Significance
The naked mole rat is a longest lived and cancer-resistant rodent. Tumor resistance in the naked mole rat is mediated by signals from the extracellular matrix component hyaluronan triggering the induction of INK4 (inhibitors of cyclin dependent kinase 4) locus expression. The human and mouse INK4 locus encodes three critical tumor-suppressor proteins, p15INK4b, ARF (alternate reading frame), and p16INK4a, which are among the most frequently mutated in cancer. Furthermore, p16INK4a is implicated in aging and senescence. Here, we show that the naked mole rat INK4 locus encodes an additional product, a hybrid between p15INK4b and p16INK4a. The novel product, named pALTINK4a/b, may contribute to tumor resistance and longevity of the naked mole rat. Understanding the regulation of the INK4 locus is critical for cancer and aging research.
Keywords: naked mole rat, INK4, p16, p15
Abstract
The naked mole rat (Heterocephalus glaber) is a long-lived and tumor-resistant rodent. Tumor resistance in the naked mole rat is mediated by the extracellular matrix component hyaluronan of very high molecular weight (HMW-HA). HMW-HA triggers hypersensitivity of naked mole rat cells to contact inhibition, which is associated with induction of the INK4 (inhibitors of cyclin dependent kinase 4) locus leading to cell-cycle arrest. The INK4a/b locus is among the most frequently mutated in human cancer. This locus encodes three distinct tumor suppressors: p15INK4b, p16INK4a, and ARF (alternate reading frame). Although p15INK4b has its own ORF, p16INK4a and ARF share common second and third exons with alternative reading frames. Here, we show that, in the naked mole rat, the INK4a/b locus encodes an additional product that consists of p15INK4b exon 1 joined to p16INK4a exons 2 and 3. We have named this isoform pALTINK4a/b (for alternative splicing). We show that pALTINK4a/b is present in both cultured cells and naked mole rat tissues but is absent in human and mouse cells. Additionally, we demonstrate that pALTINK4a/b expression is induced during early contact inhibition and upon a variety of stresses such as UV, gamma irradiation-induced senescence, loss of substrate attachment, and expression of oncogenes. When overexpressed in naked mole rat or human cells, pALTINK4a/b has stronger ability to induce cell-cycle arrest than either p15INK4b or p16INK4a. We hypothesize that the presence of the fourth product, pALTINK4a/b of the INK4a/b locus in the naked mole rat, contributes to the increased resistance to tumorigenesis of this species.
The naked mole rat (Heterocephalus glaber) is notable for being the longest-lived rodent, with an observed maximum lifespan of over 30 y (1). Additionally, despite being kept in captivity since 1974, there has been no reported incidence of neoplasia in these animals (2, 3). Naked mole rats remain healthy until almost the end of their lives, showing resistance to multiple age-related diseases (4). Understanding the mechanisms responsible for tumor resistance of this unusual rodent may lead to the discovery of novel targets to cancer therapy that cannot be found in cancer-prone laboratory species.
In tissue culture, naked mole rat fibroblasts display hypersensitivity to contact inhibition, termed early contact inhibition (ECI) (5). ECI is directly linked to the cancer resistance of naked mole rats. The signal triggering ECI comes from an extracellular matrix component, hyaluronan, which, in the naked mole rat, is characterized by an extremely high molecular weight (6). Removal of high molecular weight hyaluronan abrogates ECI and makes naked mole rat cells susceptible to malignant transformation (6). ECI is associated with an increase in p16INK4a expression, and cell lines that spontaneously lost expression from the INK4a/b (inhibitors of cyclin dependent kinase 4) locus no longer display ECI (5).
Deletion or silencing of the INK4a/b locus is the hallmark of human cancer (7, 8). This small (<50 kb) locus is remarkable because, in mammals, it encodes three distinct tumor suppressors: p16INK4a, p15INK4b, and p19ARF (p14ARF in human) (9). These three proteins coordinate a signaling network that depends on the activities of the retinoblastoma protein (RB) and the p53 tumor suppressor protein. The p16INK4a and p15INK4b proteins are cyclin-dependent kinase inhibitors that directly inhibit the binding of cyclins to their target cyclin-dependent kinases (10). p16INK4a is involved in establishing replicative senescence, oncogene-induced senescence, and stress-induced premature senescence (11, 12), plays a role in stem-cell aging (9, 13), and acts as a barrier to induced pluripotency (14). Many genes involved in senescence are also associated with age-related disease (15, 16). Indeed, evidence has accumulated to support the role of the locus in atherosclerosis and other aging-related pathologies (17–20). Differences between p15INK4b and p16INK4a include expression profile and stability, with p15INK4b having a lower overall half-life (21, 22). ARF (alternate reading frame) protein p19ARF (in mice) also has a tumor-suppressing function (23): it acts to repress the oncogene MDM2, which in turn binds p53, repressing its activation (24). ARF has other functions in regulating the cell cycle, independently of p53 (25).
The structure of the INK4a/b locus is complex and has undergone several modifications during its evolutionary history (26). The emergence of separate p16INK4a and p15INK4b genes is the result of an ancient duplication event (27). They are structurally very similar, featuring several ankyrin repeats. The ARF is a completely different protein produced by using an alternative first exon that splices to the second exon of p16INK4b in a different frame (28). The first exon of ARF was acquired before the INK4a/b duplication event (29). The whole-genome sequencing of the naked mole rat revealed that the INK4a/b locus was altered by early translation termination events (30), suggesting that it may have undergone specific evolutionary changes to increase cancer resistance in this species. This genomic finding complemented our earlier results that INK4a/b was required for ECI and cancer resistance.
Because of the importance of INK4a/b in cancer resistance, we decided to characterize the expression of its products. Unexpectedly, we found that, in the naked mole rat, the INK4a/b locus produced a novel hybrid protein isoform from the INK4b promoter that splices p15INK4b exon 1 to p16INK4a exon 2. This isoform, which we named pALTINK4a/b, is unique to the naked mole rat and is not observed in human or mouse. pALTINK4a/b is present in cultured cells and in naked mole rat tissues in vivo. pALTINK4a/b is activated by a variety of stresses, similar to p15INK4b and p16INK4a, but has higher capacity to induce cell-cycle arrest. We hypothesize that the presence of the additional, more potent, cyclin-dependent kinase inhibitor pALTINK4a/b contributes to the cancer resistance of the naked mole rat.
Results
Identification of the Novel INK4 Isoform in the Naked Mole Rat.
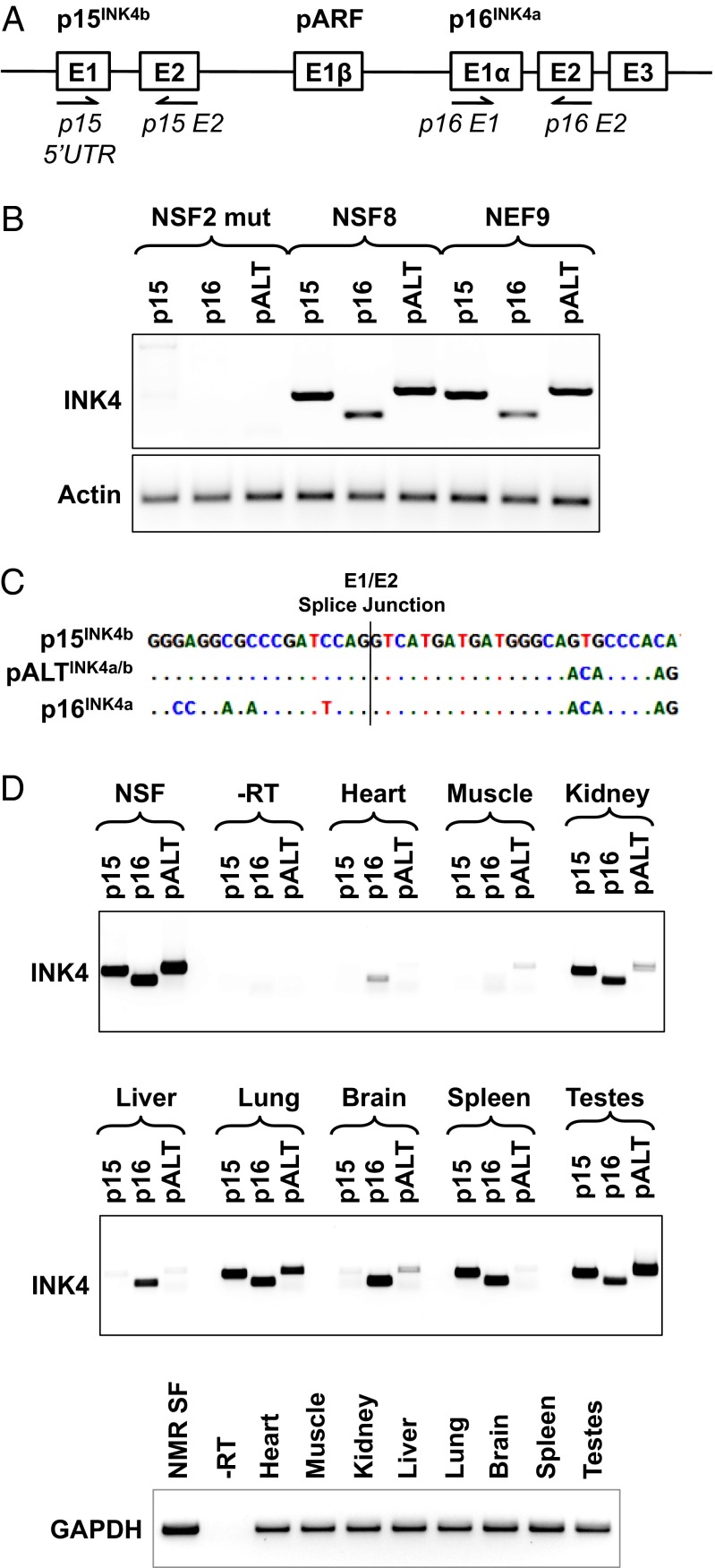
Previous work in naked mole rat fibroblasts had indicated that the INK4 locus was an important mediator of the tumor resistance of this species (5). To examine the expression of INK4 products, we designed two pairs of primers (Fig. 1A) to amplify p15INK4b and p16INK4a cDNAs from three independent lines of naked mole rat fibroblasts. We observed the expected p16INK4a, p15INK4b products in primary skin fibroblasts (NSF8) and embryonic fibroblasts (NEF9). The cell line NSF2 mut, which spontaneously lost the early contact inhibition phenotype, did not express any INK4 products (Fig. 1B), consistent with our previous observations (5). Unexpectedly, we observed a previously unidentified transcript comprised of exon 1 of p15INK4b and exons 2 and 3 followed by the 3′ UTR from p16INK4a (Fig. 1B). The junction between the exon 1 of p15INK4b and exon 2 of p16INK4a corresponded to the canonical splice junction (Fig. 1C). We named this isoform pALTINK4a/b. In the fibroblasts, the three isoforms were expressed at roughly similar levels, but the direct comparison was difficult because the primer sequences were similar but not identical. We next examined the expression of the three INK4 products in a panel of naked mole rat tissues obtained from a 1-y-old animal. We observed that pALTINK4a/b was expressed in several tissues, with the highest levels present in lung and testes, indicating that pALTINK4a/b is produced in vivo. The expression of various INK4 products appeared to be tissue-specific (Fig. 1D), suggesting that different tissues rely on various combinations of INK4 products for cell-cycle control. It is important to note that the testes were collected from a nonbreeding male, which was characterized by suppressed spermatogenesis, explaining the high levels of INK4 products in the testes. Consistent with the PCR data, the pALTINK4a/b product was also detected in the RNA-seq dataset that we previously reported (30). In that dataset, which combined seven tissues, pALTINK4a/b appeared to be less abundant than p15INK4b but more abundant than p16INK4a (Table S1).
Fig. 1.
The INK4a/b locus of the NMR produces an alternative protein isoform pALTINK4a/b composed of the first exon of p15INK4b and the second and third exons of p16INK4a. (A) Schematic of the INK4a/b locus showing the location of the primers used to amplify the three INK4a/b transcripts. (B) RT-PCR products corresponding to the three INK4a/b transcripts amplified from RNA isolated from three lines of naked mole rat fibroblasts, with primers shown in A and Table S2; pALTINK4a/b was amplified with primers p15-5UTR and p16-E2. NSF2 mut is the line of spontaneously mutated naked mole rat fibroblasts that lost INK4a/b locus expression and do not show ECI; NSF8 are primary naked mole rat skin fibroblasts; NEF9 are naked mole rat embryonic fibroblasts. (C) Sequences of the Exon1/Exon2 splice junctions in p15, p16, and pALT. (D) RT-PCR showing INK4a/b products amplified from naked mole rat tissues. GAPDH was used to show that equal amounts of RNA template were used for each tissue. The reactions were repeated at least three times, and representative gels are shown.
To further characterize the pALT product and exclude a possibility that it is the result of a PCR template switch, we transfected human fibroblasts with pCMV vectors containing naked mole rat p15INK4b, p16INK4a, or pALTINK4a/b cDNAs, or with a mix of the p15INK4b and p16INK4a plasmids. We then performed RT-PCR analysis using primer pairs specific to p15INK4b, p16INK4a, and pALTINK4a/b (Fig. S1). As expected, the cells expressing only one gene showed one respective band whereas the mix showed a p15INK4b and p16INK4a, but no pALTINK4a/b, band. If template switching had occurred, we would have expected to detect the pALTINK4a/b product in the mixed transfection. This experiment further verifies that the pALTINK4a/b transcript is a genuine gene product and not a result of a PCR artifact.
We also tested mouse and human cells for the presence of a similar alternative splice product. Mouse skin fibroblasts were induced by UV irradiation to produce p16INK4a, and RNA was extracted from these cells. Replicatively senescent human skin fibroblasts served as human cell lines expressing p16INK4a. Several primer pairs (p15 5′ UTR and p16 3′ UTR) for each species were used, and no hybrid products between the p15 and p16 were detected. The canonical p16INK4a product was detected normally in these extracts (Fig. S2). Although this result does not eliminate the possibility that a pALTINK4a/b-like transcript may occur under some conditions in mouse and human, it strongly suggests that this isoform is unique to the naked mole rat.
pALTINK4a/b Is Induced by High Cell Density and a Stress.
To determine the biological significance of pALTINK4a/b, we examined the expression of pALTINK4a/b, p15INK4b, and p16INK4a in naked mole rat cells under various conditions that lead to the induction of the INK4a/b locus. We used four lines of naked mole rat fibroblasts derived from four individual animals and analyzed expression using quantitative PCR (qPCR).
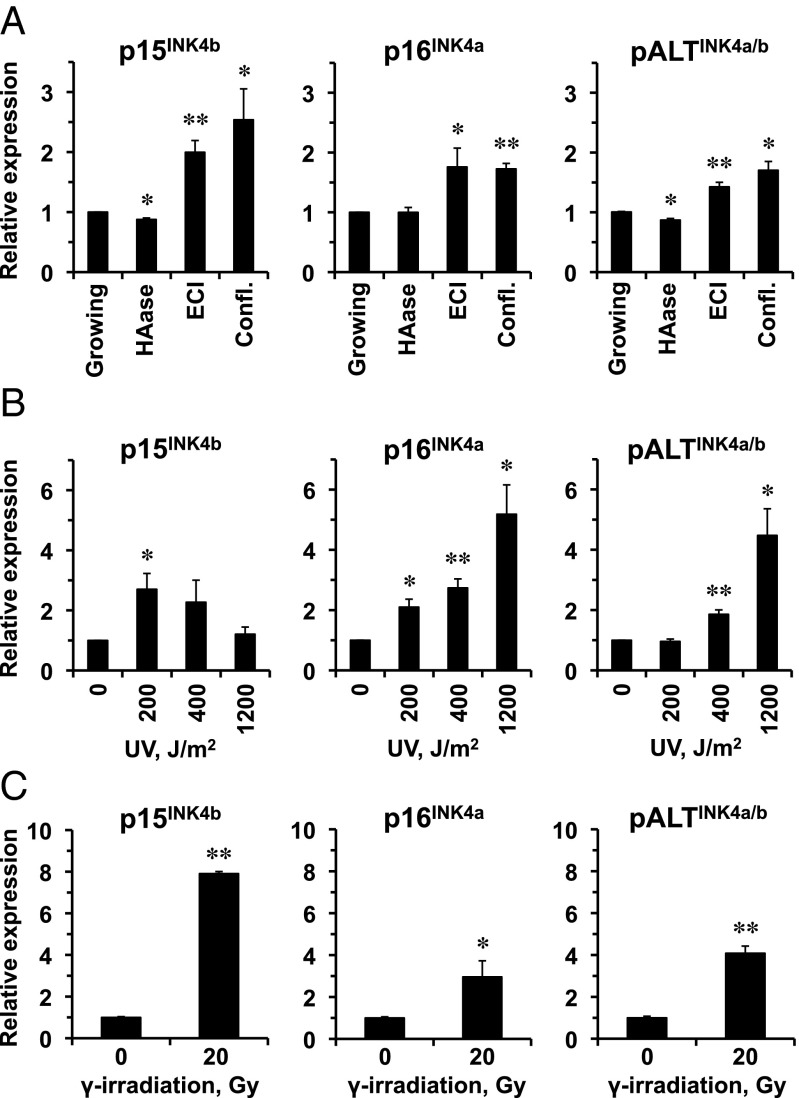
First, we examined the induction of p15INK4b, p16INK4a, and pALTINK4a/b under different growth conditions: logarithmically growing cells; logarithmically growing cells in the presence of an enzyme, hyaluroinidase (HAase), that breaks down hyaluronan, leading to more rapid cell growth; cells in ECI; and cells that entered confluence in the presence of HAase. We found that all three transcripts were up-regulated in cells growth-arrested by either confluence or ECI (Fig. 2A). Furthermore, in growing cells, HAase treatment significantly reduced p15INK4b and pALTINK4a/b expression. This result indicates that pALTINK4a/b responds to hyaluronan-induced ECI and cell density.
Fig. 2.
pALTINK4a/b expression is induced by ECI, confluence, and senescence. (A) Naked mole rat fibroblasts were passaged under four different conditions. “Growing” cells were passaged at low cell density to ensure continuous cell proliferation. “HAase” cells were passaged at low cell density in the presence of hyaluronidase. “ECI” cells were kept in the same plate without passaging for 10–14 d until they stopped proliferation and entered the ECI state. “Confl.” cells were cultured in the presence of hyaluronidase until they reached confluence at high cell density. RNA was extracted from the cells, and expression from INK4a/b locus was analyzed by qPCR. (B) Naked mole rat cells were treated with indicated doses of UV-C and analyzed for expression of INK4a/b products as described above. (C) Naked mole rat cells were treated with 20 Gy of γ-irradiation, kept for 30 d until they developed senescent morphology, and analyzed for expression of INK4a/b products as described above. Four lines of low-passage naked mole rat skin fibroblasts were used for each experiment. Error bars indicate SD; *P < 0.05; **P < 0.005; Student’s t test.
Next, we tested the induction of p15INK4b, p16INK4a, and pALTINK4a/b under stress. Ultraviolet-C (UV-C) exposure induces pyrimidine dimer formation in DNA and is known to induce INK4a expression in human melanocytes and keratinocytes (31). UV-C irradiation induced all three transcripts but with different kinetics. p15INK4b was induced by lower doses of UV but declined at the highest dose whereas p16INK4a and pALTINK4a/b showed similar patterns of induction that increased with the UV dose (Fig. 2B). Stress-induced premature senescence triggered by γ-irradiation also induced all of the three transcripts, with the highest induction observed for p15INK4b, followed by pALTINK4a/b and weaker induction observed for p16INK4a (Fig. 2C).
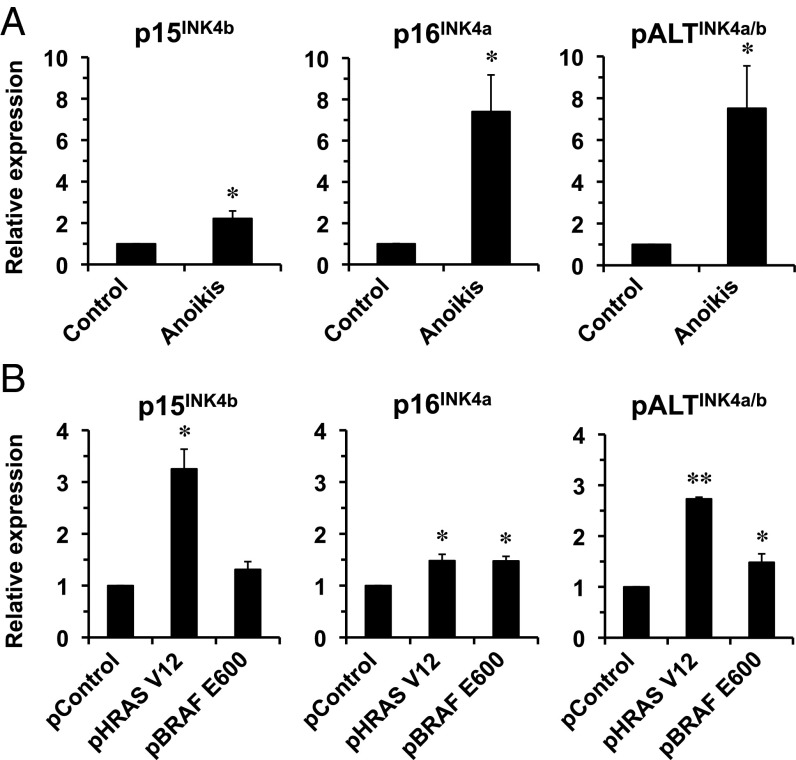
All of the three transcripts were induced to a similar extent by anoikis (Fig. 3A). We additionally tested whether oncogene overexpression known to activate p15INK4b and p16INK4a would also activate pALTINK4a/b. The cells were transfected with either a GFP plasmid or an RASV12 plasmid, or a BRAF V600E-expressing plasmid, and RNA was extracted 10 d later. RASV12 significantly activated p15INK4b, p16INK4a, and pALTINK4a/b whereas BRAF V600E induced p16INK4a and pALTINK4a/b (Fig. 3B). These results show that pALTINK4a/b is induced by oncogenic stimuli, suggesting that, similarly to p15INK4b and p16INK4a, it has a tumor-suppressor function.
Fig. 3.
pALTINK4a/b expression is induced by oncogenic stimuli. (A) Naked mole rat cells were subjected to anchorage-independent growth to trigger anoikis by placing them in plates precoated with 1% agarose for 48 h. RNA was extracted from the cells, and expression from INK4a/b locus was analyzed by qPCR. (B) Naked mole rat cells were transfected with plasmids encoding HRas V12 or BRAF E600 oncogenes or pControl plasmid encoding GFP. RNA was extracted 10 d after transfection, and expression from INK4a/b locus was analyzed by qPCR. Four lines of low-passage naked mole rat skin fibroblasts were used for each experiment. Error bars indicate SD; *P < 0.05; **P < 0.005; Student’s t test.
pALTINK4a/b Is Efficient at Inducing Cell-Cycle Arrest.
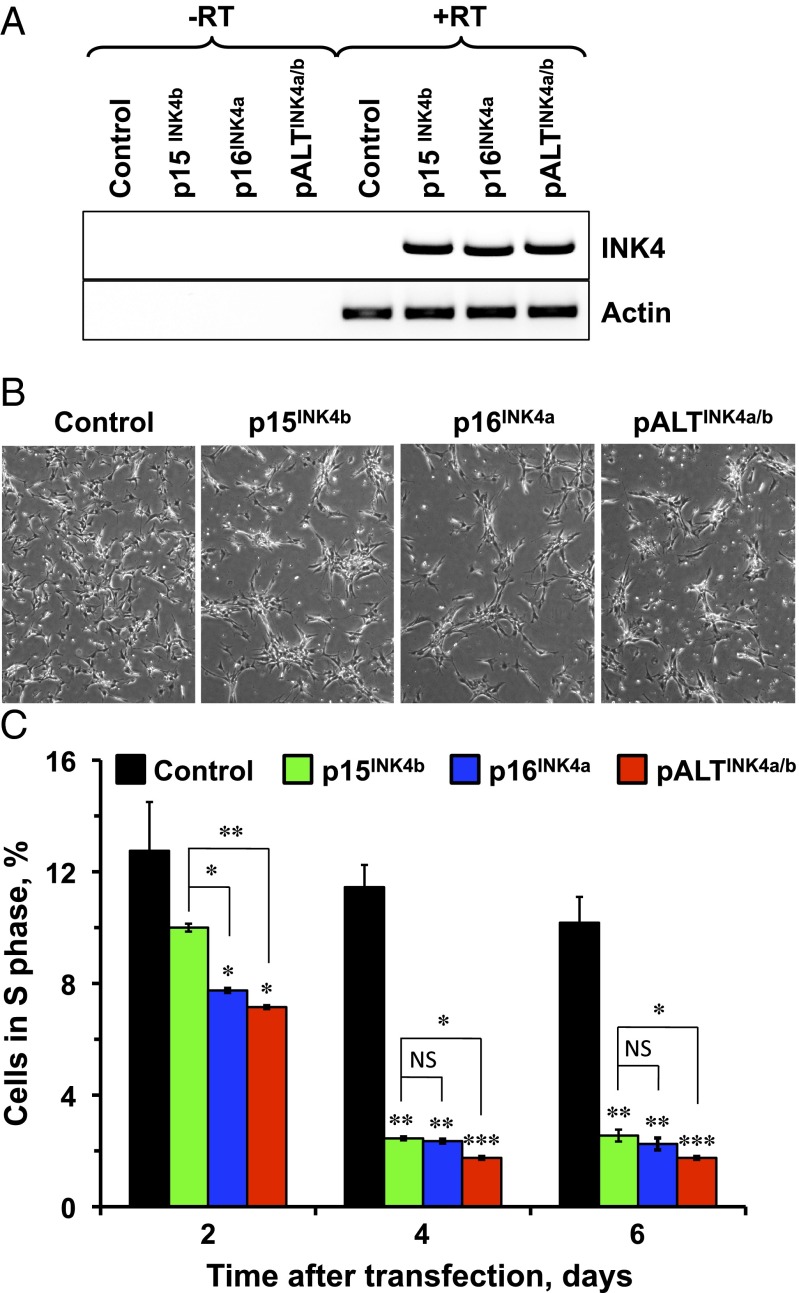
We next set out to test whether pALTINK4a/b can function to induce cell-cycle arrest. We cloned p15INK4b, p16INK4a, and pALTINK4a/b cDNAs into a pCMV6 expression vector and transiently transfected them into naked mole rat skin fibroblasts. Similar levels of expression were confirmed by semiquantitative PCR (Fig. 4A). We found that, compared with mock or GFP transfection, cells overexpressing p15INK4b, p16INK4a, or pALTINK4a/b had substantially lower rates of growth (Fig. 4B). Remarkably, pALTINK4a/b protein was significantly more capable of halting cell cycle than p15INK4b, p16INK4a (Fig. 4C). We speculate that this enhanced ability of pALTINK4a/b to mediate cell-cycle arrest may contribute to stricter cell-cycle control and ultimately increased cancer resistance in the naked mole rat.
Fig. 4.
pALTINK4a/b overexpression triggers cell-cycle arrest. Naked mole rat cells were transfected with constructs expressing HPRT (control), p15INK4b, p16INK4a, or pALTINK4a/b under the CMV promoter. (A) Expression of the INK4a/b products was verified with RT-PCR. (B) Microphotograph showing cells transfected with p15INK4b, p16INK4a, or pALTINK4a/b 6 d after transfection. Control cells continued to proliferate whereas cells transfected with INK4a/b products entered cell-cycle arrest. (C) Cell-cycle arrest induced by expression of p15INK4b, p16INK4a, or pALTINK4a/b was quantified at indicated times after transfection by staining cells with propidium iodide and quantifying the number of cells in S-phase by FACS analysis. The experiments were repeated three times, and error bars show SD; statistical significance is indicated by asterisk. Three lines of low-passage naked mole rat skin fibroblasts were used for each experiment. *P < 0.05; **P < 0.005; ***P < 0.0005, Student’s t test.
pALTINK4a/b Protects Cells from Radiation-Induced Apoptosis.
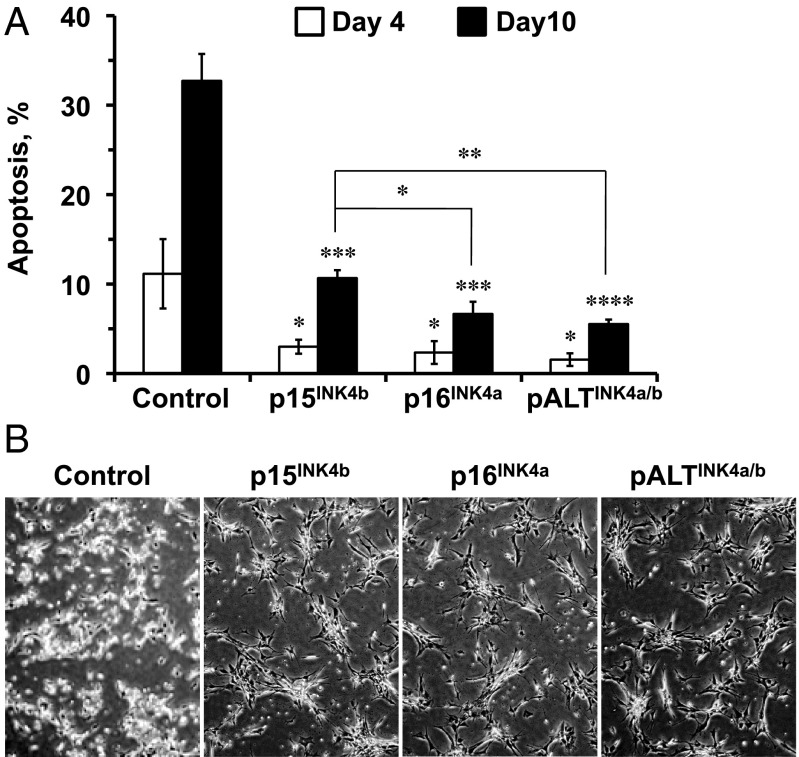
NSF2 mut fibroblasts have lost the expression of the INK4 locus (5) and are defective in cell-cycle arrest. Unlike primary fibroblasts that undergo stress-induced senescence, NSF2 mut cells undergo apoptosis in response to γ-irradiation. To test whether pALTINK4a/b can rescue the apoptotic response in NSF2 mut cells, the cells were transfected with plasmids encoding NMR INK4 proteins or a control plasmid expressing the HPRT gene. Two days after transfection, cells were subjected to 20 Gy γ-irradiation. Four or 10 d after irradiation, floating and adherent cells were harvested and analyzed by Annexin-V staining and flow cytometry. Cells transfected with the control plasmid underwent massive apoptosis, which was significantly reduced by expression of the INK4 proteins (Fig. 5). Interestingly, p16INK4a and pALTINK4a/b were significantly more effective at reducing apoptotic cell death of NSF2 mut cells (Fig. 5). There was a trend toward even lower apoptotic response in pALTINK4a/b-transfected cells compared with p16INK4a-transfected cells. This result suggests that p16INK4a and pALTINK4a/b are more efficient in inducing cell-cycle arrest and/or senescence than p15INK4b.
Fig. 5.
pALTINK4a/b overexpression attenuates γ-irradiation–induced apoptosis in NSF2 mut cells. NSF2 mut cells were transfected with plasmids encoding HPRT (control), p15INK4b, p16INK4a, or pALTINK4a/b under the CMV promoter. Two days after transfection, cells were subjected to 20 Gy γ-irradiation. (A) At four and 10 d after γ-irradiation, the cells were harvested, and the percent of apoptotic cells was analyzed by Annexin-V staining and flow cytometry. (B) Cells were photographed 10 d after γ-irradiation. Control plate shows many floating apoptotic cells whereas plates transfected with INK4a/b products show lower numbers of apoptotic bodies and many growth-arrested cells. The experiments were repeated three times, and error bars show SD; statistical significance is indicated by asterisk. *P < 0.05; **P < 0.005; ***P < 0.0005; ****P < 0.00005, Student’s t test.
Discussion
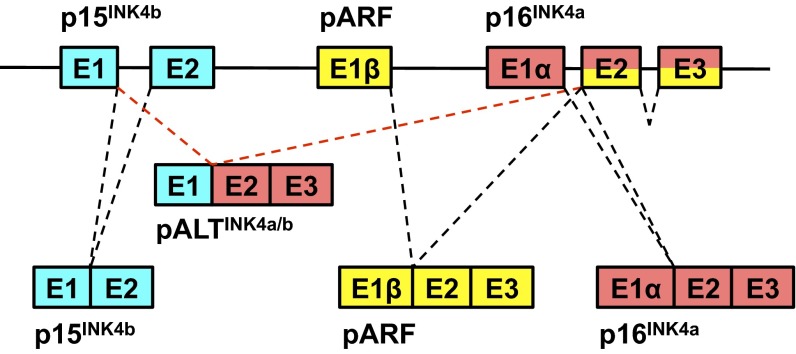
The dynamic regulation of the INK4a/b locus is of tremendous importance to tumorigenesis (32–36), stem-cell maintenance (9, 13), cellular senescence (37), and systemic aging (19, 38). Here, we have demonstrated that naked mole rat cells express a novel INK4 isoform, pALTINK4a/b, composed of the first exon of p15INK4b and the second and third exons of p16INK4a (Fig. 6). This novel isoform was detected in multiple naked mole rat tissues and in cultured cells by RT-PCR, as well as by RNA-seq. A concern that arose during this work was the possibility that the alternative splicing we had observed was an aberrant intermolecular “template switching” product of the reverse transcriptase reaction (39). We do not believe that template switching is a likely explanation for the following five independent reasons: artifacts of this nature do not often occur at canonical splice junctions whereas the isoform we observed was reproducibly found to occur between the canonical E1 and E2 junction; we could replicate our results on multiple cell lines and even extracts from tissues from live animals; we found this product independently using different primer pairs; a similar product could not be found in human or mouse cells even when the locus was strongly induced; and we could not find the ALT transcript when NSF2 mut cells were simultaneously transfected with p15INK4b and p16INK4a overexpression plasmids.
Fig. 6.
Structure of the naked mole rat INK4a/b locus. The diagram shows the genomic locus and the canonical p15INK4b, p16INK4a, and pARF transcripts, as well as the pALTINK4a/b transcript unique to the naked mole rat.
We previously showed that ECI in naked mole rat cells is associated with the induction of p16INK4a (5). Our current work shows that the entire INK4a/b locus, including the pALTINK4a/b isoform, undergoes up-regulation in response to cell crowding and buildup of hyaluronan. We show that pALTINK4a/b is induced by other stresses, such as UV- and γ-irradiation, expression of oncogenes, and anoikis. Although both p15INK4b and p16INK4a function as repressors of the cell cycle, their relative importance in different cancers has not been fully resolved. p15INK4b has been proposed to serve as a backup to p16INK4a, and mice in which both genes are mutated show higher cancer susceptibility than animals missing p16INK4a alone (40). However, only p16INK4a, but not p15INK4b, expression has been reported to be induced during the aging process (41, 42). p15INK4b and p16INK4a are induced to a different extent by partially overlapping sets of stimuli (7, 43), which likely contributes to the complex regulation of the cellular stress response. The pattern of pALTINK4a/b expression we observed in naked mole rat fibroblasts was intermediate between p15INK4b and p16INK4a, which may be explained by pALTINK4a/b and p15INK4b sharing the same promoter and the 5′ UTR and to regulatory elements located in the 3′ UTR shared with p16INK4a. Furthermore, we demonstrate that the novel isoform pALTINK4a/b is a biologically active repressor of the cell cycle. Remarkably, pALTINK4a/b was more efficient at arresting the cell cycle upon overexpression than either p15INK4b or p16INK4a, and more efficient at mediating cell-cycle arrest after γ-irradiation than p15INK4b. p16INK4a protein was found to be more stable than p15INK4b (the C-terminal segment is responsible for differential stability of INK4 proteins) (21, 22). Therefore, we hypothesize that pALTINK4a/b has similar stability to p16INK4a, combined with the expression profile intermediate between p15INK4b and p16INK4a. The presence of an additional product of the INK4a/b locus, with higher capacity to induce cell-cycle arrest, may confer an added protection to naked mole rat cells, contributing to cancer resistance of this species. It was recently shown that contact inhibition represses mTOR and thus suppresses the senescence program (44). Thus, it is tempting to speculate that ECI in naked mole rats mediated by the unusual structure of the INK4a/b locus may contribute to naked mole rat longevity, in addition to conferring cancer resistance.
Furthermore, the balance between tumor suppression and proliferation is important for longevity. For example, overly zealous tumor suppression, such as that observed in mice with an overactive p53 gene (45, 46), may limit stem-cell reserves and lead to premature aging; and very loose tumor suppression would also shorten lifespan by increasing cancer incidence. We hypothesize that an additional cyclin-dependent kinase inhibitor may provide an added layer of redundancy, allowing naked mole rats to further fine-tune their cell-cycle checkpoints for more optimal balance between growth and tumor suppression necessary to achieve longevity.
Experimental Procedures
Detailed experimental procedures including plasmid construction, qRT-PCR, cell culture, cell-growth analysis, analysis of INK4 expression, cell-cycle analysis, apoptosis analysis, and RNA-Seq are provided in SI Experimental Procedures.
Supplementary Material
Acknowledgments
This work was supported by grants from the US National Institutes of Health (to Z.D.Z., J.V., V.N.G., V.G., and A.S.), the Life Extension Foundation (to V.G. and A.S.), the Glenn Foundation for Medical Research (to V.G. and J.V.), and the Ellison Medical Foundation (to V.G. and A.S.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1418203112/-/DCSupplemental.
References
- 1.Edrey YH, Hanes M, Pinto M, Mele J, Buffenstein R. Successful aging and sustained good health in the naked mole rat: A long-lived mammalian model for biogerontology and biomedical research. ILAR J. 2011;52(1):41–53. doi: 10.1093/ilar.52.1.41. [DOI] [PubMed] [Google Scholar]
- 2.Buffenstein R. The naked mole-rat: A new long-living model for human aging research. J Gerontol A Biol Sci Med Sci. 2005;60(11):1369–1377. doi: 10.1093/gerona/60.11.1369. [DOI] [PubMed] [Google Scholar]
- 3.Delaney MA, Nagy L, Kinsel MJ, Treuting PM. Spontaneous histologic lesions of the adult naked mole rat (Heterocephalus glaber): A retrospective survey of lesions in a zoo population. Vet Pathol. 2013;50(4):607–621. doi: 10.1177/0300985812471543. [DOI] [PubMed] [Google Scholar]
- 4.Buffenstein R. Negligible senescence in the longest living rodent, the naked mole-rat: Insights from a successfully aging species. J Comp Physiol B. 2008;178(4):439–445. doi: 10.1007/s00360-007-0237-5. [DOI] [PubMed] [Google Scholar]
- 5.Seluanov A, et al. Hypersensitivity to contact inhibition provides a clue to cancer resistance of naked mole-rat. Proc Natl Acad Sci USA. 2009;106(46):19352–19357. doi: 10.1073/pnas.0905252106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tian X, et al. High-molecular-mass hyaluronan mediates the cancer resistance of the naked mole rat. Nature. 2013;499(7458):346–349. doi: 10.1038/nature12234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim WY, Sharpless NE. The regulation of INK4/ARF in cancer and aging. Cell. 2006;127(2):265–275. doi: 10.1016/j.cell.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 8.Serrano M, et al. Role of the INK4a locus in tumor suppression and cell mortality. Cell. 1996;85(1):27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- 9.Sherr CJ. Ink4-Arf locus in cancer and aging. Wiley Interdiscip Rev Dev Biol. 2012;1(5):731–741. doi: 10.1002/wdev.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Serrano M, Hannon GJ, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366(6456):704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- 11.Dai CY, Enders GH. p16 INK4a can initiate an autonomous senescence program. Oncogene. 2000;19(13):1613–1622. doi: 10.1038/sj.onc.1203438. [DOI] [PubMed] [Google Scholar]
- 12.Brookes S, Rowe J, Gutierrez Del Arroyo A, Bond J, Peters G. Contribution of p16(INK4a) to replicative senescence of human fibroblasts. Exp Cell Res. 2004;298(2):549–559. doi: 10.1016/j.yexcr.2004.04.035. [DOI] [PubMed] [Google Scholar]
- 13.Molofsky AV, et al. Increasing p16INK4a expression decreases forebrain progenitors and neurogenesis during ageing. Nature. 2006;443(7110):448–452. doi: 10.1038/nature05091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H, et al. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460(7259):1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tacutu R, Budovsky A, Yanai H, Fraifeld VE. Molecular links between cellular senescence, longevity and age-related diseases: A systems biology perspective. Aging (Albany, NY Online) 2011;3(12):1178–1191. doi: 10.18632/aging.100413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ovadya Y, Krizhanovsky V. Senescent cells: SASPected drivers of age-related pathologies. Biogerontology. 2014;15(6):627–642. doi: 10.1007/s10522-014-9529-9. [DOI] [PubMed] [Google Scholar]
- 17.Liu Y, et al. Expression of p16(INK4a) in peripheral blood T-cells is a biomarker of human aging. Aging Cell. 2009;8(4):439–448. doi: 10.1111/j.1474-9726.2009.00489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Y, et al. INK4/ARF transcript expression is associated with chromosome 9p21 variants linked to atherosclerosis. PLoS ONE. 2009;4(4):e5027. doi: 10.1371/journal.pone.0005027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burd CE, et al. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 2010;6(12):e1001233. doi: 10.1371/journal.pgen.1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuo CL, et al. Cdkn2a is an atherosclerosis modifier locus that regulates monocyte/macrophage proliferation. Arterioscler Thromb Vasc Biol. 2011;31(11):2483–2492. doi: 10.1161/ATVBAHA.111.234492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuan C, Selby TL, Li J, Byeon IJ, Tsai MD. Tumor suppressor INK4: Refinement of p16INK4A structure and determination of p15INK4B structure by comparative modeling and NMR data. Protein Sci. 2000;9(6):1120–1128. doi: 10.1110/ps.9.6.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuan C, Li J, Selby TL, Byeon IJ, Tsai MD. Tumor suppressor INK4: Comparisons of conformational properties between p16(INK4A) and p18(INK4C) J Mol Biol. 1999;294(1):201–211. doi: 10.1006/jmbi.1999.3231. [DOI] [PubMed] [Google Scholar]
- 23.Kamijo T, et al. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997;91(5):649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, Xiong Y, Yarbrough WG. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell. 1998;92(6):725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- 25.Weber JD, et al. p53-independent functions of the p19(ARF) tumor suppressor. Genes Dev. 2000;14(18):2358–2365. doi: 10.1101/gad.827300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szklarczyk R, Heringa J, Pond SK, Nekrutenko A. Rapid asymmetric evolution of a dual-coding tumor suppressor INK4a/ARF locus contradicts its function. Proc Natl Acad Sci USA. 2007;104(31):12807–12812. doi: 10.1073/pnas.0703238104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gilley J, Fried M. One INK4 gene and no ARF at the Fugu equivalent of the human INK4A/ARF/INK4B tumour suppressor locus. Oncogene. 2001;20(50):7447–7452. doi: 10.1038/sj.onc.1204933. [DOI] [PubMed] [Google Scholar]
- 28.Quelle DE, Zindy F, Ashmun RA, Sherr CJ. Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell. 1995;83(6):993–1000. doi: 10.1016/0092-8674(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 29.Kim SH, Mitchell M, Fujii H, Llanos S, Peters G. Absence of p16INK4a and truncation of ARF tumor suppressors in chickens. Proc Natl Acad Sci USA. 2003;100(1):211–216. doi: 10.1073/pnas.0135557100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim EB, et al. Genome sequencing reveals insights into physiology and longevity of the naked mole rat. Nature. 2011;479(7372):223–227. doi: 10.1038/nature10533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pavey S, Conroy S, Russell T, Gabrielli B. Ultraviolet radiation induces p16CDKN2A expression in human skin. Cancer Res. 1999;59(17):4185–4189. [PubMed] [Google Scholar]
- 32.Krimpenfort P, Quon KC, Mooi WJ, Loonstra A, Berns A. Loss of p16Ink4a confers susceptibility to metastatic melanoma in mice. Nature. 2001;413(6851):83–86. doi: 10.1038/35092584. [DOI] [PubMed] [Google Scholar]
- 33.Schneider G, Schmid RM. Genetic alterations in pancreatic carcinoma. Mol Cancer. 2003;2:15. doi: 10.1186/1476-4598-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Esteller M, et al. K-ras and p16 aberrations confer poor prognosis in human colorectal cancer. J Clin Oncol. 2001;19(2):299–304. doi: 10.1200/JCO.2001.19.2.299. [DOI] [PubMed] [Google Scholar]
- 35.Matheu A, et al. Anti-aging activity of the Ink4/Arf locus. Aging Cell. 2009;8(2):152–161. doi: 10.1111/j.1474-9726.2009.00458.x. [DOI] [PubMed] [Google Scholar]
- 36.Matheu A, et al. Increased gene dosage of Ink4a/Arf results in cancer resistance and normal aging. Genes Dev. 2004;18(22):2736–2746. doi: 10.1101/gad.310304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baker DJ, et al. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature. 2011;479(7372):232–236. doi: 10.1038/nature10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jeck WR, Siebold AP, Sharpless NE. Review: A meta-analysis of GWAS and age-associated diseases. Aging Cell. 2012;11(5):727–731. doi: 10.1111/j.1474-9726.2012.00871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cocquet J, Chong A, Zhang G, Veitia RA. Reverse transcriptase template switching and false alternative transcripts. Genomics. 2006;88(1):127–131. doi: 10.1016/j.ygeno.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 40.Krimpenfort P, et al. p15Ink4b is a critical tumour suppressor in the absence of p16Ink4a. Nature. 2007;448(7156):943–946. doi: 10.1038/nature06084. [DOI] [PubMed] [Google Scholar]
- 41.Zindy F, Quelle DE, Roussel MF, Sherr CJ. Expression of the p16INK4a tumor suppressor versus other INK4 family members during mouse development and aging. Oncogene. 1997;15(2):203–211. doi: 10.1038/sj.onc.1201178. [DOI] [PubMed] [Google Scholar]
- 42.Krishnamurthy J, et al. Ink4a/Arf expression is a biomarker of aging. J Clin Invest. 2004;114(9):1299–1307. doi: 10.1172/JCI22475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thullberg M, et al. Distinct versus redundant properties among members of the INK4 family of cyclin-dependent kinase inhibitors. FEBS Lett. 2000;470(2):161–166. doi: 10.1016/s0014-5793(00)01307-7. [DOI] [PubMed] [Google Scholar]
- 44.Leontieva OV, Demidenko ZN, Blagosklonny MV. Contact inhibition and high cell density deactivate the mammalian target of rapamycin pathway, thus suppressing the senescence program. Proc Natl Acad Sci USA. 2014;111(24):8832–8837. doi: 10.1073/pnas.1405723111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tyner SD, et al. p53 mutant mice that display early ageing-associated phenotypes. Nature. 2002;415(6867):45–53. doi: 10.1038/415045a. [DOI] [PubMed] [Google Scholar]
- 46.Maier B, et al. Modulation of mammalian life span by the short isoform of p53. Genes Dev. 2004;18(3):306–319. doi: 10.1101/gad.1162404. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.