Significance
Many infectious bacteria such as Shigella and Salmonella use type III secretion machines, also called injectisomes, to transfer virulence proteins into eukaryotic host cells. A cytoplasmic sorting platform is required for effector selection and assembly of the needle but has not been visualized in any bacteria. We combine advanced imaging and genetic techniques to visualize the frozen-hydrated diarrheal pathogen Shigella flexneri and reveal the intact type III secretion machine and its interaction with a host cell for the first time to our knowledge. The structures characterized herein provide new insights into the mechanisms underlying type III secretion and pathogenesis and also highlight the major distinctions from the evolutionarily related bacterial flagellum.
Keywords: nanomachine, injectisome, pathogen–host interaction, cryo-electron tomography, protein secretion
Abstract
Bacterial type III secretion machines are widely used to inject virulence proteins into eukaryotic host cells. These secretion machines are evolutionarily related to bacterial flagella and consist of a large cytoplasmic complex, a transmembrane basal body, and an extracellular needle. The cytoplasmic complex forms a sorting platform essential for effector selection and needle assembly, but it remains largely uncharacterized. Here we use high-throughput cryoelectron tomography (cryo-ET) to visualize intact machines in a virulent Shigella flexneri strain genetically modified to produce minicells capable of interaction with host cells. A high-resolution in situ structure of the intact machine determined by subtomogram averaging reveals the cytoplasmic sorting platform, which consists of a central hub and six spokes, with a pod-like structure at the terminus of each spoke. Molecular modeling of wild-type and mutant machines allowed us to propose a model of the sorting platform in which the hub consists mainly of a hexamer of the Spa47 ATPase, whereas the MxiN protein comprises the spokes and the Spa33 protein forms the pods. Multiple contacts among those components are essential to align the Spa47 ATPase with the central channel of the MxiA protein export gate to form a unique nanomachine. The molecular architecture of the Shigella type III secretion machine and its sorting platform provide the structural foundation for further dissecting the mechanisms underlying type III secretion and pathogenesis and also highlight the major structural distinctions from bacterial flagella.
Type III secretion systems (T3SSs) are essential virulence determinants for many Gram-negative pathogens. The injectisome, also known as the needle complex, is the central T3SS machine required to inject effector proteins from the bacterium into eukaryotic host cells (1, 2). The injectisome has three major components: an extracellular needle, a basal body, and a cytoplasmic complex (3). Contact with a host cell membrane triggers activation of the injectisome and the insertion of a translocon pore into the target cell membrane. The entire complex then serves as a conduit for direct translocation of effectors (1, 2). Assembly of a functional T3SS requires recognition and sorting of specific secretion substrates in a well-defined order by the cytoplasmic complex (4, 5). Furthermore, genes encoding the cytoplasmic complex are regulated by physical and environmental signals (6), providing temporal control of the injection of effector proteins and thereby optimizing invasion and virulence.
Significant progress has been made in elucidating T3SS structures from many different bacteria (7, 8). 3D reconstructions of purified injectisomes from Salmonella and Shigella, together with the atomic structures of major basal body proteins, have provided a detailed view of basal body architecture (9, 10). Recent in situ structures of injectisomes from Shigella flexneri, Salmonella enterica, and Yersinia enterocolitica revealed an export gate and the structural flexibility of the basal body (11, 12). Unfortunately, these in situ structures from intact bacteria (11, 12) did not reveal any evident densities related to the proposed model of the cytoplasmic complex (8, 13).
The flagellar C ring is the cytoplasmic complex in evolutionarily related flagellar systems. It is composed of flagellar proteins FliG, FliM, and FliN and plays an essential role in flagellar assembly, rotation, and switching (14). Large drum-shaped structures of the flagellar C ring have been determined in both purified basal bodies (15, 16) and in situ motors (17–19). Similarly, electron microscopy analysis in Shigella indicated that the Spa33 protein (a homolog of the flagellar proteins FliN and FliM) is localized beneath the basal body via interactions with MxiG and MxiJ and is an essential component of the putative C ring (20). Recent experimental evidence suggests that the putative C ring provides a sorting platform for the recognition and secretion of the substrates in S. enterica (5). This sorting platform consists of three proteins, SpaO, OrgA, and OrgB, which are highly conserved among other T3SSs (21) (SI Appendix, Table S1). Despite its critical roles, little is still known about the structure and assembly of the cytoplasmic sorting platform in T3SS. In this study, we choose S. flexneri as a model system to study the intact T3SS machine and its cytoplasmic complex, mainly because a wealth of structural, biochemical, and functional information is available for the S. flexneri T3SS (22).
Results and Discussion
Shigella Minicells as a Model System for Elucidating T3SS Structure.
S. flexneri is an important diarrheal pathogen that uses its Mxi–Spa T3SS to transport effector proteins into human colonocytes, consequently altering host cell signaling to promote bacterial invasion (22). The Shigella T3SS is encoded by ∼25 genes located in the mxi, spa, and ipa operons on a large 230-kb virulence plasmid. Similar to the SpaO–OrgA–OrgB complex in S. enterica (5), homologous proteins Spa33, MxiK, and MxiN in S. flexneri are known to form a high molecular weight complex required for needle formation and substrate secretion (20, 23).
To achieve high-resolution images of intact injectisomes and the cytoplasmic complex, we constructed Shigella strains that produced minicells significantly smaller than normal bacterial cells (Fig. 1A and SI Appendix, Figs. S1 and S2). The purified minicells were able to induce contact hemolysis (SI Appendix, Fig. S3), which is known to be correlated with both phagosomal membrane lysis and tied to the secretion of three Ipa proteins (IpaB, IpaC, and IpaD) (24, 25). Furthermore, our results showed that the purified minicells were able to maintain an intimate association with red blood cells (RBCs) via the T3SS needle in the absence of any added adhesins (Fig. 1B). This is consistent with a previous report that minicells from invasive Shigella strains retained the invasive phenotype (26). A recent study from Salmonella provided further evidence that the T3SS machines in minicells are competent for protein translocation into mammalian cells (27).
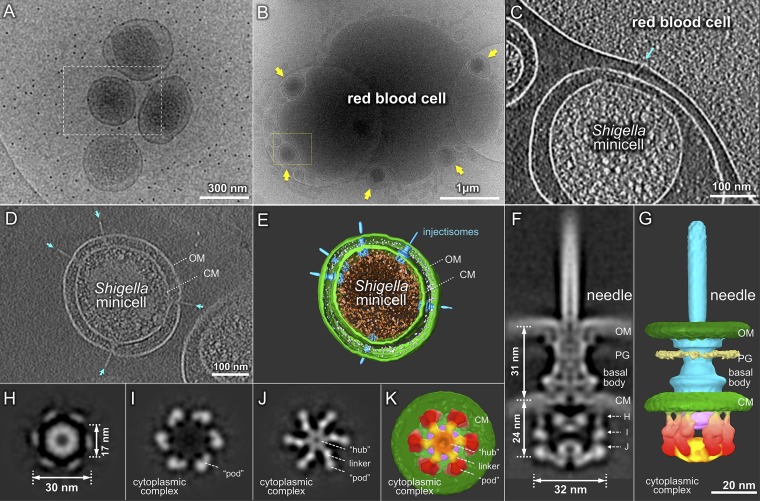
Fig. 1.
Cryo-ET of S. flexneri minicells reveals intact T3SS and its cytoplasmic structure. (A) A cryo-EM image shows tiny S. flexneri minicells with diameters of ∼0.3 μm. Purified S. flexneri minicells are able to interact intimately with an RBC (B). Yellow arrows highlight the five minicells adhering to the RBC, whose membrane is indented at each contact point. A tomographic slice reveals that an injectisome (indicated by a cyan arrow) is directly involved in the interaction with the host cell membrane (C). (D) A central slice and (E) a 3D surface rendering of a tomographic reconstruction of a typical minicell show multiple injectisomes embedded in the cell envelope, including outer membrane (OM) and cytoplasmic membrane (CM). A central section (F) and a 3D surface rendering (G) of the subtomogram average of the intact injectisome show OM, CM, peptidoglycan (PG), basal body, and needle in detail. Importantly, there is a large cytoplasmic complex that is 32 nm in diameter and 24 nm in height (F). Three cross-sections (indicated in F) of the cytoplasmic complex show sixfold symmetric features (H–J). The bottom view (K) and a side view (G) of the injectisome present the apparent discontinuity of the outer ring of six “pod-like” densities. The six pods (colored in red) are linked to the central hub (orange) by radially arranged (spoke-like) linker densities (yellow).
Cryoelectron Tomography of Shigella Minicells Reveals Intact T3SS Machines.
We exploited a newly developed direct electron detector and high-throughput cryoelectron tomography (cryo-ET) to visualize frozen-hydrated minicells derived from WT Shigella. Two different procedures were used to collect and analyze the tomographic data. Initially, low-dose tilt series were collected at 15,500× magnification and 2 × 2 binning, similar to our previous procedure (28). Subsequently, we collected tilt series in dose fractionation mode, which enabled us to correct the motion-induced image blurring (29) and effectively improve the quality of the final reconstructions. A typical 3D reconstruction of a Shigella minicell revealed multiple injectisomes embedded in an intact cell envelope (Fig. 1 D and E and Movie S1). We also observed injectisomes from Shigella minicells directly connected with a host RBC (Fig. 1 B and C and Movie S2), suggesting that the injectisomes in our minicells were intact and could mediate host cell contact.
To determine 3D structures of intact injectisomes at high resolution, subtomogram averaging was used to analyze 4,631 injectisome subtomograms (SI Appendix, Figs. S4 and S5). We determined the intact injectisome structure at 2.7 nm resolution (SI Appendix, Fig. S6) and observed a previously uncharacterized cytoplasmic complex immediately beneath the cytoplasmic membrane and basal body (Fig. 1 F–K and SI Appendix, Fig. S7). This complex contained six pod-like structures 32 nm in diameter and 24 nm in height (Fig. 1 F–K). The top portion of each pod was associated with the cytoplasmic membrane, whereas the bottom portion was connected to a distal short cylinder by six spoke-like densities (Fig. 1 F and G and SI Appendix, Fig. S7). There are relatively weak densities between the pod and the cytoplasmic membrane, perhaps suggesting that the cytoplasmic complex is loosely attached to the basal body.
The six pods are distinct from the dense, contiguous arrangement of proteins in flagellar C ring structures (15–19). Their connections with the basal body appear to be delicate, potentially explaining why they typically are not retained during purification of basal bodies (9, 10). The pods also were not resolved in a recent in situ structure from whole Yersinia cells (11). Furthermore, our initial effort in determining the injectisome structure from osmotically shocked cells failed to reveal the pod densities (SI Appendix, Figs. S5 D and H and S8), indicating that the basal body is relatively stable, whereas its interactions with the cytoplasmic complex are more delicate and can be disturbed by harsh treatment of the cells (SI Appendix, Fig. S9).
The Presence of MxiN or Spa33 Has a Dramatic Impact on T3SS Machine Structure and Function.
To define the requirements for assembling the complex cytoplasmic structure, we constructed minicell-producing strains of ∆mxiN and ∆spa33 deletion mutants of S. flexneri. Both MxiN and Spa33 are required to assemble functional needles, but not the basal body (20). As expected, we detected basal bodies, but not extracellular needles, in the minicells derived from both deletion mutant strains (Fig. 2). The bottom portion of the needle, which is inserted into the basal body, also was absent (Fig. 2 C and H). These findings indicate that both MxiN and Spa33 are important for translocation of MxiH, the major needle component. The secretion channel appeared to be closed at the base, presumably to prevent leakage (Fig. 2 C and H), reminiscent of findings for the flagellar motor of Borrelia burgdorferi (28).
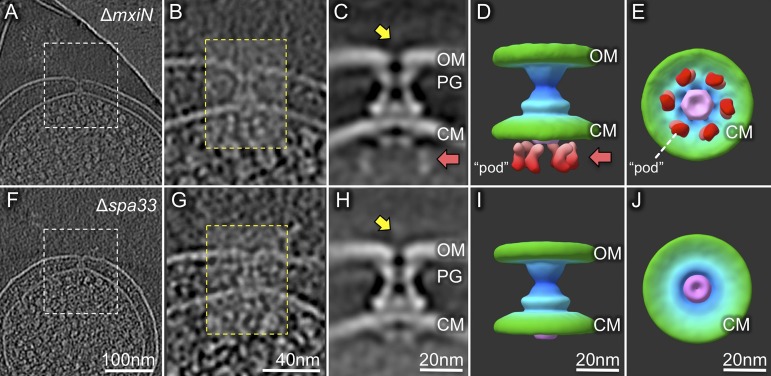
Fig. 2.
Injectisome structural differences in S. flexneri minicell mutants lacking either MxiN or Spa33. Analysis of ∆mxiN (A–E) or ∆spa33 (F–J) minicells is shown. Depicted are representative slices of cryo-ET reconstructions of a ∆mxiN minicell (A) or a ∆spa33 minicell (F), followed by the corresponding zoomed-in views (B and G), averaged structures (C and H), and two 3D surface renderings (D, E, and I–J). Both mutants lack the needle (yellow arrows) and the central hub of the cytoplasmic domain (Fig. 1 G and J). The ∆spa33 injectisomes also lack the six outer densities (pods) of the cytoplasmic domain (red arrows and red-colored densities seen in the ∆mxiN injectisomes). The predicted location of the MxiA complex is indicated in purple in the surface renderings.
In the T3SS map derived from ∆mxiN minicells, the six pods and a torus-like structure were detected beneath the basal body (Fig. 2 C–E). The torus-like structure (purple in Fig. 2), which was observed previously (11), has been proposed as a nonameric MxiA ring (13). The presence of the torus-like structure is not affected by the ∆mxiN mutation. In contrast, both the spoke-like densities and the central “hub” are absent in the ∆mxiN mutant (Fig. 2 C and D). Similar to the ATPase FliI in bacterial flagella (17), the hub is likely formed by the Spa47 ATPase. The spoke-like densities probably consist of MxiN, which interconnects the adjacent pods and the central hub. This is consistent with the previous prediction of an indeterminate number of spoke-like linkers comprised of MxiN that connect Spa47 with Spa33 (13). Apparently, the absence of MxiN significantly reduces the colocalization of the Spa47 ATPase with the remainder of the complex, thereby dramatically impacting substrate secretion.
Spa33 is thought to be a major component of the Shigella cytoplasmic complex, as demonstrated by immunogold EM labeling of Spa33 at the cytoplasmic side of purified basal bodies and biochemical evidence of Spa33 interactions with Spa47, MxiN, and MxiG (20). Our structural studies indicated that the basal bodies assembled in ∆spa33 minicells completely lack this cytoplasmic complex (Fig. 2 H–J). Furthermore, we provide structural evidence that the Spa47 ATPase fails to engage with the MxiA export gate beneath the basal body in the absence of Spa33 (Fig. 2 H–J). We infer from this that Spa33 is an essential component of the pods and provides the docking site for the Spa47 ATPase via the MxiN linkers.
Molecular Model of the T3SS Machine in Situ.
To characterize the overall architecture of the injectisome, we fitted the existing basal body structure of the Shigella T3SS (10) into the intact injectisome map described here (Fig. 3 A and B). The basal body structure matched well with the periplasmic portion of the map (Fig. 3 A and B). Compared with the existing basal body, however, the intact injectisome structure contains extra densities, including outer membrane, cytoplasmic membrane, peptidoglycan, the cytoplasmic complex, and an element associated with the outer membrane (Fig. 3A). The new element may be formed by the pilotin MxiM, which is known to interact with the ring-forming protein MxiD (30). To determine how well molecular structures could dock into our injectisome map, we built a model for MxiG based on the recent pseudoatomic model of the Salmonella PrgH (homolog of MxiG; SI Appendix, Tables S1 and S2) (31). The periplasmic domain fitted well into our map, but an additional shift of the cytoplasmic domain (MxiGC) was required to position it immediately underneath the cytoplasmic membrane (SI Appendix, Fig. S10). This alteration is apparently needed to facilitate MxiG–Spa33 interactions (20) and is also consistent with the recent model of the Yersinia YscD (homolog of MxiG; SI Appendix, Table S1) (11).
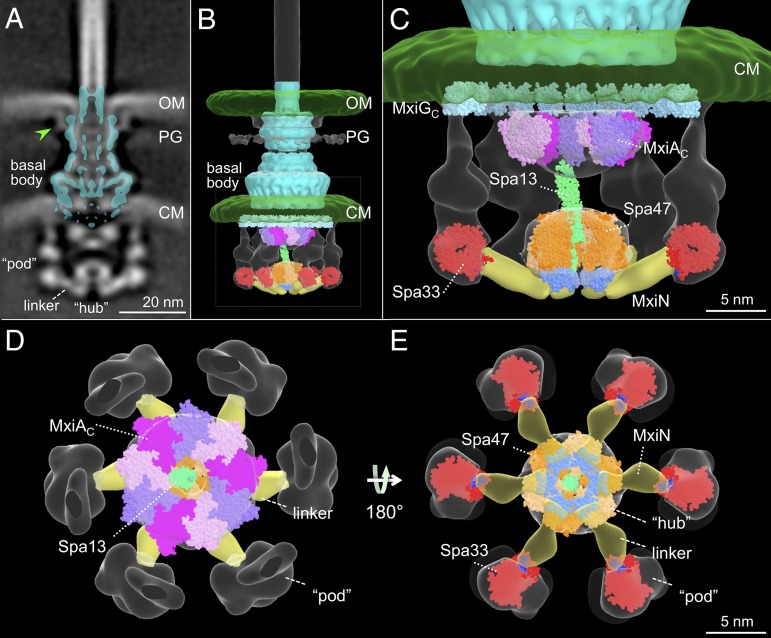
Fig. 3.
A molecular model of the T3SS injectisome. The isolated Shigella T3SS basal body (10) is fitted onto the intact injectisome map in a central section (A) and a surface rendering (B). Extra densities are apparent, including outer membrane (OM), cytoplasmic membrane (CM), peptidoglycan (PG), a large cytoplasmic complex, and a base element (green arrowhead) where it interacts with the OM. Models of several cytoplasmic proteins (MxiAC, Spa47, Spa13, Spa33) are fitted in the surface rendering map. Zoomed-in views of the cytoplasmic portion of B are shown from the side (C), top (D), and bottom (E). The Spa47 ATPase hexameric ring (the homolog of V-ATPase, PDB ID code 3J0J, orange) together with Spa13 (the homolog of FliJ, PDB ID code 3AJW, green) is docked into the central hub of the map. Spa13 is long enough to interact with the nonameric ring of MxiAC (different shades of purple), which fits well into the torus-like structure near the cytoplasmic membrane (C and D). Underneath the MxiGC ring (the homolog of PrgHC, PDB ID code 3J1W, dark green), six spa33-encoded complexes form the proposed sorting platform. We place the Spa33 homologs (FliN tetramer, PDB ID code 1YAB, red) into the bottom part of the pod. MxiN forms the spoke-like linker (yellow) that interacts with both the hydrophobic patch (blue) in Spa33 and the C-terminal domain (cyan) of Spa47 as shown in the bottom view (E).
Another key component of the cytoplasmic side of the injectisome is MxiA, which consists of a transmembrane domain and a large cytoplasmic domain (MxiAC). As MxiAC has been postulated to form a nonameric ring essential for secretion (13), we docked the crystal structure of this MxiAC nonameric ring into the torus-like structure, which extends ∼6 nm from the cytoplasmic membrane. The excellent fit supports the proposed juxtapositioning of the MxiAc nonameric ring beneath the basal body (Fig. 3 B–D and Movie S3) (13). Furthermore, in both ∆mxiN and ∆spa33 mutants, the torus-like structure remained intact (colored purple in Fig. 2), consistent with a direct linkage of the MxiAC nonameric ring with the MxiA transmembrane domain.
The main component of the hub, Spa47, energizes secretion of effector proteins and shares significant amino acid similarity with FliI and V-ATPases (SI Appendix, Table S3). By analogy with a FliI hexamer, which is positioned within the in situ flagellar motor (17), a Spa47 hexamer likely accounts for density present ∼10 nm beneath the MxiAC ring (Fig. 3A). Spa47 is also known to interact with MxiN and Spa13 (the homolog of FliJ; SI Appendix, Table S4) (20, 32, 33). Together, we built a model of the Spa47–Spa13–MxiN complex (Movie S3). The C-terminal domains of Spa47 are connected to the MxiAC ring via Spa13, whereas the N-terminal domains of Spa47 interact with MxiN (Fig. 3 B–E).
Spa33 is likely a main constituent of the pods. The C-terminal region of Spa33 is homologous to the flagellar protein FliN (SI Appendix, Table S5), which has been proposed to form a homotetramer (34, 35) at the bottom of the flagellar C ring. Recent studies showed that the Spa33 ortholog YscQ from Yersinia exists as two translation products: intact YscQ and a C-terminal fragment (36). Similar to FliN, the C-terminal domain structure of YscQ reveals an intertwined homodimer (36). Spa33 may also form a multimeric complex. The tetramer model of FliN fills well into the bottom portion of each pod density, suggesting that the C-terminal fragment of Spa33 likely forms a stable tetramer and an intact Spa33 and another protein MxiK could account for the remainder of the density of the pods (Fig. 3D), as they are known to form a high molecular weight complex (20). It remains to be resolved how these proteins are elegantly organized within the pods.
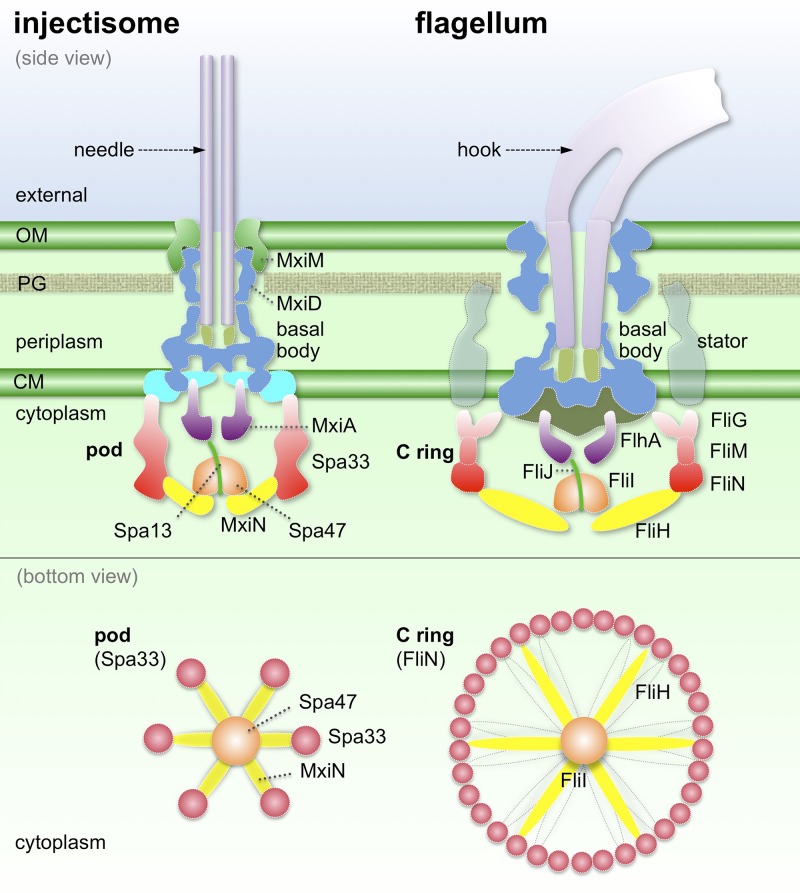
The discovery of the spa33-encoded hexameric complex and its physical connections with the ATPase highlights the similarities and differences between an injectisome and a bacterial flagellum (Fig. 4). Both systems contain an export gate powered by the proton motive force and a cytoplasmic ATPase complex (21, 37, 38). A similar integrated network and molecular mechanism are likely used for the recognition and secretion of specific substrates. Nevertheless, the six discrete pods observed in the injectisome are distinct from the flagellar C ring, which is not only indispensable for substrate secretion but also essential for flagellar rotation and switching (Fig. 4).
Fig. 4.
Comparative structural models of injectisomes and bacterial flagella. The cytoplasmic hexamer of the Spa33 complex positions the Spa47 ATPase (via MxiN) directly beneath and in line with the export gate, beginning with the MxiA cytoplasmic domain. In contrast, the flagellar C ring contains multiple copies of FliG, FliM, and FliN that are involved in secretion, rotation, and switching. FliH, the homolog of MxiN, is likely sufficiently flexible to interact with different FliN proteins at the bottom of the C ring. The outer membrane (OM), cytoplasmic membrane (CM), peptidoglycan (PG), a large cytoplasmic complex, and the basal body are colored consistently as those in previous figures.
The conserved cytoplasmic complex in S. enterica T3SS provides a sorting platform that determines the order of protein secretion (5). We propose that the hub–spoke–pod complex functions specifically as the sorting platform. The Spa47 ATPase within the complex is aligned with the central channel of the MxiA gate, suggesting that they function together to engage and secrete needle components and effector proteins. The multiple contacts we identified among components of the cytoplasmic complex are likely to be important for the integrity of the sorting platform and its functions in needle formation and subsequent substrate secretion (Fig. 4). We postulate that this platform functions in the recruitment of needle components and effectors either without or within a complex with dedicated chaperones. The sorting platform delivers these substrates to the Spa47 ATPase for chaperone dissociation and substrate unfolding, whereupon the substrates are delivered to MxiA and the export gate for secretion (Fig. 4 and Movie S3). Although many questions remain regarding recognition of chaperone/substrate complexes and the temporal control of substrate selection, our architectural definition of the Shigella T3SS sorting platform provides a structural basis for further dissecting the mechanisms underlying the T3SS-mediated secretion and pathogenesis.
Materials and Methods
Generation of ΔmxiN and Δspa33 Mutants.
Streptomycin-resistant strain S. flexneri serotype 5a (M90T-Sm) was used (39) as a parent strain to create mxiN::tetRA and spa33::tetRA mutants via lambda red recombination as described in ref. 40. Integration of the knockout cassette at the desired location was confirmed by PCR using a primer common to the tetRA cassette and one upstream from mxiN and spa33, respectively. Specific sequences used to target mxiN and spa33 are described in SI Appendix, Table S6.
Preparation of Minicell-Producing S. flexneri Strains.
Minicells of WT, ∆mxiN, and ∆spa33 S. flexneri were generated by introducing plasmid pBS58, which constitutively expresses Escherichia coli cell division genes ftsQ, ftsA, and ftsZ from a low-copy, spectinomycin-resistant plasmid (41). Bacterial cultures were grown overnight at 37 °C in Trypticase Soy Broth and fresh cultures were prepared from a 1:100 dilution and then grown at 37 °C to late log phase. Spectinomycin (100 µg/mL) was added for selection of pBS58 and 5 μg/mL of tetracycline to select for the two mutants. To enrich for minicells, the culture was centrifuged at 1,000 × g for 5 min to remove the large cells, and the supernatant fraction was further centrifuged at 20,000 × g for 10 min to collect the minicells (SI Appendix, Fig. S2).
Preparation of Osmotically Shocked Cells.
Bacteria were collected after centrifugation of 5 mL of bacterial culture in l-broth grown to early log phase with shaking at 37 °C. Osmotically shocked cells were prepared as described previously (20).
Contact Hemolysis and Initial Minicell–Host Interaction.
The hemolytic activity of RBCs was used to test the function of the S. flexneri minicells as described previously (25). Sheep RBCs were obtained from Innovative Research, Inc. RBCs were washed 3 times in PBS by centrifugation at 2,000 × g for 5 min at 4 °C and resuspended at 108/mL. We mixed 50 μL of minicells (or WT cells) with 50 μL of RBCs. The mixed culture was centrifuged at 5,000 × g for 5 min at 4 °C and incubated at 37 °C for 1 h. The cells were resuspended in PBS to disrupt bacterial attachment. All solid material was removed by centrifugation, and the released hemoglobin in the supernatant fraction was measured by absorbance at 595 nm as a measure of the hemolytic activity (SI Appendix, Fig. S3). Additionally, the resulting samples containing RBCs and attached cells were examined in a light microscope and were also used to prepare frozen-hydrated specimens.
Preparation of Frozen-Hydrated Specimens.
Bacterial cultures were mixed with 10 nm of colloidal gold particles (used as fiducial markers in image alignment) and then deposited onto freshly glow-discharged, holey carbon grids for 1 min. The grids were blotted with filter paper and rapidly frozen in liquid ethane, using a gravity-driven plunger apparatus.
Cryo-ET Data Collection and 3D Reconstructions.
The resulting frozen-hydrated specimens were imaged at −170 °C using a Polara G2 electron microscope (FEI Company) equipped with a field emission gun and a Direct Detection Camera (Gatan K2 Summit). Because the camera was recently integrated into our EM system, two tomographic packages [UCSF Tomography (42) and SerialEM (43)] and different procedures were used to collect low-dose tilt series.
For initial data collection, the microscope was operated at 300 kV with a magnification of 15,500×, resulting in an effective pixel size of 5.04 Å after 2 × 2 binning. Using UCSF Tomography software (42), low-dose, single-axis tilt series were collected from each minicell at 6–9 μm defocus with a cumulative dose of ∼60 e−/Å2 distributed over 61 images and covering an angular range of −60° to +60°, with an angular increment of 2°.
For subsequent data collection, we used SerialEM (43) to collect low-dose, single-axis tilt series with dose fractionation mode at 6 μm defocus. The microscope was operated at a magnification of 9,400×, resulting in an effective pixel size of 4.45 Å without binning and a cumulative dose of ∼60 e−/Å2 distributed over 61 stacks. Each stack contains eight images. We developed Tomoauto (a wrapper library, available at https://github.com/DustinMorado/tomoauto) to facilitate the automation of cryo-ET data processing. System scripts in the library configure and coordinate the execution of several programs essential for the processing and alignment of tilt series as well as the subsequent reconstruction of these series into tomograms. An input/output library for the MRC file format maintains the header information generated during collection throughout the processing. The main executable encompasses the following: drift correction of dose-fractionated data using dosefgpu_driftcorr (29) and the assembly of corrected sums into tilt series, automatic fiducial seed model generation by RAPTOR software (44), alignment and contrast transfer function correction of tilt series by IMOD software (45, 46), and reconstruction of tilt series into tomograms by TOMO3D software (47). The flowchart is shown in SI Appendix, Fig. S11. In total, 1,917 tomographic reconstructions were generated (SI Appendix, Table S7).
Subtomogram Averaging and Correspondence Analysis.
A general procedure of subtomogram averaging was described previously (18, 48, 49). A total of 7,824 injectisome subtomograms (400 × 400 × 400 voxels) were visually identified and then extracted from cryo-tomographic reconstructions (SI Appendix, Table S7). The initial orientation of each particle was estimated by the basal body and needle tip coordinates, thereby providing two of the three Euler angles. To accelerate image analysis, 4 × 4 × 4 binned subtomograms (100 × 100 × 100 voxels) were used for initial alignment (SI Appendix, Fig. S4). A global average of all of the extracted 4 × 4 × 4 binned subtomograms was performed after application of the two Euler angles previously determined (SI Appendix, Fig. S4B). After aligning the basal body, we generated a binary mask for cytoplasmic area (SI Appendix, Fig. S4 D and H). Relevant voxels of the aligned subtomograms were analyzed by multivariate statistical analysis and hierarchical ascendant classification (50). Class averages were computed in Fourier space to minimize the missing wedge problem of tomography. All class averages were further aligned with each other to minimize differences (SI Appendix, Fig. S5). Fourier shell correlation between the two independent reconstructions was used to estimate the resolution of the averaged structures (SI Appendix, Table S7 and Fig. S6).
3D Visualization and Molecular Modeling.
UCSF Chimera was used for 3D surface rendering of subtomogram averages and molecular modeling (51). Three isosurface maps were rendered in different contour levels—0.67 σ, 1.00 σ, and 1.66 σ, respectively (SI Appendix, Fig. S12). Because of differential mobility in different parts of the structure, we cannot be certain which of the three levels is the most appropriate. The T3SS basal body map from Shigella (EMD-1617) was then fitted into our intact T3SS map using the function “fit in map” in UCSF Chimera. We built the initial model based on refined structures from Salmonella T3SS (31): InvG [Protein Data Bank (PDB) ID code 3J1V], PrgHP (PDB ID code 3J1X), and PrgHC (PDB ID code 3J1W) (SI Appendix, Fig. S10). A large shift is required to relocate the cytoplasmic domain of MxiG (SI Appendix, Fig. S10). Structures of V-ATPase (PDB ID code 3J0J) and FliJ (PDB ID code 3AJW) were used to build the model of the Spa47–Spa13–MxiN complex. Six FliN tetramers (PDB ID code 1YAB) (35) were placed into the six ring-shaped densities at the bottom, with the hydrophobic patch interacting with the MxiN linkers.
Supplementary Material
Acknowledgments
We thank Drs. Steven J. Norris and Peter J. Christie for helpful comments on the manuscript. We thank Drs. Shawn Zheng and David Agard for the support on UCSF Tomography, Drs. David Mastronarde and Chen Xu for the support on SerialEM, and Dr. Daniel Haeusser for the pBS58 plasmid. B.H. and J.L. were supported in part by Grant R01AI087946 from the National Institute of Allergy and Infectious Diseases, Grants R01GM110243 and R01GM107629 from the National Institute of General Medical Sciences (NIGMS), and Grant AU-1714 from the Welch Foundation. W.M. was supported by Grant R01GM61074 from the NIGMS, and J.R.R. was supported by Canadian Institute of Health Research Grant MOP-102594. The direct electron detector was funded by National Institutes of Health Award S10OD016279.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the EMDataBank database, emdatabank.org (accession nos. EMD-2667, EMD-2668, and EMD-2669).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1411610112/-/DCSupplemental.
References
- 1.Cornelis GR. The type III secretion injectisome. Nat Rev Microbiol. 2006;4(11):811–825. doi: 10.1038/nrmicro1526. [DOI] [PubMed] [Google Scholar]
- 2.Galán JE, Wolf-Watz H. Protein delivery into eukaryotic cells by type III secretion machines. Nature. 2006;444(7119):567–573. doi: 10.1038/nature05272. [DOI] [PubMed] [Google Scholar]
- 3.Kubori T, et al. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science. 1998;280(5363):602–605. doi: 10.1126/science.280.5363.602. [DOI] [PubMed] [Google Scholar]
- 4.Deane JE, Abrusci P, Johnson S, Lea SM. Timing is everything: The regulation of type III secretion. Cell Mol Life Sci. 2010;67(7):1065–1075. doi: 10.1007/s00018-009-0230-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lara-Tejero M, Kato J, Wagner S, Liu X, Galán JE. A sorting platform determines the order of protein secretion in bacterial type III systems. Science. 2011;331(6021):1188–1191. doi: 10.1126/science.1201476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marteyn B, et al. Modulation of Shigella virulence in response to available oxygen in vivo. Nature. 2010;465(7296):355–358. doi: 10.1038/nature08970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abrusci P, McDowell MA, Lea SM, Johnson S. Building a secreting nanomachine: A structural overview of the T3SS. Curr Opin Struct Biol. 2014;25:111–117. doi: 10.1016/j.sbi.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diepold A, Wagner S. Assembly of the bacterial type III secretion machinery. FEMS Microbiol Rev. 2014;38(4):802–822. doi: 10.1111/1574-6976.12061. [DOI] [PubMed] [Google Scholar]
- 9.Schraidt O, Marlovits TC. Three-dimensional model of Salmonella’s needle complex at subnanometer resolution. Science. 2011;331(6021):1192–1195. doi: 10.1126/science.1199358. [DOI] [PubMed] [Google Scholar]
- 10.Hodgkinson JL, et al. Three-dimensional reconstruction of the Shigella T3SS transmembrane regions reveals 12-fold symmetry and novel features throughout. Nat Struct Mol Biol. 2009;16(5):477–485. doi: 10.1038/nsmb.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kudryashev M, et al. In situ structural analysis of the Yersinia enterocolitica injectisome. eLife. 2013;2:e00792. doi: 10.7554/eLife.00792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawamoto A, et al. Common and distinct structural features of Salmonella injectisome and flagellar basal body. Scientific Reports. 2013;3:3369. doi: 10.1038/srep03369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abrusci P, et al. Architecture of the major component of the type III secretion system export apparatus. Nat Struct Mol Biol. 2013;20(1):99–104. doi: 10.1038/nsmb.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chevance FF, Hughes KT. Coordinating assembly of a bacterial macromolecular machine. Nat Rev Microbiol. 2008;6(6):455–465. doi: 10.1038/nrmicro1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Francis NR, Sosinsky GE, Thomas D, DeRosier DJ. Isolation, characterization and structure of bacterial flagellar motors containing the switch complex. J Mol Biol. 1994;235(4):1261–1270. doi: 10.1006/jmbi.1994.1079. [DOI] [PubMed] [Google Scholar]
- 16.Thomas DR, Francis NR, Xu C, DeRosier DJ. The three-dimensional structure of the flagellar rotor from a clockwise-locked mutant of Salmonella enterica serovar Typhimurium. J Bacteriol. 2006;188(20):7039–7048. doi: 10.1128/JB.00552-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen S, et al. Structural diversity of bacterial flagellar motors. EMBO J. 2011;30(14):2972–2981. doi: 10.1038/emboj.2011.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu J, et al. Intact flagellar motor of Borrelia burgdorferi revealed by cryo-electron tomography: Evidence for stator ring curvature and rotor/C-ring assembly flexion. J Bacteriol. 2009;191(16):5026–5036. doi: 10.1128/JB.00340-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murphy GE, Leadbetter JR, Jensen GJ. In situ structure of the complete Treponema primitia flagellar motor. Nature. 2006;442(7106):1062–1064. doi: 10.1038/nature05015. [DOI] [PubMed] [Google Scholar]
- 20.Morita-Ishihara T, et al. Shigella Spa33 is an essential C-ring component of type III secretion machinery. J Biol Chem. 2006;281(1):599–607. doi: 10.1074/jbc.M509644200. [DOI] [PubMed] [Google Scholar]
- 21.Büttner D. Protein export according to schedule: Architecture, assembly, and regulation of type III secretion systems from plant- and animal-pathogenic bacteria. Microbiol Mol Biol Rev. 2012;76(2):262–310. doi: 10.1128/MMBR.05017-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schroeder GN, Hilbi H. Molecular pathogenesis of Shigella spp.: Controlling host cell signaling, invasion, and death by type III secretion. Clin Microbiol Rev. 2008;21(1):134–156. doi: 10.1128/CMR.00032-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson S, Blocker A. Characterization of soluble complexes of the Shigella flexneri type III secretion system ATPase. FEMS Microbiol Lett. 2008;286(2):274–278. doi: 10.1111/j.1574-6968.2008.01284.x. [DOI] [PubMed] [Google Scholar]
- 24.Sansonetti PJ, Ryter A, Clerc P, Maurelli AT, Mounier J. Multiplication of Shigella flexneri within HeLa cells: Lysis of the phagocytic vacuole and plasmid-mediated contact hemolysis. Infect Immun. 1986;51(2):461–469. doi: 10.1128/iai.51.2.461-469.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blocker A, et al. The tripartite type III secreton of Shigella flexneri inserts IpaB and IpaC into host membranes. J Cell Biol. 1999;147(3):683–693. doi: 10.1083/jcb.147.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hale TL, Sansonetti PJ, Schad PA, Austin S, Formal SB. Characterization of virulence plasmids and plasmid-associated outer membrane proteins in Shigella flexneri, Shigella sonnei, and Escherichia coli. Infect Immun. 1983;40(1):340–350. doi: 10.1128/iai.40.1.340-350.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carleton HA, Lara-Tejero M, Liu X, Galán JE. Engineering the type III secretion system in non-replicating bacterial minicells for antigen delivery. Nat Commun. 2013;4:1590. doi: 10.1038/ncomms2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao X, et al. Cryoelectron tomography reveals the sequential assembly of bacterial flagella in Borrelia burgdorferi. Proc Natl Acad Sci USA. 2013;110(35):14390–14395. doi: 10.1073/pnas.1308306110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10(6):584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schuch R, Maurelli AT. MxiM and MxiJ, base elements of the Mxi-Spa type III secretion system of Shigella, interact with and stabilize the MxiD secretin in the cell envelope. J Bacteriol. 2001;183(24):6991–6998. doi: 10.1128/JB.183.24.6991-6998.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bergeron JR, et al. A refined model of the prototypical Salmonella SPI-1 T3SS basal body reveals the molecular basis for its assembly. PLoS Pathog. 2013;9(4):e1003307. doi: 10.1371/journal.ppat.1003307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cherradi Y, Hachani A, Allaoui A. Spa13 of Shigella flexneri has a dual role: Chaperone escort and export gate-activator switch of the type III secretion system. Microbiology. 2014;160(Pt 1):130–141. doi: 10.1099/mic.0.071712-0. [DOI] [PubMed] [Google Scholar]
- 33.Ibuki T, et al. Common architecture of the flagellar type III protein export apparatus and F- and V-type ATPases. Nat Struct Mol Biol. 2011;18(3):277–282. doi: 10.1038/nsmb.1977. [DOI] [PubMed] [Google Scholar]
- 34.Paul K, Blair DF. Organization of FliN subunits in the flagellar motor of Escherichia coli. J Bacteriol. 2006;188(7):2502–2511. doi: 10.1128/JB.188.7.2502-2511.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown PN, Mathews MA, Joss LA, Hill CP, Blair DF. Crystal structure of the flagellar rotor protein FliN from Thermotoga maritima. J Bacteriol. 2005;187(8):2890–2902. doi: 10.1128/JB.187.8.2890-2902.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bzymek KP, Hamaoka BY, Ghosh P. Two translation products of Yersinia yscQ assemble to form a complex essential to type III secretion. Biochemistry. 2012;51(8):1669–1677. doi: 10.1021/bi201792p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Erhardt M, Namba K, Hughes KT. Bacterial nanomachines: The flagellum and type III injectisome. Cold Spring Harb Perspect Biol. 2010;2(11):a000299. doi: 10.1101/cshperspect.a000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Galán JE. Energizing type III secretion machines: What is the fuel? Nat Struct Mol Biol. 2008;15(2):127–128. doi: 10.1038/nsmb0208-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Onodera NT, et al. Genome sequence of Shigella flexneri serotype 5a strain M90T Sm. J Bacteriol. 2012;194(11):3022. doi: 10.1128/JB.00393-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pruneda JN, et al. E2∼Ub conjugates regulate the kinase activity of Shigella effector OspG during pathogenesis. EMBO J. 2014;33(5):437–449. doi: 10.1002/embj.201386386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bi E, Lutkenhaus J. FtsZ regulates frequency of cell division in Escherichia coli. J Bacteriol. 1990;172(5):2765–2768. doi: 10.1128/jb.172.5.2765-2768.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng SQ, et al. UCSF tomography: An integrated software suite for real-time electron microscopic tomographic data collection, alignment, and reconstruction. J Struct Biol. 2007;157(1):138–147. doi: 10.1016/j.jsb.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 43.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152(1):36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 44.Amat F, et al. Markov random field based automatic image alignment for electron tomography. J Struct Biol. 2008;161(3):260–275. doi: 10.1016/j.jsb.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 45.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116(1):71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 46.Xiong Q, Morphew MK, Schwartz CL, Hoenger AH, Mastronarde DN. CTF determination and correction for low dose tomographic tilt series. J Struct Biol. 2009;168(3):378–387. doi: 10.1016/j.jsb.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Agulleiro JI, Fernandez JJ. Fast tomographic reconstruction on multicore computers. Bioinformatics. 2011;27(4):582–583. doi: 10.1093/bioinformatics/btq692. [DOI] [PubMed] [Google Scholar]
- 48.Hu B, Margolin W, Molineux IJ, Liu J. The bacteriophage t7 virion undergoes extensive structural remodeling during infection. Science. 2013;339(6119):576–579. doi: 10.1126/science.1231887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu J, Wright ER, Winkler H. 3D visualization of HIV virions by cryoelectron tomography. Methods Enzymol. 2010;483:267–290. doi: 10.1016/S0076-6879(10)83014-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Winkler H. 3D reconstruction and processing of volumetric data in cryo-electron tomography. J Struct Biol. 2007;157(1):126–137. doi: 10.1016/j.jsb.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 51.Pettersen EF, et al. UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.