Abstract
Phosphinothricin-tripeptide (PTT, phosphinothricyl-alanyl-alanine) is a natural product antibiotic and potent herbicide that is produced by Streptomyces hygroscopicus ATCC 217051 and Streptomyces viridochromogenes DSM 407362. PTT has attracted widespread interest due to its commercial applications and unique phosphinic acid functional group. Despite intensive study since its discovery in 1972 (see3 for a comprehensive review), a number of steps early in the PTT biosynthetic pathway remain uncharacterized. Here we report a series of interdisciplinary experiments involving the construction of defined S. viridochromogenes mutants, chemical characterization of accumulated intermediates, and in vitro assay of selected enzymes to examine these critical steps in PTT biosynthesis. Our results indicate that early PTT biosynthesis involves a series of heretofore undescribed catalyses, including a highly unusual reaction for carbon bond cleavage. In sum, we define a more complex pathway for early PTT biosynthesis that includes biochemically unprecedented and chemically interesting steps.
Keywords: Streptomyces viridochromogenes, phosphinothricin, biosynthesis, bialaphos, phosphonate metabolism
The bioactive portion of the PTT (also known as bialaphos, 1) tripeptide is phosphinothricin (PT, 2). PT is the only known naturally occurring compound to incorporate a direct carbon to phosphorus to carbon (C-P-C) bond motif. The phosphinic acid functional group comprised by this bonding arrangement allows PT to act as a glutamine synthetase inhibitor, where it mimics the tetrahedral transition state of the catalytic intermediate γ-glutamyl phosphate4. PTT is a member of a growing family of secondary metabolites containing phosphonate or phosphinate functional groups. This family includes fosfomycin (3), a clinically utilized antibacterial5, and fosmidomycin (4), a potent antimalarial agent currently undergoing field trials6.
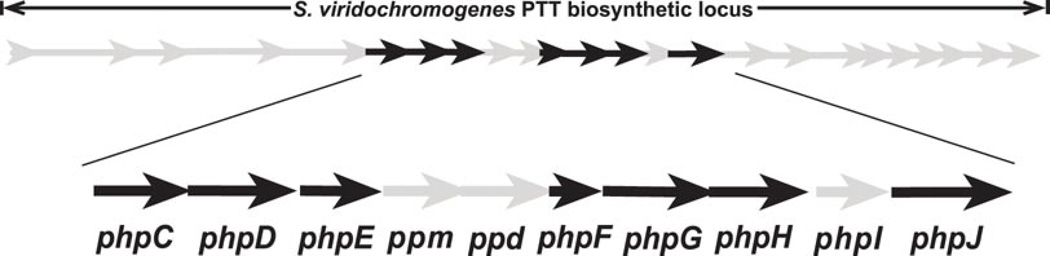
PTT has become a model for the biosynthesis of phosphonic acid antibiotics based on numerous studies3, leading to a pathway that is regarded as mostly solved. It should be noted, however, that many of the PTT non-producing mutants analyzed to define the pathway were generated by random chemical mutagenesis and some mutations were not rigorously mapped to individual genes. Accordingly, the resulting biosynthetic model is based upon a patchwork of chemical, enzymatic, and genetic data. Further, though many PTT biosynthetic genes were previously cloned and sequenced, the full gene cluster was not completely sequenced from either producer until very recently, when the cluster from S. viridochromogenes was isolated and characterized7, 8. Sequence analysis of the cluster revealed that only about half of the genes identified had rigorous genetic or biochemical data establishing their roles in PTT biosynthesis. Some of the remaining genes were assigned tentative biosynthetic roles based upon sequence homology, but others could not be confidently assigned despite their location in the core of the PTT gene cluster (Fig. 1a).
Figure 1.
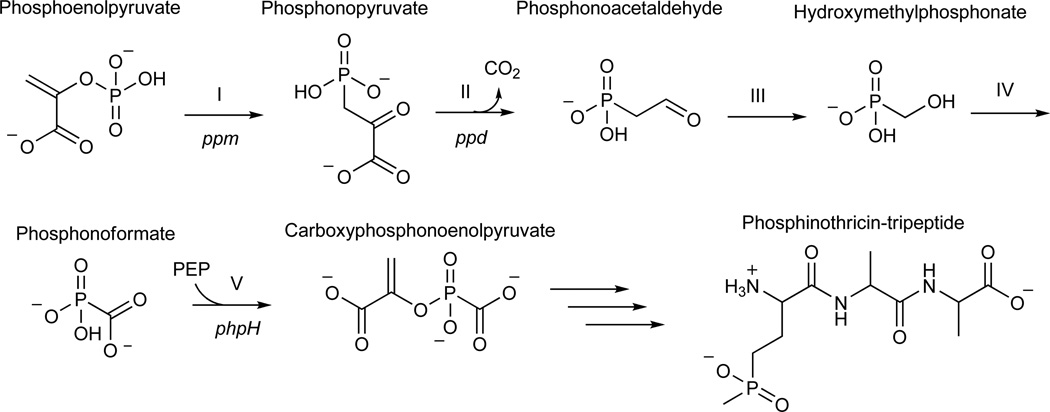
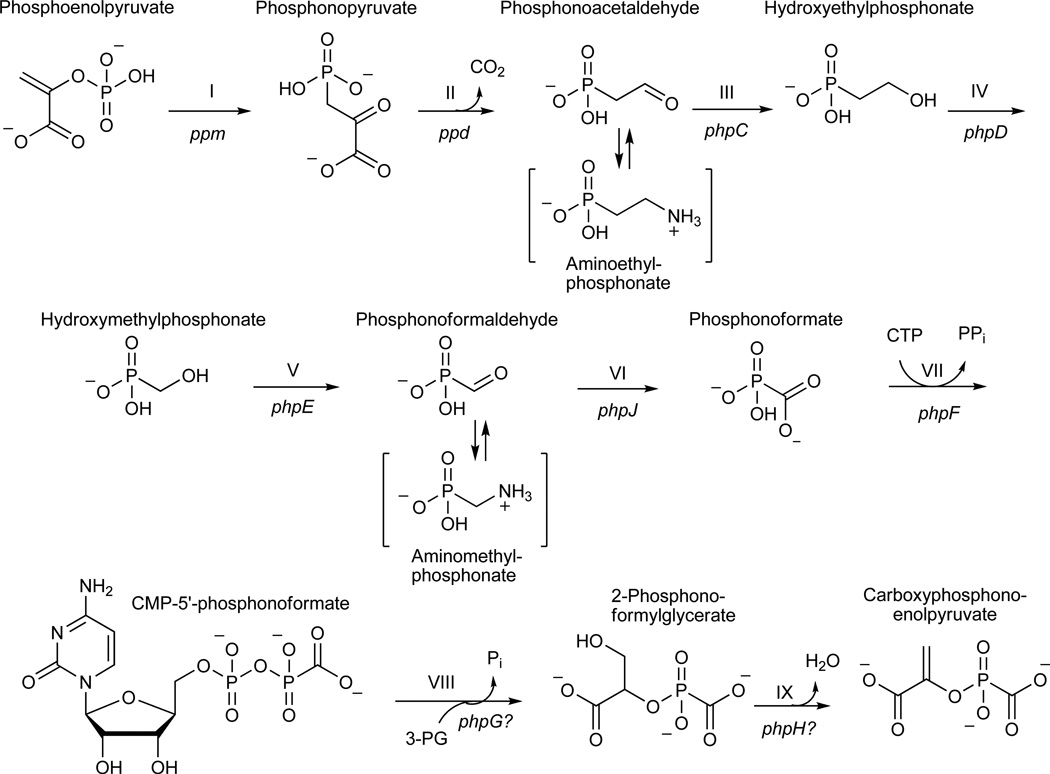
Map of PTT biosynthetic genes and model for early steps in PTT biosynthesis. (a) Detail of genes in the core of the PTT biosynthetic gene cluster. Genes shown in grey were not studied in this work because they have experimentally established roles in PTT biosynthesis or because homology has allowed roles to be assigned by inference. The open reading frames in black did not have experimentally assigned roles, except for phpH, where this gene is expected to function in CPEP synthesis. (b) Model for carboxyphosphonoenolpyruvate biosynthesis from phosphonoacetaldehyde via the intermediates hydroxymethylphosphonate and phosphonoformate (adapted from3). Steps referred to in the text are denoted by roman numerals, and S. viridochromogenes genes thought to be involved in each step7 are indicated where possible.
The first two steps in PTT biosynthesis are well established9–11, but the chemical logic of the previously proposed pathway (Fig. 1b) suggested that a number of other reactions found early in the pathway could be biochemically interesting. These include the unusual cleavage of phosphonoacetaldehyde (5) into hydroxymethylphosphonate (HMP, 6) (step III) and the subsequent oxidation of hydroxymethylphosphonate directly into phosphonoformate (7) (step IV). In addition, the utilization of phosphonoformate to directly replace the phosphate of phosphoenolpyruvate (8), thereby producing the intermediate carboxyphosphonoenolpyruvate (CPEP, 9) (step V)12, is also unexpected. This is because the enzyme that is reported to be involved, CPEP synthase, shares significant identity with enolase and related proteins13. Because these enzymes usually carry out dehydration reactions, the proposed reaction is biochemically inconsistent with the expected activity the protein14. Based upon these observations, the early steps in PTT biosynthesis clearly warranted further investigation.
Previous analyses indicated that location of the group of genes comprised of phpC, phpD, and phpE in the PTT gene cluster suggested that they could have roles in the conversion of phosphonoacetaldehyde to phosphonoformate (Fig 1b, steps III and IV) 7, 15. The role of phpC was unknown and its homology to type III (iron-dependent) alcohol dehydrogenases did not suggest an obvious function for the gene in PTT biosynthesis. To determine its role, we constructed and characterized a ΔphpC mutant of S. viridochromogenes (WM6547). Initial bioassay results indicated that the ΔphpC mutant retained the ability to produce PTT when grown in liquid culture or on ISP2 (a glucose-yeast-malt medium) plates. However, the amount of PTT produced appeared to be less than that produced by the parent strain (Supplementary Fig. 1). When concentrated culture supernatants were examined by 31P NMR, the characteristic peak of PTT at 42 ppm was not observed. Instead, a strong resonance (δ 17.2) that was identified as 2-aminoethylphosphonate (AEP, 10) (by spiking the sample with authentic standard) was observed. AEP was also identified in the sample by LC/MS (Table 1, Supplementary Fig. 2).
Table 1.
LC/MS data acquired for early PTT biosynthetic intermediates
| Sample | Compound | Formula | Retention time (min)a |
Ion detected |
Calculated (m/z) |
Measured (m/z)b |
|---|---|---|---|---|---|---|
| WM6492 (ΔphpC) | AEP | C2H8NO3P | 22.5/ 22.5 | [M−H]− | 124.0169 | 124.0168/ 124.0169 |
| PhpC in vitro | HEP | C2H7O4P | 15.8/ 16.4 | [M−H]− | 125.0009 | 125.0010/ 125.0009 |
| WM6601 (ΔphpD) | HEP | C2H7O4P | 16.2/ 16.4 | [M−H]− | 125.0009 | 125.0009/ 125.0009 |
| PhpD in vitro | HMP | CH5O4P | 15.7/ 16.9 | [M−H]− | 110.9853 | 110.9852/ 110.9853 |
| WM6602 (ΔphpE) | HMP | CH5O4P | 17.6/ 16.9 | [M−H]− | 110.9853 | 110.9852/ 110.9853 |
| WM6488 (ΔphpJ) | AMPn | CH6NO3P | 22.1/ 22.8 | [M−H]− | 110.0013 | 110.0011/ 110.0013 |
| PhpF in vitro | CMP-5’-PF | C10H15N3O12P2 | 9.6 | [M+H]+ | 432.0204 | 432.0203 |
| PhpF in vitro | CMP-5’-PF, NH4+ | C10H15N3O12P2 | 9.6 | [M+NH4]+ | 449.0469 | 449.0468 |
Retention times and measured mass to charge ratio (m/z) values are listed such that the first value represents our experimental results and the second value is derived from the analysis of a corresponding authentic standard (if available).
Although AEP was not known or expected to be an early PTT intermediate, we reasoned that it could represent a derivative of the biosynthetic intermediate phosphonoacetaldehyde because the recovery of amino derivatives of carbonyl-containing PTT biosynthetic intermediates from blocked mutants is not unprecedented. Phosphinoalanine (11) and aminoethylphosphinic (12) acid, likely degradation products of the established intermediate phosphinopyruvate (13), have been isolated from a S. hygroscopicus non-producing mutant16. Further, previous groups working on phosphonate biosynthesis have utilized AEP as a physiological equivalent of phosphonoacetaldehyde17,18. The conversions of carbonyls to amino groups, as well as the reverse reactions, are likely carried out by aminotransferases. This is supported by the observation that many of these enzymes often display relaxed substrate specificity19.
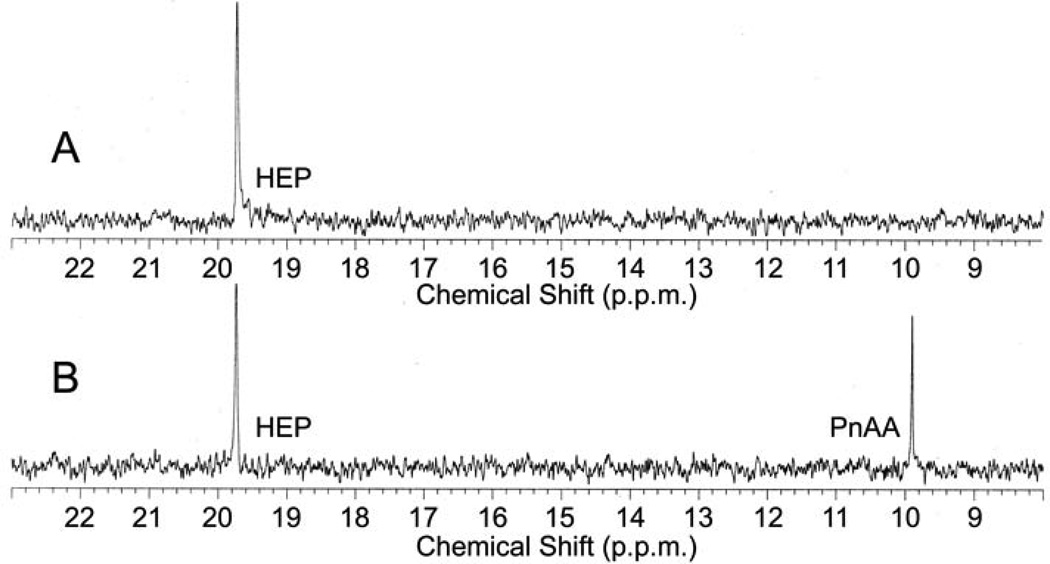
If AEP is a phosphonoacetaldehyde derivative, then phosphonoacetaldehyde should be the substrate for the alcohol dehydrogenase encoded by phpC. To test if PhpC acts as a phosphonoacetaldehyde reductase, we purified a histidine-tagged PhpC fusion after over-expression in Streptomyces lividans. His-PhpC catalyzed the in vitro reduction of phosphonoacetaldehyde to hydroxyethylphosphonate (HEP, 14) with either NADH (15) or NADPH (16) as a cofactor using 31P NMR spectroscopy (Fig. 2) and LC/MS (Table 1, Supplementary Fig. 3) to monitor the reaction. The reaction proceeds to apparent completion in the presence of NADH, but to a lesser extent with NADPH, suggesting that NADH is the preferred cofactor. These data support the hypothesis that the in vivo accumulation of aminoethylphosphonate results from a side reaction, and further suggests that the reduction of phosphonoacetaldehyde to hydroxyethylphosphonate is step III (Fig. 3) of the PTT biosynthetic pathway.
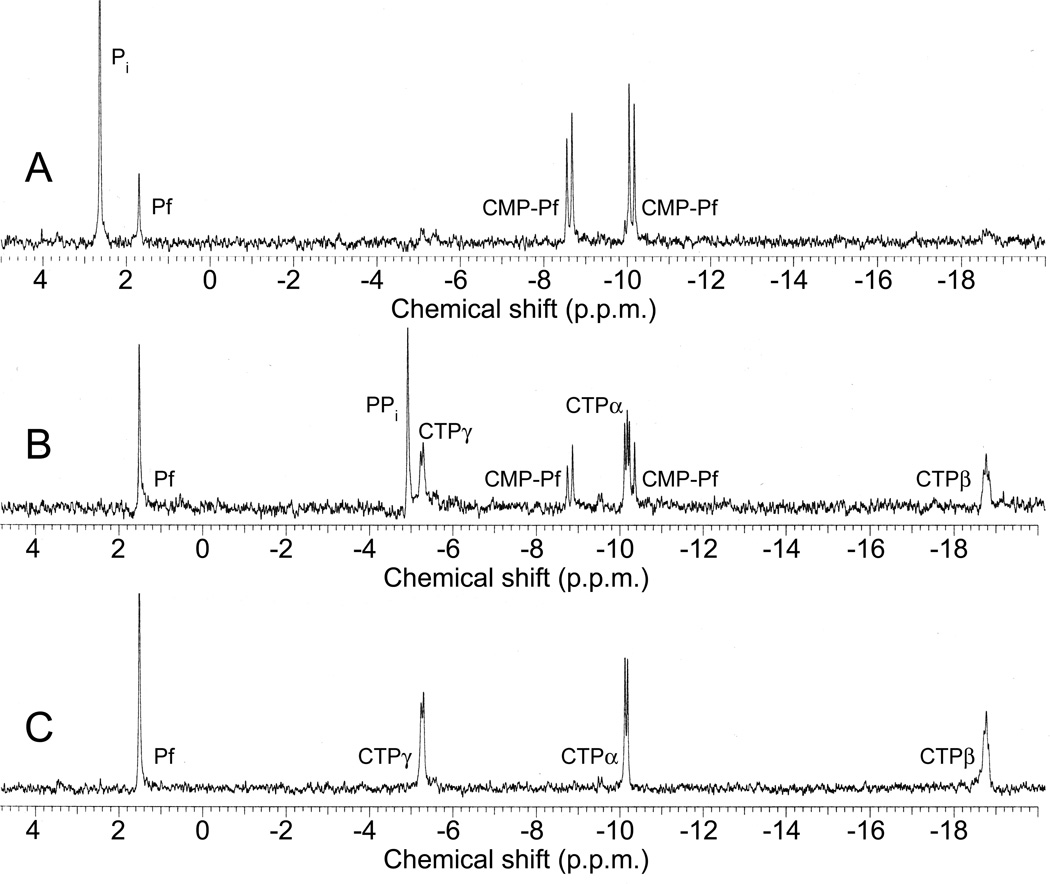
Figure 2.
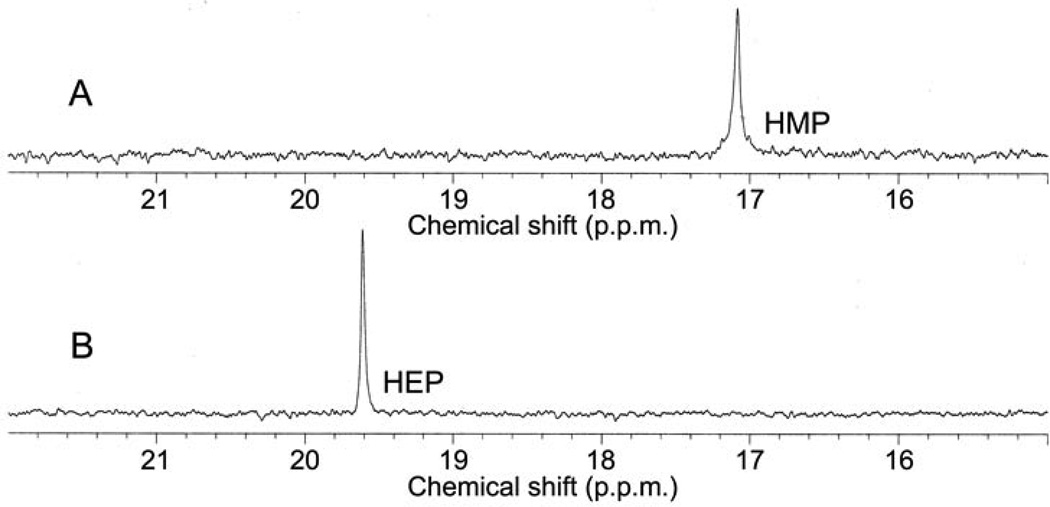
In vitro reconstitution of early PTT biosynthetic reactions monitored by 31P NMR. (a) 31P NMR results of the His-PhpC catalyzed reduction of phosphonoacetaldehyde (PnAA, δ 9.9) to hydroxyethylphosphonate (HEP, δ 9.7) in the presence of (A) NADH and (B), the same reaction performed in the presence of NADPH. (b) 31P NMR results confirming the direct in vitro conversion of hydroxyethylphosphonate to hydroxymethylphosphonate by PhpD. (A), an assay performed in the presence of Rosetta(DE3)pLysS/pJVD26 extract containing PhpD, showing the conversion of hydroxyethylphosphonate into hydroxymethylphosphonate (HMP, δ 17.1). (B) a control assay containing hydroxyethylphosphonate (HEP, δ 19.6) and Rosetta(DE3)pLysS extract. (c) 31P NMR results of assays establishing the CTP-phosphonoformate nucleotidyltransferase activity of His-PhpF. (A) A reaction containing His-PhpF, inorganic pyrophosphatase, CTP and phosphonoformate showing the accumulation of phosphate (δ 2.6), and the conversion of substrates into CMP-5’-phosphonoformate (δ −8.6, −10.1). The top of the phosphate peak is not shown to fit the spectrum to the figure. (B), the same reaction as in A in the absence of pyrophosphatase showing the partial conversion of phosphonoformate and CTP into CMP-5’-phosphonoformate (δ −8.6, −10.1) and pyrophosphate (δ −4.7) and (C) a control assay containing heat-inactivated His-PhpF, CTP (δ −5.1, −9.97, −18.6) and phosphonoformate (δ 1.7), The putative α, β, and γ designations of the CTP phosphorus nuclei have been assigned by comparison to those analogously found in ATP 30.
Figure 3.
An alternative pathway for carboxyphosphonoenolpyruvate biosynthesis from phosphonoacetaldehyde supported by data presented in this work. Steps referred to in the text are indicated by roman numerals and are annotated with their respective genetic determinants. Bracketed intermediates were identified from blocked mutants and likely represent modifications of the corresponding aldehyde intermediates.
The discovery of hydroxyethylphosphonate as a biosynthetic intermediate is interesting because the compound has previously been isolated from the culture broth of a PTT non-producing S. hygroscopicus mutant20, but the compound has largely been regarded as the product of a side reaction3. We found data to directly establish hydroxyethylphosphonate as a true biosynthetic intermediate after we characterized a ΔphpD mutant (WM6601). The mutant failed to produce PTT under any conditions tested and 31P NMR analysis of culture supernatants from this mutant revealed the presence of hydroxyethylphosphonate (δ 19.1). The accumulation of this compound was additionally confirmed by LC/MS (Table 1, Supplementary Fig. 4). The secretion of hydroxyethylphosphonate by the ΔphpD mutant suggested that this intermediate might be the substrate for PhpD. To test this, we over-expressed PhpD in E. coli and examined its ability to convert hydroxyethylphosphonate to another phosphonate via LC/MS and 31P NMR spectroscopy. Our NMR results (Fig. 2b) demonstrated that extracts from cells carrying a PhpD expression plasmid, but not from the plasmid-free host, convert hydroxyethylphosphonate (δ 19.6) into hydroxymethylphosphonate (δ 17.1). These results were confirmed by LC/MS (Table 1, Supplementary Fig. 5).
The direct conversion of 2-hydroxyethylphosphonate into hydroxymethylphosphonate (Fig. 3, step IV) by PhpD is worthy of special attention because the reaction has few, if any, biochemical precedents. PhpD is not homologous to any protein of known function in the current Genbank release7 and therefore we cannot propose a mechanism based upon bioinformatic analysis. It is difficult to envision a simple chemical mechanism for the reaction, but our initial biochemical results firmly establish hydroxyethylphosphonate-cleavage activity to PhpD.
Hydroxymethylphosphonate, the product of the PhpD reaction, is a recognized PTT biosynthetic intermediate that was first isolated during the analysis of a genetically uncharacterized non-producing mutant of S. hygroscopicus20. During our characterization of a ΔphpE mutant (WM6602), we found that hydroxymethylphosphonate (HMP, δ 16.4) accumulated in culture broth after 31P NMR analysis, with LC/MS independently verifying the compound (Table 1, Supplementary Fig. 6) in the sample. Bioassays of liquid cultures indicated that the ΔphpE mutant does not produce PTT; however, weak bioactivity was noted when the strain was assayed after growth on ISP2 plates (Supplementary Fig. 1) The phpE gene encodes a protein with similarity to members of the 2-hydroxyacid alcohol dehydrogenase family of enzymes, which, taken together with our data, suggests that PhpE is an alcohol dehydrogenase whose substrate is hydroxymethylphosphonate (Fig. 3, step V). Initial attempts to demonstrate this activity after over-expression of the protein in both E. coli and S. lividans failed (data not shown).
If PhpE is a hydroxymethylphosphonate dehydrogenase, then its expected product would be phosphonoformaldehyde (17). Therefore, to produce the known intermediate phosphonoformate, the activity of an aldehyde dehydrogenase would likely be required. Sequence homology suggests that phpJ encodes an aldehyde dehydrogenase that could have this function8. We found that a ΔphpJ mutant (WM6548) produced PTT in both liquid and plate-grown cultures. Nevertheless, 31P NMR analysis of culture supernatants from this mutant indicated the accumulation of a compound with a strong resonance (δ 9.8) corresponding to aminomethylphosphonate (AMPn, 18). Two additional small peaks were also noted in the spectrum; one corresponding to hydroxymethylphosphonate (δ 16.4), the other consistent with PTT (δ 43.3). LC/MS analysis independently confirmed the presence of aminomethylphosphonate in the sample (Table 1, Supplementary Fig. 7). Given the proposed activity of PhpJ, we expected to find phosphonoformaldehyde in the culture broth of the phpJ mutant, not aminomethylphosphonate. These results, however, are reminiscent of the accumulation of aminoethylphosphonate by the phpC mutant. Thus, we surmise that non-specific aminotransferases similar to those that convert phosphonoacetaldehyde into aminoethylphosphonate perform the analogous reaction on phosphonoformaldehyde, giving aminomethylphosphonate. Our genetic results therefore suggest that step VI (Fig. 3) of PTT biosynthesis is the oxidation of phosphonoformaldehyde to phosphonoformate.
Our characterization of a phpF mutant suggested that it is involved in the next step of PTT biosynthesis. The gene encodes a nucleotidyltransferase homolog, and although phpF has been sequenced in both S. hygroscopicus and S. viridochromogenes, a proposed role for it in PTT biosynthesis has never been suggested. We found that a ΔphpF mutant (WM6546) fails to produce PTT after growth in MYG broth, though low level PTT biosynthesis was noted when the strain was grown on ISP2 plates (Supplementary Fig. 1). 31P NMR analysis of concentrated culture supernatants from this mutant revealed 2 unique peaks (δ 1.1, 4.4), which correspond to phosphonoformate and phosphite (19), respectively. In addition, a minor peak corresponding to hydroxymethylphosphonate (S/N~9, δ 16.7) was also detected. Phosphite is a known product of the spontaneous decarboxylation of phosphonoformate3, thus its presence in a sample containing phosphonoformate is not surprising. These data suggested that PhpF plays an important role in PTT biosynthesis, probably acting after the formation of phosphonoformate.
To verify that PhpF acts after the synthesis of phosphoformate, we performed a chemical feeding experiment. The ΔphpF mutant was unable to produce PTT when grown in MYG broth supplemented with 0.5 mM phosphonoformate, using a bioassay to score for antibiotic production. Conversely, both the ΔphpD and ΔphpE mutants were able to convert this compound to PTT, indicating that it can be taken up and metabolized by strains used in our study. Therefore phpD and phpE act before phosphonoformate is produced in the PTT pathway, whereas phpF acts after phosphonoformate production.
To address the role of phpF biochemically, we purified a histidine-tagged PhpF fusion by nickel-affinity chromatography after over-expression in E. coli. Nucleotidyltransferase activity was examined by 31P NMR spectroscopy in an assay that included His-PhpF, CTP (20), phosphonoformate and inorganic pyrophosphatase (Fig. 2c). In these reactions, a small portion of unreacted phosphonoformate (δ 1.7) remains and a significant amount of phosphate (δ 2.6) has accumulated. Further, two new peaks are observed [δ −8.6 (d, J=29.9 Hz), −10.1 (d, J=29.6 Hz)]. The spin-spin coupling (J) observed for this new product is much broader than that seen for CTP (Fig. 2c), suggesting that the compound is unlikely to be another phosphoanhydride (such as CDP, 21). Similar, but lower-level activity was also observed when UTP (22) and dCTP (23) were used as co-substrates with phosphonoformate (Supplemental Fig. 8); however, because these reactions never approached completion we assume that CTP is the in vivo substrate.
In the preceding assay we included inorganic pyrophosphatase to drive the reaction to completion, because we assumed that pyrophosphate (PPi, 24) would be displaced from CTP during catalysis. To verify this, we performed an assay in the absence of pyrophosphatase, which resulted in the partial conversion of the substrates to product with the concomitant accumulation of pyrophosphate (δ −4.7) (Fig. 2c). To show that the reactions described above are dependent upon His-PhpF, a control assay using heat inactivated His-PhpF was performed. The 31P NMR assay results indicated that activity of the protein was abolished and peaks corresponding to the unreacted substrates phosphonoformate (δ 1.7) and the phosphoanhydride of CTP [δ −5.1 (d, J= 14.1 Hz), −9.97 (d, J=15.3 Hz), −18.6] were observed.
Our in vitro experiments with His-PhpF strongly suggested that the enzyme catalyzes the displacement of the β and γ phosphate of CTP (as pyrophosphate) by phosphonoformate to produce a novel intermediate: cytidine monophosphate-5’-phosphonoformate (CMP-5’-PF, 25). LC/MS analysis of the PhpF-catalyzed reaction detected CMP-5’-PF and a mass corresponding to an ammonium adduct (26) of the product (Table 1, Supplementary Fig. 9). To further confirm the structure of CMP-5’-PF, tandem mass spectrometry (MS2) was performed, providing additional supporting data (see Supplementary Data).
Our identification of the CMP-5’-PF intermediate is important for two reasons. First, it establishes that CPEP biosynthesis probably doesn’t occur as shown in Fig. 1b. Second, CMP-5’-phosphonoformate can be viewed as an activated intermediate primed for transfer of the phosphonoformate group to an appropriate acceptor molecule. This fits well with our recent suggestion that the PhpG and PhpH proteins could work in concert to produce CPEP in vivo via a reaction series analogous to the phosphoglycerate mutase and enolase reactions of glycolysis7. PhpG is a close homolog of an autophosphorylating phosphoglycerate mutase from the archaeon Sulfolobus solfatericus21. The Sulfolobus enzyme has been shown to catalyze the transfer of the γ-phosphate of ATP (27) directly to an active site serine, creating a phosphoprotein intermediate. This phosphate group is subsequently transferred to 3-phosphoglycerate (28), yielding 2-phosphoglycerate (29) as a product. We suggest that PhpG catalyzes an analogous reaction using the activated CMP-5’-phosphonoformate intermediate to donate phosphonoformate to the active site of the enzyme. The enzyme bound phosphonoformate could then be donated to a compound such as 3-phosphoglycerate, yielding a phosphonoformylated intermediate that would then serve as the substrate for the PhpH enolase, creating CPEP (Fig. 3).
We tested the above hypotheses regarding the functions of phpG and phpH by mutational analysis. Bioassays of a ΔphpG mutant (WM6549) indicated that PTT was not produced by liquid-grown cultures, although the strain did exhibit some bioactivity when grown on solid ISP2 medium. The ΔphpH mutant (WM6604) failed to produce PTT under all conditions tested. Unfortunately, no significant accumulation of phosphonates was observed in either strain based upon 31P NMR analysis. Nevertheless, because PTT production is eliminated in the phpH mutant and strongly reduced in the phpG mutant, it seems likely that both genes are involved in the biosynthesis of this antibiotic. Neither mutant was able to convert phosphonoformate to PTT in chemical feeding experiments; thus, both phpG and phpH clearly act after the formation of phosphonoformate in the PTT pathway. Because we failed to obtain additional data to support our proposed roles for phpG and phpH, this portion of our model remains speculative. However, based upon the evidence presented above, we believe this model to be more credible than that previously proposed.
During our analysis of PTT biosynthesis, we expected our mutants to be deficient for biosynthesis of the antibiotic. We were surprised to find that many of our mutants can produce PTT to some extent, despite using strains that were rigorously validated by both PCR and DNA hybridization techniques. We suggest that the apparent bypass of some of our mutations occurs when other cellular enzymes can substitute at some low level for PTT pathway-specific enzymes. The observation that most bypassed genes deleted in this study encode proteins that are similar to common metabolic enzymes supports this idea. Further, we surmise that the bypass phenotype could be directly responsible for the failure of previous efforts to identify mutants in the new steps that we described here because similar phenomena have not been documented in any other reports on PTT biosynthesis. This observation demonstrates the importance of using a directed approach for pathway analyses because, despite the biosynthetic bypasses, our mutants still secreted sufficient quantity of biosynthetic intermediates to allow identification. Although not fully blocked for PTT biosynthesis, the mutants clearly possess strong physiological bottlenecks for intermediate conversions.
Finally, our data have implications for the biosynthesis of other phosphonic acids. For example, the biosynthetic pathways of fosfomycin and PTT were long thought to share only the phosphoenolpyruvate phosphomutase and phosphonopyruvate decarboxylase catalyzed reactions, implying that phosphonoacetaldehyde would be the last common intermediate between the pathways22. It was recently shown that a PhpC homolog is required for fosfomycin biosynthesis, and hydroxyethylphosphonate is now believed to be a biosynthetic intermediate in this pathway23, 24. These results suggest that the reduction of phosphonoacetaldehyde by enzymes similar to PhpC may represent a common step in other phosphonate biosynthetic pathways as well.
Materials and Methods
Bacterial strains, plasmids, and culture conditions
The bacterial strains and plasmids used in this study are listed in Supplementary Table 1. Details on gene cloning can be found in Supplementary Methods and the oligonucleotides used in the cloning experiments are listed in Supplementary Table 2. Escherichia coli strains were grown on TYE solid medium or Luria-Bertani (LB) liquid medium supplemented with antibiotics where appropriate25. All E. coli strains were grown at 37°C for DNA cloning and manipulation; conditions for protein over expression experiments are specifically noted in the Supplementary Methods. Antibiotic concentrations used for cloning and plasmid maintenance in E. coli were chloramphenicol (Cm) 12 µg/mL, apramycin (Apr) 30 or 50 µg/mL, ampicillin (Amp) 100 µg/mL. Streptomyces viridochromogenes strains were grown on ISP2 (Difco, Sparks, MD) solid medium or in MYG liquid medium17 with antibiotics added where appropriate. S. viridochromogenes strains were incubated at 30 °C, unless specifically noted. Antibiotics used in ISP2 medium were thiostrepton (Thio) 50 µg/mL; streptomycin (Str) 20 or 100 µg/mL; nalidixic acid (Nal) 25 µg/mL; and Apr 30 µg/mL. Antibiotic concentrations used in Streptomyces liquid cultures are noted where appropriate. Intergenic conjugations between E. coli donor strains and S. viridochromogenes recipients were carried out essentially as described in26, except that 2 µl of the donor/recipient mix was spotted to the conjugation plates with a micropipettor instead of using a pin replicator. S. viridochromogenes spores were obtained by spreading MYG grown cultures to SMMS medium27 for confluence. After green spores developed, they were harvested by rolling sterile 3 mm glass beads over the plates. Beads were gathered and washed with TX buffer28 to create spore suspensions. Bioassays for PTT production were performed against both sensitive and resistant strains of Bacillus subtilis as previously described7. Production in liquid cultures was measured by disk diffusion and production on solid medium was assayed by applying pieces of inoculated agar directly to the assay plate.
Generation of Streptomyces viridochromogenes mutants
The mutants used in this study were constructed using a selection/counterselection scheme that resulted in the generation of unmarked deletions of genes from the chromosome (Supplementary Fig. 10). All putative mutants were initially screened by PCR and were later confirmed by Southern blot. A detailed description of the procedures used for mutagenesis is located in Supplementary Methods.
Identification of biosynthetic intermediates accumulated by Streptomyces viridochromogenes mutants
Starter cultures (15 mL MYG in 125 mL Erlenmeyer flask w/sterile glass beads to provide mechanical disruption of mycelial clumps) of S. viridochromogenes mutants were inoculated from ISP2 plates and grown for 2 days with agitation. Cultures were homogenized to break up mycelial clumps and 200 µL of each was used to inoculate 100 mL MYG medium in 500 mL baffled flasks. These cultures were incubated for 4 days with continuous shaking, after which mycelia were removed from the culture supernatant via centrifugation. Culture supernatants were concentrated in vacuo and subsequently analyzed by 31P NMR as outlined above. In general, due to the complex mixture of compounds in the exhausted culture media, peaks in the range of phosphate and phosphate esters (δ 5 to δ −20) are not reported. In addition, shifts in the phosphonate/phosphinate range (~δ 8 to 45) are only reported if they have a signal-to-noise (S/N) ratio >7:1. Phosphonate intermediates were identified by the addition of authentic standards to the samples and not by resonance peak value alone because 31P NMR phosphonate signals are strongly pH dependent29. Coupled liquid chromatography/mass spectrometry (LC/MS) was used as an independent line of evidence to verify the identity of the accumulated intermediates. Samples (50 µL concentrated supernatant) were directly injected onto a 2.1 mm × 100 mm Atlantis Hydrophilic Interaction Chromatography (HILIC) silica column (Waters, Milford, MA), operated at 200 µL/min on a Thermo Surveyor HPLC (ThermoFisher Scientific, San Jose, CA). Samples were separated during a 30 minute gradient from 20:80 A:B to 60:40 A:B where A was 100 mM ammonium acetate + 0.1 % acetic acid and B was acetonitrile. The eluent was directly infused onto a linear ion trap/Fourier-Transfrom hybrid mass spectrometer (LTQ-FT, ThermoFisher Scientific, San Jose, CA) where Fourier-Transform mass spectrometry (FT-MS) data were acquired in negative ion mode at 50,000 resolving power. Phosphonate intermediates were identified by accurate mass analysis, retention time comparison and comparison of tandem MS fragmentation patterns against authentic standards.
31P NMR methods
All NMR experiments were performed at the Varian Oxford Center for Excellence in NMR laboratory at the University of Illinois, Urbana-Champaign. Biological and enzymatic samples were routinely assayed for the presence of phosphonates using 1H-decoupled 31P NMR. Samples were suspended in 20% D2O and resonance peak values are reported against an 85% phosphoric acid standard (δ 0). Spectra were acquired on either a Varian Unity 500 spectrometer equipped with a 5 mm Nalorac Quad probe manually tuned for phosphorus at 202.28 Mhz or on a Varian Inova 600 spectrometer equipped with a 5 mm Varian 600DB AutoX probe with ProTune accessory tuned for phosphorus at 242.79 Mhz.
Other methods
See Supplementary Methods for details on strain construction, protein overexpression, enzyme assay conditions, and the sources and synthesis of phosphonic acids used in this study.
Supplementary Material
Acknowledgements
This work was supported by National Institute of General Medical Sciences grants GM59334 and GM067725 and National Institute of Health Chemical Biology Interface Training Grant 5T32 GM070421. The authors wish to thank A. Salyers (University of Illinois, Urbana-Champaign) for the gift of Bacteroides fragilis NCTC9343 genomic DNA and M.J. Thomas (University of Wisconsin, Madison) for plasmids pOJ260 and pKC1139. We also thank V. Mainz and P. Molitor for NMR advice and C. Wenger for developing custom data analysis software for MS (University of Illinois, Urbana-Champaign).
Footnotes
Accession codes
Sequence data from the PTT biosynthetic gene cluster that was used in designing the experiments reported here was derived solely from previously available sequence (GenBank accession number AY632421). Sequence data generated in this study that corrects apparent sequencing errors in S. hygroscopicus PTT biosynthetic genes (found in GenBank accessions AB029917.1 and D37878.1) were deposited in GenBank under accession numbers EF486265 and EF486266 respectively.
Author Contributions
W.W.M. and J.A.V.B. designed most experiments and wrote the manuscript. J.A.V.B. conducted all microbiological, genetic and molecular biological experiments. NMR analyses were conducted by J.A.V.B. and G.L.. G.L., J.E.V. and W.A.V. designed and carried out chemical syntheses and analyses. P.M.T. and N.L.K. designed and conducted the mass spectrometry experiments.
References
- 1.Seto H, et al. Studies on the biosynthesis of bialaphos (SF-1293). 1. Incorporation of 13C- and 2H-labeled precursors into bialaphos. The Journal of antibiotics. 1982;35(12):1719–1721. doi: 10.7164/antibiotics.35.1719. [DOI] [PubMed] [Google Scholar]
- 2.Bayer E, et al. Phosphinothricin und Phosphinothricyl-Alanyl-Alanin. Helvetica Chimica Acta. 1972;55:224–239. doi: 10.1002/hlca.19720550126. [DOI] [PubMed] [Google Scholar]
- 3.Thompson CJ, Seto H. In: Genetics and Biochemistry of Antibiotic Production. Vining LC, Stuttard C, editors. Newton, MA: Butterworth- Heinemann; 1995. pp. 197–222. [Google Scholar]
- 4.Abell LM, Villafranca JJ. Investigation of the mechanism of phosphinothricin inactivation of Escherichia coli glutamine synthetase using rapid quench kinetic technique. Biochemistry. 1991;30:6135–6141. doi: 10.1021/bi00239a008. [DOI] [PubMed] [Google Scholar]
- 5.Bailey RR. Brief overview of single-dose therapy for uncomplicated urinary tract infections. Chemotherapy. 1990;36(Suppl 1):27–30. doi: 10.1159/000238812. [DOI] [PubMed] [Google Scholar]
- 6.Missinou MA, et al. Fosmidomycin for malaria. Lancet. 2002;360:1941–1942. doi: 10.1016/S0140-6736(02)11860-5. [DOI] [PubMed] [Google Scholar]
- 7.Blodgett JA, Zhang JK, Metcalf WW. Molecular cloning, sequence analysis, and heterologous expression of the phosphinothricin tripeptide biosynthetic gene cluster from Streptomyces viridochromogenes DSM 40736. Antimicrob Agents Chemother. 2005;49:230–240. doi: 10.1128/AAC.49.1.230-240.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwartz D, et al. Biosynthetic gene cluster of the herbicide phosphinothricin tripeptide from Streptomyces viridochromogenes Tü494. Applied and Environmental Microbiology. 2004;70:7093–7102. doi: 10.1128/AEM.70.12.7093-7102.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hidaka T, Hidaka M, Seto H. Studies on the biosynthesis of bialaphos (SF-1293). 14. Nucleotide sequence of phosphoenolpyruvate phosphomutase gene isolated from a bialaphos producing organism, Streptomyces hygroscopicus and its expression in Streptomyces lividans. The Journal of antibiotics. 1992;45(12):1977–1980. doi: 10.7164/antibiotics.45.1977. [DOI] [PubMed] [Google Scholar]
- 10.Nakashita H, Kozuka K, Hidaka T, Hara O, Seto H. Identification and expression of the gene encoding phosphonopyruvate decarboxylase of Streptomyces hygroscopicus. Biochimica et biophysica acta. 2000;1490(1–2):159–162. doi: 10.1016/s0167-4781(99)00249-3. [DOI] [PubMed] [Google Scholar]
- 11.Schwartz D, Recktenwald J, Pelzer S, Wohlleben W. Isolation and characterization of the PEP-phosphomutase and the phosphonopyruvate decarboxylase genes from the phosphinothricin tripeptide producer Streptomyces viridochromogenes Tü494. FEMS microbiology letters. 1998;163(2):149–157. doi: 10.1111/j.1574-6968.1998.tb13039.x. [DOI] [PubMed] [Google Scholar]
- 12.Hidaka T, et al. Studies on the Biosynthesis of Bialaphos (SF-1293). 11. Biochemical Mechanism of C-P Bond Formation of Bialaphos: Use of Gene Manipulation for the Analysis of the C-P Bond Formation Step. Agricultural and Biological Chemistry. 1990;54:2121–2125. [PubMed] [Google Scholar]
- 13.Lee SH, Hidaka T, Nakashita H, Seto H. The carboxyphosphonoenolpyruvate synthase-encoding gene from the bialaphos-producing organism Streptomyces hygroscopicus. Gene. 1995;153(1):143–144. doi: 10.1016/0378-1119(94)00832-d. [DOI] [PubMed] [Google Scholar]
- 14.Babbitt PC, et al. The enolase superfamily: a general strategy for enzyme-catalyzed abstraction of the alpha-protons of carboxylic acids. Biochemistry. 1996;35:16489–16501. doi: 10.1021/bi9616413. [DOI] [PubMed] [Google Scholar]
- 15.Hara O, et al. The bialaphos biosynthetic genes of Streptomyces viridochromogenes : cloning, heterospecific expression, and comparison with the genes of Streptomyces hygroscopicus. Journal of General Microbiology. 1991;137:351–359. doi: 10.1099/00221287-137-2-351. [DOI] [PubMed] [Google Scholar]
- 16.Seto H, et al. Studies on the biosynthesis of bialaphos (SF-1293). Part 3. Production of phosphinic acid derivatives, MP-103, MP-104 and MP-105, by a blocked mutant of Streptomyces hygroscopicus SF-1293 and their roles in the biosynthesis of bialaphos. Biochemical and biophysical research communications. 1983;111(3):1008–1014. doi: 10.1016/0006-291x(83)91400-6. [DOI] [PubMed] [Google Scholar]
- 17.Alijah R, Dorendorf J, Talay S, Pühler A, Wohlleben W. Genetic analysis of the phosphinothricin-tripeptide biosynthetic pathway of Streptomyces viridochromogenes Tü494. Applied Microbiology and Biotechnology. 1991;34(6):749–755. doi: 10.1007/BF00169345. [DOI] [PubMed] [Google Scholar]
- 18.Imai S, Seto H, Ogawa H, Satoh A, Otake N. Studies on the biosynthesis of fosfomycin. Conversion of 2-hydroxyethylphosphonic acid and 2-aminoethylphosphonic acid to fosfomycin. Agricultural and Biological Chemistry. 1985;49:873–874. [Google Scholar]
- 19.Taylor PP, Pantaleone DP, Senkpeil RF, Fotheringham IG. Novel biosynthetic approaches to the production of unnatural amino acids using transaminases. Trends in Biotechnology. 1998;16:412–418. doi: 10.1016/s0167-7799(98)01240-2. [DOI] [PubMed] [Google Scholar]
- 20.Imai S, et al. Studies on the biosynthesis of bialaphos (SF-1293). 4. Production of phosphonic acid derivatives, 2-hydroxyethylphosphonic acid, hydroxymethylphosphonic acid and phosphonoformic acid by blocked mutants of Streptomyces hygroscopicus SF-1293 and their roles in the biosynthesis of bialaphos. The Journal of antibiotics. 1984;37(11):1505–1508. doi: 10.7164/antibiotics.37.1505. [DOI] [PubMed] [Google Scholar]
- 21.Potters MB, et al. Phosphoprotein with phosphoglycerate mutase activity from the archaeon Sulfolobus solfataricus. Journal of Bacteriology. 2003;185:2112–2121. doi: 10.1128/JB.185.7.2112-2121.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seto H, et al. Studies on the Biosynthesis of Fosfomycin. 2. Conversion of 2-Hydroxypropylphosphonic Acid to Fosfomycin by Blocked Mutants of Streptomyces wedmorensis. Journal of Antibiotics. 1991;44:1286–1288. doi: 10.7164/antibiotics.44.1286. [DOI] [PubMed] [Google Scholar]
- 23.Woodyer RD, Li GY, Zhao HM, van der Donk WA. New insight into the mechanism of methyl transfer during the biosynthesis of fosfomycin. Chemical Communications. 2007:359–361. doi: 10.1039/b614678c. [DOI] [PubMed] [Google Scholar]
- 24.Woodyer RD, et al. Heterologous production of fosfomycin and identification of the minimal biosynthetic gene cluster. Chemistry and Biology. 2006;13:1171–1182. doi: 10.1016/j.chembiol.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 25.Wanner BL. In: Gene expression in bacteria using TnphoA and TnphoA' elements to make and switch phoA, lacZ(op) and lacZ(pr) fusions. Adolph KW, editor. Orlando, FL: Academic; 1994. [Google Scholar]
- 26.Martinez A, et al. Genetically modified bacterial strains and novel bacterial artificial chromosome shuttle vectors for constructing environmental libraries and detecting heterologous natural products in mutiple expression hosts. Applied and environmental microbiology. 2004;70:2452–2463. doi: 10.1128/AEM.70.4.2452-2463.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keiser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. Norwich, England: John Innes Centre; 2000. [Google Scholar]
- 28.Hirsch CF, Ensign JC. Heat activation of Streptomyces viridochromogenes spores. Journal of Bacteriology. 1976;126:24–30. doi: 10.1128/jb.126.1.24-30.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vidal G, Thiaudiere E, Canioni P, Gallis JL. Aminomethylphosphonate and 2-aminoethylphosphonate as 31P-NMR pH markers for extracellular and cytosolic spaces in the isolated perfused rat liver. NMR in Biomedicine. 2000;13:289–296. doi: 10.1002/1099-1492(200008)13:5<289::aid-nbm647>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 30.Vasavada KV, Ray BD, Nageswara Rao BD. 31P NMR lineshapes of β-P (ATP) in the presence of Mg2+ and Ca2+: Estimate of exchange rates. Journal of Inorganic Biochemistry. 1984;21:323–335. doi: 10.1016/0162-0134(84)85054-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.