Significance
“Ribosomal scanning” is the generally accepted hypothesis for explaining how eukaryotic 40S ribosomal subunits find initiation codons. Some recently described phenomena cannot be explained by the ribosomal scanning hypothesis, however. Here we show that 43S ribosomal complexes recruited to locations downstream of a reporter gene can direct translation of the reporter independent of the 5′ end, suggesting that 43S ribosomal complex recognizes the initiation codon by “RNA looping” of the intervening mRNA segment between the ribosome recruiting site and the initiation codon. Moreover, we provide a mathematical model for the RNA looping hypothesis. The RNA looping hypothesis provides a logical explanation for translational augmentation by translation-enhancing elements located upstream and/or downstream of a protein-coding region.
Keywords: RNA looping, translation initiation, ribosome scanning, eukaryotic mRNA
Abstract
Eukaryotic translation initiation commences at the initiation codon near the 5′ end of mRNA by a 40S ribosomal subunit, and the recruitment of a 40S ribosome to an mRNA is facilitated by translation initiation factors interacting with the m7G cap and/or poly(A) tail. The 40S ribosome recruited to an mRNA is then transferred to the AUG initiation codon with the help of translation initiation factors. To understand the mechanism by which the ribosome finds an initiation codon, we investigated the role of eIF4G in finding the translational initiation codon. An artificial polypeptide eIF4G fused with MS2 was localized downstream of the reporter gene through MS2-binding sites inserted in the 3′ UTR of the mRNA. Translation of the reporter was greatly enhanced by the eIF4G-MS2 fusion protein regardless of the presence of a cap structure. Moreover, eIF4G-MS2 tethered at the 3′ UTR enhanced translation of the second cistron of a dicistronic mRNA. The encephalomyocarditis virus internal ribosome entry site, a natural translational-enhancing element facilitating translation through an interaction with eIF4G, positioned downstream of a reporter gene, also enhanced translation of the upstream gene in a cap-independent manner. Finally, we mathematically modeled the effect of distance between the cap structure and initiation codon on the translation efficiency of mRNAs. The most plausible explanation for translational enhancement by the translational-enhancing sites is recognition of the initiation codon by the ribosome bound to the ribosome-recruiting sites through “RNA looping.” The RNA looping hypothesis provides a logical explanation for augmentation of translation by enhancing elements located upstream and/or downstream of a protein-coding region.
Translation initiation is complex process in which more than 10 kinds of proteins participate (1). During the first event of translation initiation, it is believed that the 43S preinitiation complex, composed of 40S ribosome, eIF3, eIF5, eIF1, eIF1A, and ternary complex (eIF2-GTP initiator tRNA), is recruited to a 5′ cap structure at the end of mRNA via a preexisting mRNA-eIF4F (eIF4E, eIF4A, and eIF4G) complex through a protein–protein interaction between eIF4G and eIF3 (1). The resulting 40S ribosomal subunit-containing complex, called the 43S preinitiation complex, moves to the initiation codon. Although most of the eukaryotic mRNAs use the cap structure at the 5′ end when recruiting the 40S ribosome, some mRNAs use a specialized RNA element, the internal ribosome entry site (IRES), for recruiting the 40S ribosome to mRNA (2, 3).
eIF4G protein plays a pivotal role in both cap-dependent and IRES-dependent translations, not only for ribosome recruitment, but also for initiation codon selection. eIF4G is a scaffold protein that links the 43S ribosomal complex and mRNA. In the cap-dependent translation, eIF4G is loaded onto mRNA as a protein complex with eIF4E (cap-binding protein) and eIF4A (RNA helicase) (4). eIF4G also participates in translational enhancement by the poly(A) tail through an interaction with poly(A)-binding protein (PABP) that binds to the poly(A) tail. Moreover, eIF4G plays pivotal roles in IRES-dependent translation of picornaviral mRNAs through direct interactions with the IRES elements (5, 6). In cap-dependent translation, eukaryotic mRNAs generally stick to the first-AUG rule—that is, the AUG codon nearest the 5′ end (ribosome recruiting site) is usually selected as an authentic initiation codon if it has a good Kozak context (7). Moreover, substantial studies indicate that the AUG codon located proximal to a ribosome recruiting site is preferentially selected as the translation start codon in IRES-dependent translation (8). In other words, the ribosome recruiting site generally determines where translation initiation occurs. However, recent studies have shown that approximately 40% of mRNAs do not follow the first AUG rule and instead use downstream AUGs as main initiation codons, indicating that initiation site selection is a complex process (9–13).
To understand the mechanism searching for the translational initiation codon by a ribosome, we investigated the function of eIF4G in finding the translational initiation codon. We found that a modified eIF4G containing the RNA-binding domain of MS2 coat protein can associate with the translational machinery, and that tethering of the modified eIF4G at the 3′ UTR of mRNA greatly stimulates translation of upstream genes. We also found that the eIF4G, tethered to the 3′ UTR of dicistronic mRNAs, stimulates translation of the second cistron. In addition, insertion of encephalomyocarditis virus (EMCV) IRES at the 3′ UTR of mRNA stimulates translation, similar to the tethering of eIF4G to the 3′ UTR. Moreover, we mathematically modeled the effect of distance between the cap structure and initiation codon on translation efficiency of mRNAs. Our experimental data and theoretical analyses suggest that the finding of an initiation codon by the 43S ribosomal complex attached to a recruiting site of an mRNA occurs through “mRNA looping” between the ribosome recruiting site and the initiation codon.
Results
Recruitment of the 40S Ribosome to a Site Downstream of the Reporter Gene Directs Translation.
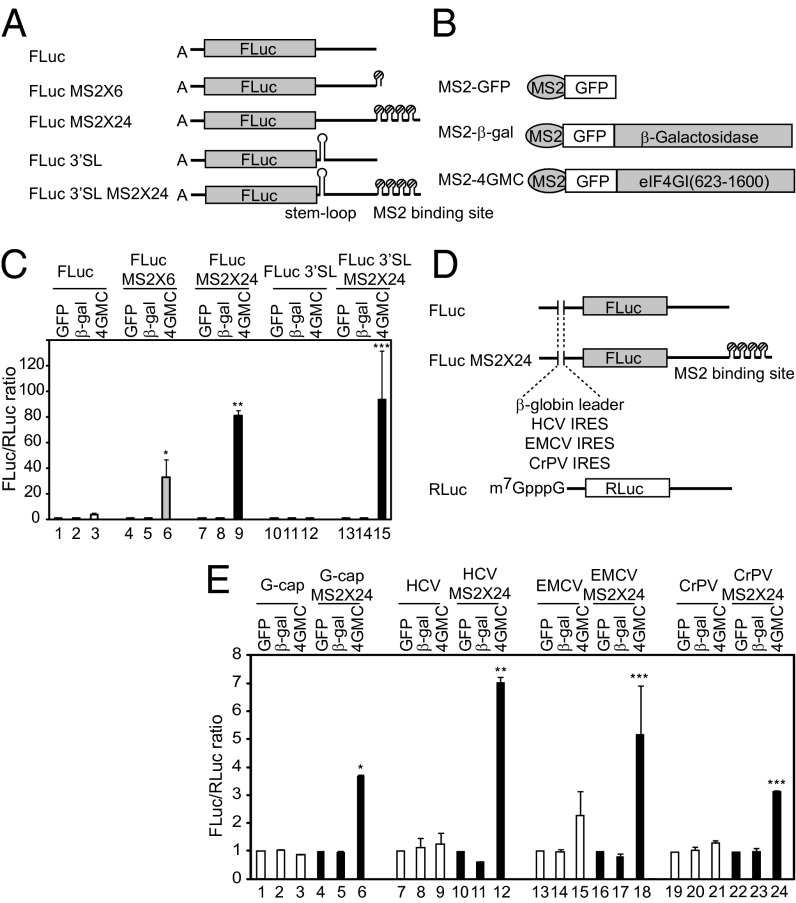
To explore the mechanism of how a ribosome finds a translation initiation site, we devised artificial mRNAs and a modified eIF4G (Fig. 1 A and B and Fig. S1A). A modified eIF4G (MS2-4GMC) is composed sequentially of MS2, GFP, and middle- to C-terminal domains of eIF4G with a deletion of the N-terminal domain containing eIF4E- and PABP-binding sites. This fusion protein could form a 43S ribosomal complex including a 40S subunit and eIF3, as indicated by coimmunoprecipitation of MS2-4GMC, eIF3b (a component of eIF3), and RpS6 (a 40S ribosomal protein) (Fig. S1B). This indicates that the 40S ribosomal subunit, which is associated with MS2-4GMC, can be recruited to the MS2-binding sites on artificial mRNAs; however, MS2-4GMC is not able to recruit the 40S ribosomal subunit to the cap structure at the 5′ end, because the domain required for eIF4E-binding is deleted. When A-capped RNAs containing MS2-binding sites were introduced into HEK293T cells expressing MS2-4GMC protein, expression of the reporter gene was greatly increased in a manner depending on the number of MS2-binding sites (Fig. 1C, lanes 6 and 9). Expression of negative control proteins, MS2-GFP and MS2-β-gal (Fig. 1B), had no effect on reporter gene expression (Fig. 1C, lanes 1 and 2, 4 and 5, and 7 and 8). The enhancement of reporter gene expression by MS2-4GMC depended on the presence of an MS2-binding site on the reporter RNAs (Fig. 1C, compare lanes 6 and 9 with lane 3). The augmentation of gene expression by MS2-4GMC was not attributable to a change in mRNA levels (Fig. S1C). These data lead us to conclude that the 40S ribosomal subunit recruited to a downstream region can participate in translation of the upstream gene.
Fig. 1.
eIF4G tethered at the 3′ UTR of mRNA augments translation of an upstream reporter gene. (A) FLuc represents a reporter RNA containing the firefly luciferase gene as a reporter. MS2-binding sites (6 or 24 copies) were inserted into the reporter RNA to generate FLuc MS2 × 6 and FLuc MS2 × 24, respectively. A stable stem-loop was inserted downstream of the stop codon of reporter RNAs, FLuc and FLuc MS2 × 24, to generate FLuc 3′SL and FLuc 3′SL MS2 × 24, respectively. (B) Schematic diagram of MS2 fusion proteins. (C) The translation efficiencies of FLuc (lanes 1–3), FLuc MS2 × 6 (lanes 4–6), and FLuc MS2 × 24 (lanes 7–9) were determined by measuring firefly luciferase activity in cells expressing MS2-GFP (lanes 1, 4, and 7), MS2-GFP-β-galactosidase (lanes 2, 5, and 8), and MS2-GFP-eIF4G (lanes 3, 6, and 9). Firefly luciferase activity was normalized to Renilla luciferase activity. Error bars reflect SD in three independent experiments. *P < 0.025 compared with lane 3; **P < 9 × 10−6 compared with lane 3; ***P < 0.005 compared with lane 12. (D) Schematic diagram of reporter mRNAs used in E. Each reporter contains β-globin leader (G-cap), HCV IRES (HCV), EMCV IRES (EMCV), or CrPV IRES (CrPV) at the 5′ UTR. m7G-capped reporter RNA containing the Renilla luciferase gene served as a control for mRNA transfection efficiency. (E) Luciferase activity was measured using the extracts from cells transfected with reporters containing m7G-capped β-globin leader sequence (lanes 1–6), HCV IRES (lanes 7–12), EMCV IRES (lanes 13–18), or CrPV IRES (lanes 19–24). Ratios of firefly luciferase activity to Renilla luciferase activity were normalized to the ratios obtained from cells transfected with effecter MS2-GFP (lanes 1, 4, 7, 10, 13, 16, 19, and 22). *P < 9.8 × 10−5 compared with lane 3; **P < 0.003 compared with lane 9; ***P < 0.01 compared with lane 15 (EMCV IRES) or lane 21 (CrPV IRES).
To explore the possibility that translation of the reporter gene by the downstream-recruited 40S ribosomal subunit occurred by “backward scanning” of the ribosome (3′ to 5′ migration of the ribosome) (14), we generated reporter RNAs containing a strong stem-loop structure (which should block the putative backward scanning) between the reporter gene and the MS2-binding sites (FLuc 3′SL and FLuc 3′SL MS2 × 24 in Fig. 1A). This stem-loop structure did not inhibit translation by ribosomes recruited downstream of the reporter gene (Fig. 1C, compare lane 15 with lane 9). Of note, a threefold to fourfold increase in reporter gene expression by MS2-4GMC was observed after introduction of a reporter RNA lacking the MS2-binding site into MS2-4GMC–producing cells, as reported previously (15) (Fig. 1C, lane 3). This finding may be attributed to weak binding of MS2-4GMC to the reporter RNA through cryptic binding sites in the mRNA (16).
Interestingly, a modified eIF4G tethered to the reporter gene enhanced translation, even when the A-capped 5′ UTR of the reporter gene was replaced by a G-capped RNA, the EMCV IRES, the hepatitis C virus (HCV) IRES, or cricket paralysis virus (CrPV) IRES (Fig. 1 D and E). Transfer of eIF4G to the 5′ UTR might not be needed for translational enhancement by downstream-tethered eIF4G, because translational enhancement occurred even when HCV IRES and CrPV IRES, which do not require eIF4G for translation initiation, were located at the 5′ UTR of the reporter RNA (Fig. 1E). These results indicate that the 40S ribosome recruited to downstream of the reporter gene can find the initiation codon without the help of any initiation factor associated with the 5′ UTR, given that the function of CrPV IRES does not require an translation initiation factor (17).
To more precisely investigate the end dependency of translation stimulation by eIF4G tethering, we generated artificial reporter RNAs, which have a stable stem-loop blocking translation from the 5′ end (Fig. S2A). We also prepared reporter RNAs with lengthened 5′ UTRs, to preclude the possibility that the stem-loop interferes with a ribosome’s ability to find an initiation codon owing to space limitations (Fig. S2A). The reporters harboring a stable stem-loop at the 5′ end also enhanced translation (by 7- to 10-fold) by tethering of eIF4G (Fig. S2 B and C, compare lanes 12 and 10), but the degree of stimulation was lower than that of reporters without a stem-loop (Fig. S2 B and C, compare lanes 6 and 12). The reduced translational enhancement by the downstream-tethered MS2-4GMC on a reporter with the 5′ stem-loop might be attributed to a putative blockade of interaction between eIF4F and the 5′ end by the stem-loop (16).
5′ End-Independent Translation Stimulation by eIF4G Tethered Downstream of a Reporter Gene.
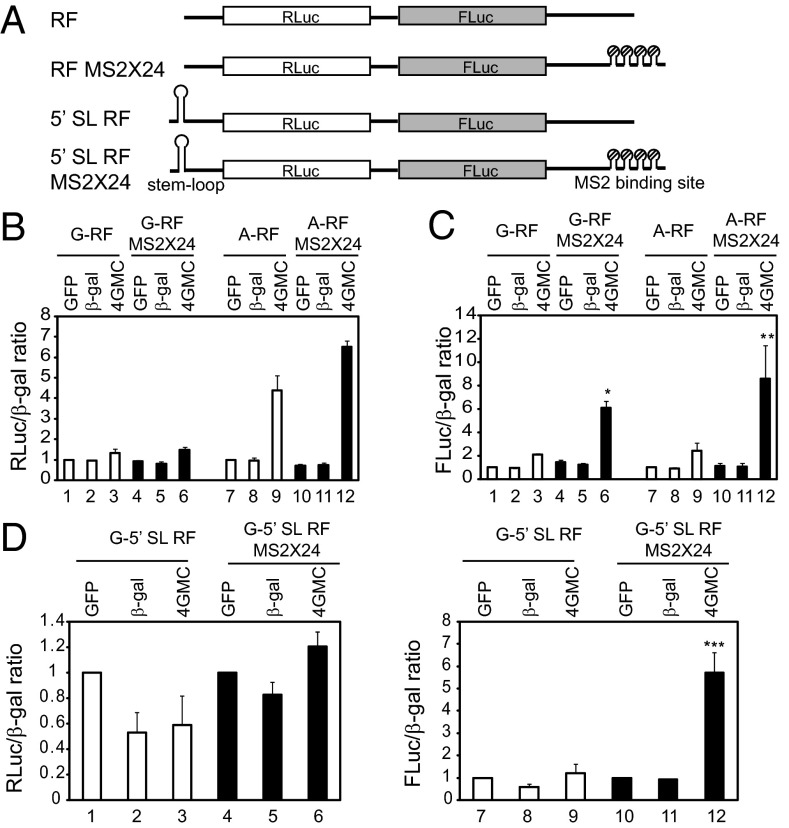
To demonstrate end-independent stimulation of translation by eIF4G tethering, we generated dual reporter RNAs harboring MS2-binding sites downstream of the second cistron (Fig. 2A). In this system, the firefly luciferase gene (FLuc) is translated end-independently, and the Renilla luciferase gene (RLuc) is translated 5′ end-dependently and/or end-independently (Fig. 2A).
Fig. 2.
5′ end-independent translational activation by eIF4G tethered downstream of a reporter gene. (A) Dual reporters contain Renilla luciferase gene followed by firefly luciferase gene. Twenty-four copies of MS2 sequence exist in reporter mRNA RF MS2 × 24. (B and C) Translation efficiencies from m7G-capped (G-RF and G-RF MS2 × 24) and A-capped (A-RF and A-RF MS2 × 24) dual reporters shown in A. Renilla and firefly luciferase activities in each set were normalized to the β-galactosidase activity from a plasmid cotransfected as a transfection efficiency control. Normalized Renilla luciferase activity (B) or firefly luciferase activity (C) of reporter without an MS2 sequence from cells overproducing MS2-GFP proteins were set to 1 (lane 1 for lanes 1–6 and lane 7 for lanes 7–12). (D) Translation efficiencies from m7G-capped dual reporters harboring a stable stem-loop (5′ SL RF and 5′ SL RF MS2 × 24). *P < 0.012 compared with lane 3; **P < 0.04 compared with lane 9; ***P < 0.025 compared with lane 9.
The translation of firefly luciferase, which is encoded at the second cistron, was greatly increased (up to ninefold) when dual reporters harboring an MS2-binding site were transfected to MS2-4GMC–overexpressing cells, regardless of the presence of m7G-cap structure (Fig. 2C, lanes 6 and 12). In contrast, translation of Renilla luciferase, which is encoded in the first cistron of the dual reporter mRNA without an MS2-binding site, was enhanced in the MS2-4GMC–expressing cells when A-capped mRNA was used (Fig. 2B, compare lane 9 with lane 7), similar to the A-capped monocistronic mRNA (Fig. 1C). A weak but noticeable translational enhancement of Renilla luciferase at the first cistron was observed when MS2-4GMC was tethered downstream of the reporter genes in the A-capped mRNAs (Fig. 2B, compare lane 12 with lane 9; P < 0.04). This indicates that the ribosomes recruited to the 3′ UTR of dicistronic mRNAs contribute, albeit weakly, to translation of the first cistron in the A-capped reporter.
The discrepancy in translational enhancement between the first and second cistrons by the MS2-4GMC tethered to the downstream of reporter genes likely is attributed to two factors:
-
i)
The firefly luciferase gene at the second cistron is closer to the tethering site compared with the Renilla luciferase gene at the first cistron. It is conceivable that the ribosome bound to an mRNA may be more able to find the initiation codon located closer to it than the initiation codon located farther away from it.
-
ii)
The basal translation level of the first cistron in an G-capped mRNA, which is attributed to the 5′ end-dependent translation mechanism, is much higher than that of the second cistron (Fig. S3). This also explains why first cistron of m7G-capped reporter showed weak translational enhancement by tethering of MS2-4GMC (Fig. 2B, compare lane 6 with lane 3).
Translation efficiencies of firefly luciferase at the second cistron were similar irrespective of the structure of the 5′ end of mRNAs (m7G-capped or A-capped), even though translation efficiencies of Renilla luciferase at the first cistron were clearly different (Fig. S3). This finding indicates that translation augmentation of the second cistron is largely independent of translation from the 5′ end.
To reduce the influence of putative ribosomes reaching the second gene after translation of the first gene, we generated dual reporters containing a stable stem-loop at the 5′ end of mRNA (Fig. 2A). Insertion of a stem-loop inhibited translation from Renilla luciferase (end-dependent translation) by 50-fold (Fig. S3A), but the did not significantly affect the translation efficiency of firefly luciferase (Fig. S3B). Importantly, translation from the second cistron (firefly luciferase) was still stimulated to a similar extent when MS2-4GMC protein was tethered at the 3′ UTR of a reporter (Fig. 2D, lane 12). Moreover, the translation stimulation from firefly luciferase was not the result of ribosome backward scanning, as demonstrated by the finding that insertion of a stable stem-loop just downstream of the firefly luciferase gene did not hamper translation enhancement from firefly luciferase gene (Fig. S4). Based on the foregoing data, we suggest that ribosomes recruited by eIF4G to downstream of a gene can enhance translation of the upstream gene independent of the 5′ end.
EMCV IRES Enhances Translation of Monocistronic mRNAs from Downstream of a Reporter Gene.
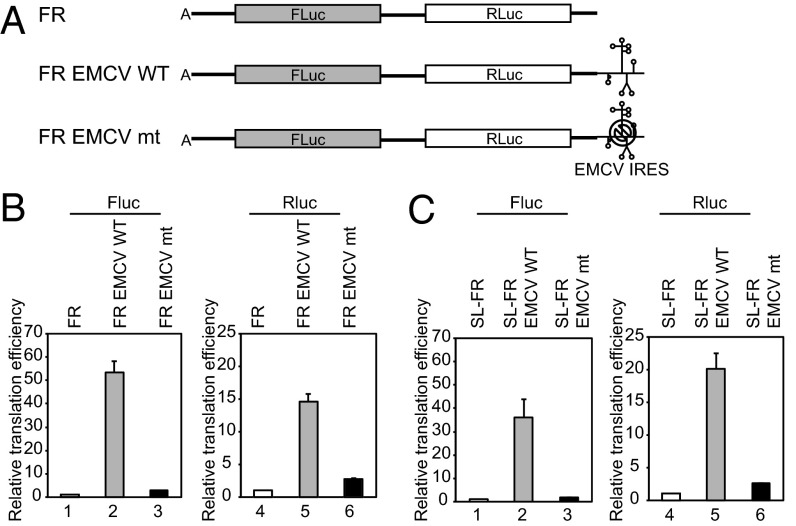
Our data lead an interesting question whether an IRES, whose activity depends on eIF4G, can enhance translation of upstream gene similarly to the tethering of eIF4G. For this purpose, we constructed artificial mRNAs containing Renilla luciferase with or without the EMCV IRES element at the 3′ UTR (Fig. 3A, Rluc EMCV WT and Rluc, respectively). To further investigate the role of eIF4G in the EMCV IRES-dependent translation, we constructed an additional artificial mRNA containing a mutation at the eIF4G-binding site in the EMCV IRES (Fig. 3A, Rluc EMCV mt). This mutation (adenine to uracil substitution at 724th nucleotide of EMCV IRES) strongly reduces the binding of eIF4G to EMCV IRES (18). Reporter RNAs were subjected to in vitro translation with nuclease-untreated rabbit reticulocyte lysates (RRLs) at physiological KCl condition (150 mM).
Fig. 3.
EMCV IRES stimulates translation of upstream genes. (A) Reporters contain Renilla luciferase followed by WT or mt EMCV IRES with two different kinds of 5′ UTRs. (B) A-capped reporters with or without a stable stem-loop were subjected to in vitro translation with nuclease-untreated RRLs in the presence of 150 mM KCl. Relative translation efficiencies of the reporters harboring a vector sequence (lanes 1–6) or 15 copies of β-globin leader (lanes 7–12) at the 5′ UTR are depicted. The Rluc activity of reporter without EMCV IRES was set to 1 for comparison.
The EMCV IRES at the 3′ end very strongly augmented the translation of A-capped reporter mRNA (Rluc EMCV WT) (Fig. 3B, compare lane 2 with lane 1). To our surprise, the level of translational augmentation by a downstream EMCV IRES at the 3′ UTR (Fig. 3B) was similar to that by an upstream EMCV IRES at the 5′ UTR (Fig. S5A). The mutant EMCV IRES also weakly stimulated translation of the reporter, albeit to a much smaller extent than the WT EMCV IRES (compare lane 3 with lane 1 in Fig. 3B). Translation of the m7G-capped mRNA with WT EMCV IRES (G-Rluc EMCV WT) downstream of the reporter gene was approximately twofold higher than that without EMCV IRES (G-Rluc) (Fig. S5B, compare lane 11 with lane 10). The difference in translation efficiencies of the mRNAs was greater at higher salt concentration in both A-capped and m7G-capped reporters (Fig. S5 B and C).
To test whether translation stimulation by downstream EMCV IRES is end-dependent, we generated reporters containing a stable stem-loop at the 5′ end of mRNAs (Fig. 3B, SL-Rluc, SL-Rluc EMCV WT, and SL-Rluc EMCV mt). In addition, we inserted 15 copies of β-globin leader into the 5′ UTR of some mRNAs to test the effect of 5′ UTR length on translation enhancement by the EMCV IRES at the 3′ UTR [(15)Rluc, SL-(15)Rluc, (15)Rluc EMCV WT, SL-(15)Rluc EMCV WT, (15)Rluc EMCV mt, and SL-(15)Rluc EMCV mt]. When mRNAs with and without a stable stem-loop at the 5′ UTR were subjected to in vitro translation, partial inhibition of translational augmentation was observed from the mRNA with a stem-loop structure at the 5′ end (Fig. 3B, compare lanes 2 and 5). However, this inhibitory effect disappeared when the 5′ UTR was extended to 800 nt by adding 15 copies of β-globin leader to provide room for ribosome landing between the stem-loop and the initiation codon (Fig. 3B, compare lanes 8 and 11). The foregoing data indicate that the EMCV IRES can augment the translation of upstream genes in an end-independent manner.
EMCV IRES Enhances Translation of Dicistronic mRNAs from Downstream of a Reporter Gene.
To further test whether the EMCV IRES can stimulate translation of upstream gene in an end-independent manner, we constructed dicistronic reporters containing the firefly luciferase gene (FLuc) followed by a spacer and the Renilla luciferase gene (RLuc) with and without the EMCV IRES (Fig. 4A). A-capped RNAs were synthesized in vitro and subjected to in vitro translation at 150 mM KCl. Translational enhancement from the addition of downstream EMCV IRES was approximately 50-fold from the first cistron (FLuc) and 15-fold from the second cistron (RLuc) (Fig. 4B, compare lanes 2 and 5 with lanes 1 and 4, respectively). When a stable stem-loop structure was added to the 5′ end of mRNA, translational augmentation of the first gene by EMCV IRES was decreased (compare lane 2 in Fig. 4C with lane 2 in Fig. 4B), but that of the second gene was increased (compare lane 5 in Fig. 4C with lane 5 in Fig. 4B). These results indicate that translation of the second cistron occurs independent of the first cistron, and that both end-dependent translation (first cistron) and end-independent translation (second cistron) of dicistronic mRNAs are augmented by EMCV IRES at the 3′ UTR.
Fig. 4.
5′ end-independent translation enhancement by the EMCV IRES residing downstream of a gene. (A) Dual reporters contain firefly luciferase gene (Fluc) followed by Renilla luciferase gene (Rluc). WT or mt EMCV IRES resides at the 3′ UTR of reporters. (B and C) A-capped reporters were subjected to in vitro translation with nuclease-untreated RRLs in the presence of 150 mM KCl. 5′ end-dependent (Fluc, lanes 1–3 in B and C) and 5′ end-independent (Rluc, lanes 4–6 in B and C) translations were measured and are depicted in graphs. Firefly and Renilla luciferase activities of FR reporter without EMCV IRES were set to 1 for comparison.
Translational Stimulation of Upstream Genes by EMCV IRES at the 3′ UTR of Reporters in Vivo.
To investigate the effect of EMCV IRES at the 3′ UTR on the translation of upstream reporter genes in living cells, we synthesized monocistronic and dicistronic mRNAs (Figs. 3 and 4), transfected them into HEK293T cells, and analyzed luciferase activity. The EMCV IRES inserted downstream of reporter genes augmented the expression of reporter genes in the cells in both monocistronic and dicistronic contexts (compare lanes 2 and 5 with lanes 1 and 4 in Fig. S6 A and C). These data indicate that translation of reporter genes were augmented by the EMCV IRES inserted downstream of the genes in both in vitro and in vivo systems.
Theoretical Perspectives on RNA Looping in Translation.
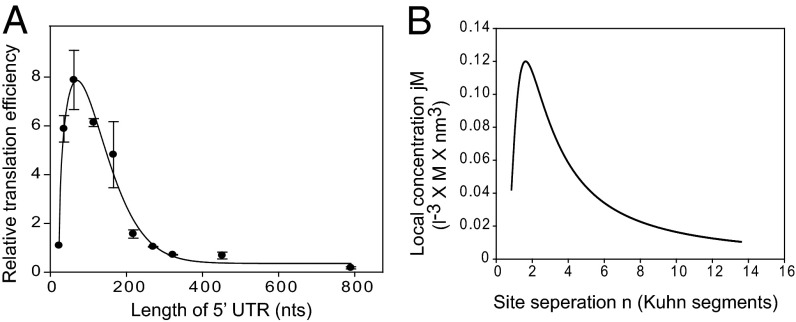
If translational initiation occurs by RNA looping, then translation efficiency should be governed by the distance between the 40S ribosome recruiting site (e.g., the cap structure at the 5′ end of an mRNA) and the initiation codon (Fig. S7A). To address this aspect, we analyzed the effect of 5′ UTR length on translation using the plasmids described by Chappell et al. (9) (Fig. 5A). We found that the optimal length for the most efficient translation was ∼70 nt, much longer than the length required for association of the 40S ribosome to the 5′ end of an mRNA. Moreover, translation efficiency decreased precipitously beyond the optimal length rather than decreasing linearly as predicted by scanning (Fig. 5A). Similar data were reported previously by Mauro et al. (9), who suggested “ribosomal tethering or clustering” as an underlying mechanism; however, the authors made no attempt to analyze the data theoretically using a mathematical model and did not confirm their hypothesis using other experimental approaches.
Fig. 5.
Comparison of the theoretically calculated probability of collision of two objects on a string with the experimentally observed translation efficiencies of mRNAs with various 5′ UTR lengths. (A) Relative translation efficiencies of mRNAs harboring 5′ UTRs of various lengths. Reporter DNAs, kindly provided by Dr. Vincent Mauro, The Scripps Research Institute, La Jolla, CA, were transfected into HEK293T cells, and luciferase activity was measured at 24 h after transfection. Similar data were reported previously by Chappell et al. (9). Error bars indicate the SD in three independent experiments. (B) Graph depicting the relationship between the local concentration, jM, and the distance, n (number of Kuhn segments), predicted by the equation in Fig. S7B. Because we cannot perform the experiments required to obtain the parameter d, we used d = 0 in the calculation (19). The peak of the graph occurred at n = 1.62.
Our empirical observation of the effect of 5′ UTR length on translation and theoretical analysis of RNA looping model revealed that the profile of translation efficiencies of mRNAs depending on 5′ UTR length (Fig. 5A) is very similar to that of the mathematically predicted collision probability of two objects associated on a string (mRNA) (Fig. 5B) (19). In fact, there is a very strong linear correlation (R2 = 0.76) between the local concentration predicted by the mathematical formula shown in Fig. S7B and the relative translational efficiency measured empirically (Fig. S7C). Our experimental data and theoretical analysis strongly suggest that a large proportion, if not 100%, of cap-dependent translation events are executed by RNA looping.
Discussion
Recognition of a start codon by the 40S ribosome is a crucial step in translation initiation. Several translational-enhancing elements [i.e., cap structure, poly(A), and IRES elements] are known to facilitate translation initiation. We have attempted to identify the mechanism through which the 40S ribosome finds the initiation codon on an mRNA by investigating the molecular function of eIF4G recruited to various translational-enhancing elements. Here we provide several lines of evidence indicating that the 43S ribosomal complex recruited to downstream of a reporter gene can recognize the initiation codon without scanning.
When eIF4G was tethered at the 3′ UTR of mRNA, translation of an upstream gene was stimulated (Fig. 1). This translational enhancement may be achieved by two possible mechanisms: direct recognition of the initiation codon by the ribosome recruited downstream of a gene, and transfer of the ribosome to the 5′ end of mRNA, followed by scanning or by other mechanism(s) (see below). A large portion of 40S ribosomes recruited to a site downstream of a reporter could stimulate translation even when a possible processive transfer of ribosomes by “backward scanning” was blocked by a stable stem-loop (Fig. 1 and Fig. S4). Moreover, translation of the second cistron of a dicistronic mRNA, which occurs only in a 5′ end-independent manner, was accomplished by the 40S ribosomes recruited to the downstream of a reporter gene through eIF4G proteins tethered to MS2-binding sites (Fig. 2). Considering the foregoing data together, we conclude that translational enhancement by ribosomes recruited to downstream of a gene can occur, at least in part, without transference of the ribosome to the 5′ end, plausibly through direct recognition of the initiation codon by the 43S ribosomal complexes.
We further investigated the effect of the 40S ribosome recruited to downstream of a gene using the EMCV IRES as a natural translation enhancer. The same translation-enhancing effect as for the eIF4G artificially tethered to the downstream of a gene via MS2-binding sites was observed from the EMCV IRES (Figs. 3 and 4). It was previously reported that the picornavirus IRES can stimulate translation of upstream genes in an eIF4G-dependent manner (20); however, the authors made no attempt to investigate whether the translation enhancement can occur in a 5′ end-independent manner. Moreover, they speculated that translational enhancement by the downstream IRES may occur by transference of eIF4G bound to the IRES element to the RNA 5′ end in cis (20). In contrast, here we show that 40S ribosomes recruited to downstream of a reporter gene can commence translation in a 5′ end-independent manner, by inserting a stem-loop structure at the 5′ end of the reporter mRNA (Fig. 3) or by analyzing translation of a reporter gene at the second cistron of a dicistronic mRNA containing a downstream EMCV IRES (Fig. 4).
Translational stimulation of upstream genes by the element at the 3′ UTR has been widely studied in plant viruses (21). These elements, generally known as cap-independent translation elements (CITEs), mediate translation of uncapped plant viral mRNAs. Interestingly, some CITEs interact with eIF4G (or eIF4F), and the binding of eIF4G to the CITEs is crucial for their translation stimulation function (22). Some viral RNAs require interactions between the CITEs and the 5′ UTRs. In contrast, in some cases the 5′ UTRs are dispensable for CITE function-enhancing cap-independent translation (21–23). Apart from plant viruses, several mammalian translation-enhancing elements functioning downstream of genes have been reported recently (13, 24); for example, histone H4 mRNA recruits eIF4E directly to the coding region of mRNA, and renders 80S complex formation at the AUG located upstream of the eIF4E-recruiting site in a cap-independent manner (13). Another example is c-myc mRNA containing A-rich element (ARE) in the 3′ UTR where an RNA-binding protein, AUF1, associates (24). The AUF1 interacts directly with eIF4G protein (25), indicating that the 40S ribosome is possibly recruited to the 3′ UTR of c-myc mRNA via the protein bridge of eIF4G. These reports indicate that translational enhancement of upstream genes by ribosomes recruited to downstream of the genes occurs in many natural mRNAs. The most common example of a downstream enhancing element is the poly(A) tail at the 3′ ends of mRNAs existing in most eukaryotic mRNAs. The poly(A) tail enhances translation through an interaction with PABP, which in turn interacts with eIF4G. It has been shown that the poly(A) tail enhances translation of mRNAs even without an m7G cap structure and facilitates internal initiation when 5′ end-dependent translation is blocked (12).
These reports and the data obtained from the present study indicate that the 43S ribosomal complex recruited to the downstream of a gene can find the initiation codon in a 5′ end-independent manner. If this is the case, then how can the 43S ribosomal complex associated with an mRNA downstream of a gene find the initiation codon in a 5′ end-independent manner? A logical mechanistic assumption of the communication between the ribosome bound to the downstream ribosome recruiting site and the initiation codon is through looping of the intervening RNA region separating the elements. The concept of looping of the intervening polymer has been well documented in various biological processes (19). This concept explains the mechanism of interaction of two components located on a string, and has been adopted to explain the regulatory mechanisms of gene expression, such as lac repression, transcription enhancement, anti-termination, and splicing enhancement, that require interactions of protein complexes bound to two separate elements on the same DNAs or RNAs (19).
The RNA looping model for recognition of initiation codons by the 43S ribosomal complexes bound to ribosome recruiting sites is reasonable not only for explaining the activities of downstream translation-enhancing elements described above, but also for explaining translational enhancement by the m7G cap structure of some mRNAs. For instance, translation of some mRNAs containing highly structured 5′ UTRs with several noninitiating AUGs, which can hardly be explained by the scanning model, occurs in a cap-dependent manner (26, 27). Translation of these mRNAs can be explained by the RNA looping model, because base-by-base inspection of the 5′ UTR by a scanning ribosome is not needed to find the initiation codon if the ribosome bound near the 5′ cap structure directly recognizes the initiation codon through RNA looping. We recently reported that at least 20–30% of m7G cap-dependent translation of mammalian mRNAs occurs without base-by-base scanning of mRNAs, by using dissected mRNAs composed of m7G-capped leader RNAs associated with uncapped reporter RNAs through double-stranded RNA bridges (28). The RNA looping model explains how the 40S ribosome associated at the 5′ cap structure on the leader RNA can be transferred to the initiation codon on the reporter RNA that is noncovalently connected with the leader RNA. Moreover, analyses of the effect of 5′ UTR length on translation, which was empirically tested, and the theoretically predicted collision probability of two objects (the initiation codon and the 40S ribosome recruited to the cap structure) associated on a string (mRNA) strongly suggest that a large proportion, if not 100%, of cap-dependent translation occurs through RNA looping instead of linear scanning of a 40S ribosome through the 5′ UTR.
In conclusion, translational enhancement by RNA looping may participate in various processes that augment mRNA translation (Fig. S8), including the following:
-
i)
Cap-dependent translation. According to the looping model, different parts of an mRNA collide with the 43S ribosomal complex on the cap until the initiation codon encounters the ribosome (Fig. S8A). The probability of a ribosome-initiation codon collision depends on the length, structure, and flexibility of the 5′ UTR and accessibility of the AUG.
-
ii)
IRES-dependent translation. RNA looping may act through a mechanism similar to that involved in cap-dependent translation to regulate the initiation of IRES-dependent translation by helping the preinitiation complex assembled on an IRES element find the initiation codon (Fig. S8B). Of interest, in the context of the proposed working model, we found that the EMCV IRES augments translation of an upstream gene when attached downstream of a reporter gene (Figs. 3 and 4). It is likely that translation of the upstream gene is executed by ribosomes loaded on the EMCV IRES through RNA looping of the intervening sequence.
-
iii)
Poly(A)-dependent translation. Translational enhancement by a poly(A) tail by RNA looping is likely (Fig. S8C). Preiss and Hentze analyzed the effect of the poly(A) tail on translation and reported that the 5′-proximal AUG codon (5′ AUG) is preferentially used as the start codon when an mRNA contains both a 5′ cap and a poly(A) tail, whereas the internal AUG codon is preferentially used as the start codon when an mRNA contains only a poly(A) tail without the cap structure (12). The latter finding suggests that the poly(A) tail itself is capable of functioning as a ribosome recruiting site (Fig. S8C), a situation similar to the tethering of eIF4G downstream of a reporter gene (Fig. 1). Even though the authors did not speculate about the mechanisms underlying their results, these phenomena can be explained by RNA looping between the poly(A) and the alternate initiation codons. Without the 5′ cap structure, only the probability of collision between the AUGs and the 40S ribosome recruited to the poly(A) tail governs translation initiation; thus, the internal AUG, which is closer to the poly(A), is preferentially used. In the case of 5′-capped and poly(A)-tailed mRNA, in contrast, communication between the 5′ cap structure and 3′-poly(A) via an RNA-protein complex consisting of cap-eIF4E-eIF4G-PABP-poly(A) preferentially facilitates translation from the 5′ AUG by the 40S ribosome recruited to the 3′ end by shortening the effective distance between the 40S ribosome and the 5′ AUG.
Materials and Methods
In vitro translation reactions were performed in RNase-untreated RRLs. The conditions for the in vitro translation are described in detail in SI Materials and Methods. Reagents, plasmid construction procedures, DNA/RNA transfection of HEK293T cells, and reporter activity assays are also described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We are indebted to Drs. Nahum Sonenberg and Yuri Svitkin (McGill University) for their critical reading of the manuscript and helpful discussions. We also thank Dr. V. P. Mauro (The Scripps Research Institute) for providing plasmid pGL3c containing various copies of β-globin leader. This research was supported by the Korean National Research Foundation’s Biotechnology R&D Program (Grant 2013076206) and Basic Research Laboratory (Grant 20100019706), funded by the Ministry of Science, ICT, and Future Planning.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1416883112/-/DCSupplemental.
References
- 1.Jackson RJ. Alternative mechanisms of initiating translation of mammalian mRNAs. Biochem Soc Trans. 2005;33(Pt 6):1231–1241. doi: 10.1042/BST0331231. [DOI] [PubMed] [Google Scholar]
- 2.Jang SK, et al. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol. 1988;62(8):2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334(6180):320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 4.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11(2):113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kolupaeva VG, Pestova TV, Hellen CU, Shatsky IN. Translation eukaryotic initiation factor 4G recognizes a specific structural element within the internal ribosome entry site of encephalomyocarditis virus RNA. J Biol Chem. 1998;273(29):18599–18604. doi: 10.1074/jbc.273.29.18599. [DOI] [PubMed] [Google Scholar]
- 6.de Breyne S, Yu Y, Unbehaun A, Pestova TV, Hellen CU. Direct functional interaction of initiation factor eIF4G with type 1 internal ribosomal entry sites. Proc Natl Acad Sci USA. 2009;106(23):9197–9202. doi: 10.1073/pnas.0900153106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kozak M. The scanning model for translation: An update. J Cell Biol. 1989;108(2):229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jang SK. Internal initiation: IRES elements of picornaviruses and hepatitis C virus. Virus Res. 2006;119(1):2–15. doi: 10.1016/j.virusres.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 9.Chappell SA, Edelman GM, Mauro VP. Ribosomal tethering and clustering as mechanisms for translation initiation. Proc Natl Acad Sci USA. 2006;103(48):18077–18082. doi: 10.1073/pnas.0608212103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsuda D, Mauro VP. Determinants of initiation codon selection during translation in mammalian cells. PLoS ONE. 2010;5(11):e15057. doi: 10.1371/journal.pone.0015057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herbreteau CH, et al. HIV-2 genomic RNA contains a novel type of IRES located downstream of its initiation codon. Nat Struct Mol Biol. 2005;12(11):1001–1007. doi: 10.1038/nsmb1011. [DOI] [PubMed] [Google Scholar]
- 12.Preiss T, Hentze MW. Dual function of the messenger RNA cap structure in poly(A) tail-promoted translation in yeast. Nature. 1998;392(6675):516–520. doi: 10.1038/33192. [DOI] [PubMed] [Google Scholar]
- 13.Martin F, et al. Cap-assisted internal initiation of translation of histone H4. Mol Cell. 2011;41(2):197–209. doi: 10.1016/j.molcel.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 14.Matsuda D, Dreher TW. Close spacing of AUG initiation codons confers dicistronic character on a eukaryotic mRNA. RNA. 2006;12(7):1338–1349. doi: 10.1261/rna.67906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hundsdoerfer P, Thoma C, Hentze MW. Eukaryotic translation initiation factor 4GI and p97 promote cellular internal ribosome entry sequence-driven translation. Proc Natl Acad Sci USA. 2005;102(38):13421–13426. doi: 10.1073/pnas.0506536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rajagopal V, Park EH, Hinnebusch AG, Lorsch JR. Specific domains in yeast translation initiation factor eIF4G strongly bias RNA unwinding activity of the eIF4F complex toward duplexes with 5′ overhangs. J Biol Chem. 2012;287(24):20301–20312. doi: 10.1074/jbc.M112.347278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jan E, et al. Initiator Met-tRNA–independent translation mediated by an internal ribosome entry site element in cricket paralysis virus-like insect viruses. Cold Spring Harb Symp Quant Biol. 2001;66:285–292. doi: 10.1101/sqb.2001.66.285. [DOI] [PubMed] [Google Scholar]
- 18.Clark AT, Robertson ME, Conn GL, Belsham GJ. Conserved nucleotides within the J domain of the encephalomyocarditis virus internal ribosome entry site are required for activity and for interaction with eIF4G. J Virol. 2003;77(23):12441–12449. doi: 10.1128/JVI.77.23.12441-12449.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rippe K. Making contacts on a nucleic acid polymer. Trends Biochem Sci. 2001;26(12):733–740. doi: 10.1016/s0968-0004(01)01978-8. [DOI] [PubMed] [Google Scholar]
- 20.Jünemann C, et al. Picornavirus internal ribosome entry site elements can stimulate translation of upstream genes. J Biol Chem. 2007;282(1):132–141. doi: 10.1074/jbc.M608750200. [DOI] [PubMed] [Google Scholar]
- 21.Miller WA, Wang Z, Treder K. The amazing diversity of cap-independent translation elements in the 3′-untranslated regions of plant viral RNAs. Biochem Soc Trans. 2007;35(Pt 6):1629–1633. doi: 10.1042/BST0351629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Z, Treder K, Miller WA. Structure of a viral cap-independent translation element that functions via high-affinity binding to the eIF4E subunit of eIF4F. J Biol Chem. 2009;284(21):14189–14202. doi: 10.1074/jbc.M808841200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meulewaeter F, Van Montagu M, Cornelissen M. Features of the autonomous function of the translational enhancer domain of satellite tobacco necrosis virus. RNA. 1998;4(11):1347–1356. doi: 10.1017/s135583829898092x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liao B, Hu Y, Brewer G. Competitive binding of AUF1 and TIAR to MYC mRNA controls its translation. Nat Struct Mol Biol. 2007;14(6):511–518. doi: 10.1038/nsmb1249. [DOI] [PubMed] [Google Scholar]
- 25.Lu JY, Bergman N, Sadri N, Schneider RJ. Assembly of AUF1 with eIF4G-poly(A) binding protein complex suggests a translation function in AU-rich mRNA decay. RNA. 2006;12(5):883–893. doi: 10.1261/rna.2308106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andreev DE, et al. Differential contribution of the m7G-cap to the 5′ end-dependent translation initiation of mammalian mRNAs. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dmitriev SE, et al. Efficient translation initiation directed by the 900-nucleotide-long and GC-rich 5′ untranslated region of the human retrotransposon LINE-1 mRNA is strictly cap-dependent rather than internal ribosome entry site-mediated. Mol Cell Biol. 2007;27(13):4685–4697. doi: 10.1128/MCB.02138-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paek KY, Park SM, Hong KY, Jang SK. Cap-dependent translation without base-by-base scanning of an messenger ribonucleic acid. Nucleic Acids Res. 2012;40(15):7541–7551. doi: 10.1093/nar/gks471. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.