 (s, CHO), 2.72 (dt, J = 14.6, 7.5, PCH2CH2), 2.06 (dt, J = 17.7, 7.5, PCH2CH2); 31P (202 MHz, D2O): δ 35.6 ppm. HR-MS (ESI+): found 139.0162, calculated 139.0160 (1.4 ppm). 31P NMR analysis of a stock of the 3-OPP mixture with an internal standard of Pi allowed the determination of the 3-OPP concentration.
Enzyme kinetics
The kinetics of substrate oxidation by wild type PhnY enzyme and site specific mutants were determined by monitoring the reduction of NAD+ using procedures described previously (Borisova et al., 2011). Enzyme (0.1–5 µM depending on the construct) was incubated with NAD+ in 50 mM HEPES, pH 7.5 supplemented with 10 mM MgSO4 for 5 min at 30° C. The reaction was initiated by addition of phosphonate/phosphate substrate. To obtain Michaelis-Menten kinetics, one substrate was maintained at saturating levels while the other substrate was varied; rates were obtained in triplicate. The Cys291Ala and Glu254Ala mutant enzymes were assayed at various substrate and enzyme concentrations. One mutant, Glu385Ala, displayed substrate inhibition and its kinetic parameters were obtained by fitting to the appropriate equation, v = kcat[E][S]/[Km,s +[S]+([S]2/Ki,s)], where [E] is the enzyme concentration, [S] is the substrate concentration, and Ki,s is the inhibition constant of the substrate. A similar procedure was used for the kinetic studies with by GAPDH (0.02–20 µM depending on the substrate) except that the buffer used was 50 mM HEPES pH 8.0 supplemented with 5 mM DTT and the cofactor used was NADP+.
Supplementary Material
Highlights.
Characterization of the oxidase involved in phosphonoacetate formation.
Crystallographic and kinetic data establish rationale for phosphonate tolerance.
First observation of a covalent thiohemiacetal intermediate.
ACKNOWLEDGEMENTS
We thank Drs. Keith Brister and Joseph Brunzelle at the LS-CAT (Sector-21) beamline at Argonne National Laboratory for assistance with X-ray diffraction data collection, and Prof. John A. Gerlt (University of Illinois) for stimulating discussions and S. mutans genomic DNA. Use of the Advanced Photon Source, an Office of Science User Facility operated for the U.S. Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the U.S. DOE under Contract No. DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant 085P1000817). NMR spectra were recorded on a 600 MHz instrument purchased with support from NIH S10 RR028833. This work was supported by the National Institutes of Health (GM P01 077596 to W.A.V. and S.K.N.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION CODES
The crystallographic coordinates and maps have been deposited in the Protein Data Bank with the following accession codes: PhnY-apo:4I3T PhnY-PnAA:4I3U PhnY-PnAA-NAD+:4I3V PhnY-PnA-NAD+:4I3X PhnY-G3P-NAD+:4I3W.
The authors declare no competing financial interests.
AUTHOR CONTRIBUTIONS
V.A., S.C.P., and S.K.N. designed research, V.A., S.C.P., J.H.C. and J.R.C. performed research, and S.A.B. contributed new analytical tools/reagents, V.A., S.C.P., W.A.V., and S.K.N. analyzed data, V.A., S.C.P., W.A.V., and S.K.N. wrote the paper.
SUPPORTING INFORMATION
Supplemental Figures S1–S8.
REFERENCES
- Agarwal V, Borisova SA, Metcalf WW, van der Donk WA, Nair SK. Structural and mechanistic insights into C-P bond hydrolysis by phosphonoacetate hydrolase. Chem Biol. 2011;18:1230–1240. doi: 10.1016/j.chembiol.2011.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisova SA, Christman HD, Metcalf ME, Zulkepli NA, Zhang JK, van der Donk WA, Metcalf WW. Genetic and biochemical characterization of a pathway for the degradation of 2-aminoethylphosphonate in Sinorhizobium meliloti 1021. J Biol Chem. 2011;286:22283–22290. doi: 10.1074/jbc.M111.237735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisova SA, Circello BT, Zhang JK, van der Donk WA, Metcalf WW. Biosynthesis of rhizocticins, antifungal phosphonate oligopeptides produced by Bacillus subtilis ATCC6633. Chem Biol. 2010;17:28–37. doi: 10.1016/j.chembiol.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobessi D, Tete-Favier F, Marchal S, Branlant G, Aubry A. Structural and biochemical investigations of the catalytic mechanism of an NADP-dependent aldehyde dehydrogenase from Streptococcus mutans. J Mol Biol. 2000;300:141–152. doi: 10.1006/jmbi.2000.3824. [DOI] [PubMed] [Google Scholar]
- Cooley NA, Kulakova AN, Villarreal-Chiu JF, Gilbert JA, McGrath JW, Quinn JP. Phosphonoacetate biosynthesis: in vitro detection of a novel NADP(+)-dependent phosphonoacetaldehyde-oxidizing activity in cell-extracts of the marine Roseovarius nubinhibens ISM. Mikrobiologiia. 2011;80:329–334. [PubMed] [Google Scholar]
- Costas AM, White AK, Metcalf WW. Purification and characterization of a novel phosphorus-oxidizing enzyme from Pseudomonas stutzeri WM88. J Biol Chem. 2001;276:17429–17436. doi: 10.1074/jbc.M011764200. [DOI] [PubMed] [Google Scholar]
- D'Ambrosio K, Pailot A, Talfournier F, Didierjean C, Benedetti E, Aubry A, Branlant G, Corbier C. The first crystal structure of a thioacylenzyme intermediate in the ALDH family: new coenzyme conformation and relevance to catalysis. Biochemistry. 2006;45:2978–2986. doi: 10.1021/bi0515117. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Hedstrom L. Serine protease mechanism and specificity. Chem Rev. 2002;102:4501–4524. doi: 10.1021/cr000033x. [DOI] [PubMed] [Google Scholar]
- Hove-Jensen B, McSorley FR, Zechel DL. Physiological role of phnP-specified phosphoribosyl cyclic phosphodiesterase in catabolism of organophosphonic acids by the carbon-phosphorus lyase pathway. J Am Chem Soc. 2011;133:3617–3624. doi: 10.1021/ja1102713. [DOI] [PubMed] [Google Scholar]
- Hove-Jensen B, McSorley FR, Zechel DL. Catabolism and detoxification of 1-aminoalkylphosphonic acids: N-acetylation by the phnO gene product. PLoS One. 2012;7:e46416. doi: 10.1371/journal.pone.0046416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamat SS, Williams HJ, Raushel FM. Intermediates in the transformation of phosphonates to phosphate by bacteria. Nature. 2011;480:570–573. doi: 10.1038/nature10622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim A, Benning MM, OkLee S, Quinn J, Martin BM, Holden HM, Dunaway-Mariano D. Divergence of chemical function in the alkaline phosphatase superfamily: structure and mechanism of the P-C bond cleaving enzyme phosphonoacetate hydrolase. Biochemistry. 2011;50:3481–3494. doi: 10.1021/bi200165h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim AD, Baker AS, Dunaway-Mariano D, Metcalf WW, Wanner BL, Martin BM. The 2-aminoethylphosphonate-specific transaminase of the 2-aminoethylphosphonate degradation pathway. J Bacteriol. 2002;184:4134–4140. doi: 10.1128/JB.184.15.4134-4140.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleywegt GJ, Brunger AT. Checking your imagination: applications of the free R value. Structure. 1996;4:897–904. doi: 10.1016/s0969-2126(96)00097-4. [DOI] [PubMed] [Google Scholar]
- Langendorf CG, Key TL, Fenalti G, Kan WT, Buckle AM, Caradoc-Davies T, Tuck KL, Law RH, Whisstock JC. The X-ray crystal structure of Escherichia coli succinic semialdehyde dehydrogenase; structural insights into NADP+/enzyme interactions. PLoS One. 2010;5:e9280. doi: 10.1371/journal.pone.0009280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Swindells MB. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model. 2011;51:2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- Marchal S, Branlant G. Evidence for the chemical activation of essential cys-302 upon cofactor binding to nonphosphorylating glyceraldehyde 3-phosphate dehydrogenase from Streptococcus mutans. Biochemistry. 1999;38:12950–12958. doi: 10.1021/bi990453k. [DOI] [PubMed] [Google Scholar]
- Marchal S, Rahuel-Clermont S, Branlant G. Role of glutamate-268 in the catalytic mechanism of nonphosphorylating glyceraldehyde-3-phosphate dehydrogenase from Streptococcus mutans. Biochemistry. 2000;39:3327–3335. doi: 10.1021/bi9914208. [DOI] [PubMed] [Google Scholar]
- Martinez A, Osburne MS, Sharma AK, DeLong EF, Chisholm SW. Phosphite utilization by the marine picocyanobacterium Prochlorococcus MIT9301. Environ Microbiol. 2012;14:1363–1377. doi: 10.1111/j.1462-2920.2011.02612.x. [DOI] [PubMed] [Google Scholar]
- McGrath JW, Chin JP, Quinn JP. Organophosphonates revealed: new insights into the microbial metabolism of ancient molecules. Nat Rev Microbiol. 2013;11:412–419. doi: 10.1038/nrmicro3011. [DOI] [PubMed] [Google Scholar]
- McRee DE. XtalView/Xfit--A versatile program for manipulating atomic coordinates and electron density. J Struct Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- McSorley FR, Wyatt PB, Martinez A, Delong EF, Hove-Jensen B, Zechel DL. PhnY and PhnZ comprise a new oxidative pathway for enzymatic cleavage of a carbon-phosphorus bond. J Am Chem Soc. 2012;134:8364–8367. doi: 10.1021/ja302072f. [DOI] [PubMed] [Google Scholar]
- Metcalf WW, Griffin BM, Cicchillo RM, Gao J, Janga SC, Cooke HA, Circello BT, Evans BS, Martens-Habbena W, Stahl DA, et al. Synthesis of methylphosphonic acid by marine microbes: a source for methane in the aerobic ocean. Science. 2012;337:1104–1107. doi: 10.1126/science.1219875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf WW, van der Donk WA. Biosynthesis of phosphonic and phosphinic acid natural products. Annu Rev Biochem. 2009;78:65–94. doi: 10.1146/annurev.biochem.78.091707.100215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morais MC, Zhang W, Baker AS, Zhang G, Dunaway-Mariano D, Allen KN. The crystal structure of Bacillus cereus phosphonoacetaldehyde hydrolase: insight into catalysis of phosphorus bond cleavage and catalytic diversification within the HAD enzyme superfamily. Biochemistry. 2000;39:10385–10396. doi: 10.1021/bi001171j. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Lebedev A, Wilson KS, Dodson EJ. Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr D Biol Crystallogr. 1999;55:247–255. doi: 10.1107/S090744499801405X. [DOI] [PubMed] [Google Scholar]
- Nair SK, van der Donk WA. Structure and mechanism of enzymes involved in biosynthesis and breakdown of the phosphonates fosfomycin, dehydrophos, and phosphinothricin. Arch Biochem Biophys. 2011;505:13–21. doi: 10.1016/j.abb.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Borek D, Majewski W, Minor W. Multiparametric scaling of diffraction intensities. Acta Crystallogr A. 2003;59:228–234. doi: 10.1107/s0108767303005488. [DOI] [PubMed] [Google Scholar]
- Peck SC, Kim SY, Evans BS, van der Donk WA. Stereochemistry of hydride transfer by group III alcohol dehydrogenases involved in phosphonate biosynthesis. Medchemcomm. 2012;3:967–970. doi: 10.1039/C2MD20009K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck SC, van der Donk WA. Phosphonate biosynthesis and catabolism: a treasure trove of unusual enzymology. Curr Opin Chem Biol. 2013;17:580–588. doi: 10.1016/j.cbpa.2013.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Miller SJ, Hurley TD. Coenzyme isomerization is integral to catalysis in aldehyde dehydrogenase. Biochemistry. 2003;42:7100–7109. doi: 10.1021/bi034182w. [DOI] [PubMed] [Google Scholar]
- Perrakis A, Sixma TK, Wilson KS, Lamzin VS. wARP: improvement and extension of crystallographic phases by weighted averaging of multiple-refined dummy atomic models. Acta Crystallogr D Biol Crystallogr. 1997;53:448–455. doi: 10.1107/S0907444997005696. [DOI] [PubMed] [Google Scholar]
- Rudinskas AJ, Hullar TL. Pyridoxal phosphate. 5. 2-Formylethynylphosphonic acid and 2-formylethylphosphonic acid, potent inhibitors of pyridoxal phosphate binding and probes of enzyme topography. J Med Chem. 1976;19:1367–1371. doi: 10.1021/jm00234a003. [DOI] [PubMed] [Google Scholar]
- Shao Z, Blodgett JA, Circello BT, Eliot AC, Woodyer R, Li G, van der Donk WA, Metcalf WW, Zhao H. Biosynthesis of 2-hydroxyethylphosphonate, an unexpected intermediate common to multiple phosphonate biosynthetic pathways. J Biol Chem. 2008;283:23161–23168. doi: 10.1074/jbc.M801788200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White AK, Metcalf WW. Microbial metabolism of reduced phosphorus compounds. Annu Rev Microbiol. 2007;61:379–400. doi: 10.1146/annurev.micro.61.080706.093357. [DOI] [PubMed] [Google Scholar]
- Woodyer RD, Li G, Zhao H, van der Donk WA. New insight into the mechanism of methyl transfer during the biosynthesis of fosfomycin. Chem Commun (Camb) 2007:359–361. doi: 10.1039/b614678c. [DOI] [PubMed] [Google Scholar]
- Zhang G, Dai J, Lu Z, Dunaway-Mariano D. The phosphonopyruvate decarboxylase from Bacteroides fragilis. J Biol Chem. 2003;278:41302–41308. doi: 10.1074/jbc.M305976200. [DOI] [PubMed] [Google Scholar]
- Zwart PH, Afonine PV, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, McKee E, Moriarty NW, Read RJ, Sacchettini JC, et al. Automated structure solution with the PHENIX suite. Methods Mol Biol. 2008;426:419–435. doi: 10.1007/978-1-60327-058-8_28. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.