Significance
Thaumarchaeota are among the most abundant microbial cells in the ocean, but to date, complete genome sequences for marine Thaumarchaeota are lacking. Here, we report the 1.23-Mbp genome of the pelagic ammonia-oxidizing thaumarchaeon “Candidatus Nitrosopelagicus brevis” str. CN25. We present the first proteomic data, to our knowledge, from this phylum, which show a high proportion of proteins translated in oligotrophic conditions. Metagenomic fragment recruitment using data from the open ocean indicate the ubiquitous presence of Ca. N. brevis-like sequences in the surface ocean and suggest Ca. N. brevis as a model system for understanding the ecology and evolution of pelagic marine Thaumarchaeota.
Keywords: nitrification, marine metagenomics, genome streamlining, archaea
Abstract
Thaumarchaeota are among the most abundant microbial cells in the ocean, but difficulty in cultivating marine Thaumarchaeota has hindered investigation into the physiological and evolutionary basis of their success. We report here a closed genome assembled from a highly enriched culture of the ammonia-oxidizing pelagic thaumarchaeon CN25, originating from the open ocean. The CN25 genome exhibits strong evidence of genome streamlining, including a 1.23-Mbp genome, a high coding density, and a low number of paralogous genes. Proteomic analysis recovered nearly 70% of the predicted proteins encoded by the genome, demonstrating that a high fraction of the genome is translated. In contrast to other minimal marine microbes that acquire, rather than synthesize, cofactors, CN25 encodes and expresses near-complete biosynthetic pathways for multiple vitamins. Metagenomic fragment recruitment indicated the presence of DNA sequences >90% identical to the CN25 genome throughout the oligotrophic ocean. We propose the provisional name “Candidatus Nitrosopelagicus brevis” str. CN25 for this minimalist marine thaumarchaeon and suggest it as a potential model system for understanding archaeal adaptation to the open ocean.
Planktonic archaea are widespread in the marine environment. Below the photic zone, archaea can constitute greater than 30% of total bacterioplankton (1), making them among the most abundant cells in the ocean. The majority of pelagic archaea belong to the recently described phylum Thaumarchaeota (2, 3), also known as the Marine Group I archaea (4). In addition to representing large fractions of marine metagenomic datasets (5), metatranscriptomic data suggest that thaumarchaeal cells are metabolically active, with thaumarchaeal transcripts ranking as the most abundant in diverse marine environments (6–8). The metabolic activity of marine Thaumarchaeota has important implications for global biogeochemical cycles via their role in nitrogen remineralization, carbon fixation (9), and production of the greenhouse gas nitrous oxide (N2O) (10).
At present there are six pure cultures of Thaumarchaeota: one from a marine aquarium [Nitrosopumilus maritimus SCM1 (11, 12)], two from an estuary in the northeast Pacific [PS0 and HCA1 (13)], and three from soil [Nitrosphaera viennensis (14) and Nitrosotalea devanaterra strains Nd1 and Nd2 (15)]. Of these isolates, N. maritimus, N. viennensis, and N. devanaterra are able to grow as chemolithoautotrophic ammonia oxidizers. Beyond these organisms, much of our knowledge of the genomic inventory (16–18), physiology, and biogeochemical activity of Thaumarchaeota has come from the characterization of enriched mixed cultures (19, 20) or uncultivated single cells (21, 22). Common genomic features in all sequenced representatives include a modified 3-hydroxypropioinate/4-hydroxybutryrate pathway for carbon fixation (23), an electron transport chain enriched in copper-centered metalloproteins, and lack of an identifiable homolog to hydroxylamine oxidoreductase (18, 24), an Fe-rich decaheme protein that catalyzes the second step of ammonia oxidation in all ammonia-oxidizing bacteria (25).
Given the tropical aquarium and estuarine origins of existing marine isolates, the extent to which their physiology and genomic features are representative of Thaumarchaeota in the open ocean is uncertain. In terms of physiology, N. maritimus grows chemolithoautotrophically, with ammonia as its sole energy source and bicarbonate as its sole carbon source. However, mixotrophy has been proposed for both N. viennensis and the marine isolates PS0 and HCA1 on the basis of growth stimulation when organic acids are added to the media (13, 14). In terms of genome content, metagenomic recruitment to N. maritimus is poor relative to that of single-cell genomes obtained from the open ocean (21).
Here, we present the closed genome of a marine ammonia-oxidizing Thaumarchaeota assembled from a low-diversity metagenome of an enrichment culture originating from the open ocean and previously described as CN25 (26). We mapped peptides collected from early stationary phase cells to translations of the CN25 genome’s predicted ORFs to produce the first global proteome, to our knowledge, from a marine thaumarchaeon. Finally, we used the genome to probe existing marine metagenomic and metatranscriptomic datasets to understand the relative distribution of CN25 and N. maritimus-like genomes in the ocean.
Results and Discussion
Cultivation, Genome Sequencing, and Global Proteome.
Previous fluorescent in situ hybridization characterization of the CN25 enrichment culture indicated that in late exponential phase, 90–95% of the cells are archaeal (26), and scanning electron microscopy shows the culture is dominated by rod-shaped cells with a diameter of 0.17–0.26 µm (mean 0.15 ± 0.02 µm; n = 50 cells) and length of 0.6–1.0 µm (0.78 ± 0.25 µm; SI Appendix, Fig. S1). A growth temperature optimum of ∼22 °C (SI Appendix, Fig. S2) suggests physiological adaptation to subtropical surface ocean temperatures compared with a temperature optimum of 30 °C for N. maritimus (12).
Consistent with earlier fluorescent in situ hybridization data, 93.3% of the 49.6 million Illumina HiSeq reads from this low-diversity metagenome were less than 45% GC (guanine-cytosine) content, with the remaining reads falling into two low-coverage bins of ∼50% and 65% GC content. A phylogenetic analysis indicated the archaeal reads were found in the low GC cluster. Assembly (via the Celera Assembler; wgs-assembler.sourceforge.net) of the low GC content bin resulted in five contigs at 40× coverage. Manual inspection of the sequence data, followed by PCR amplification and direct Sanger sequencing, resolved the genome into a single chromosome with a GC content of 33% (SI Appendix, Table S1 and Fig. S3).
At 1.23 Mbp, the closed CN25 genome is one of the smallest genomes of any free-living cell (Fig. 1, Table 1, and SI Appendix, Fig. S4). It encodes for 1,445 predicted protein-coding genes, one rRNA operon, and 42 tRNA genes. No extrachromosomal elements were identified. We propose the provisional name “Candidatus Nitrosopelagicus brevis” str. CN25 (Ca. N. brevis). The genus name refers to the organism’s water column habitat and its ability to oxidize ammonia to nitrite. The species name refers both to the organism’s affiliation with a clade of shallow water Thaumarchaeota (26) and its small genome.
Fig. 1.
The 1.23-Mbp genome and proteome of Ca. N. brevis str. CN25. The outermost ring is the position along the genome in thousands of nucleotide base pairs and annotations of the 15 most abundant proteins in the proteome, plus ammonia monooxygenase subunit a (amoA). The second ring (histogram) is the relative abundance of protein spectral counts detected in a global proteome. The third and fourth rings (blue and cyan) indicate predicted ORFs on the plus and minus strands, respectively. The fifth ring (red) indicates the location of putative genomic island regions (IR). The sixth or innermost ring (green) is GC anomaly based on a 2,000-bp moving average. Key to protein annotations: 1. conserved domain protein (T478_1299); 2. ATP synthase (T478_1372); 3. conserved domain protein (T478_1300); 4. translation elongation factor EF-1 (T478_0861); 5. AAA family ATPase (T478_0115); 6. RNA polymerase subunit A (rpoA, T478_0275); 7. RNA polymerase subunit B (rpoB, T478_0274); 8. alcohol dehydrogenase (T478_1333); 9. putative glutamate dehydrogenase (T478_1059); 10. putative malate dehydrogenase (T478_0268); 11. conserved hypothetical protein (T478_0572); 12. oxidoreductase, short chain dehydrogenase (T478_0869); 13. ATP synthase alpha/beta chain T478_1371); 14. flavodoxin (T478_0486); 15. putative acetyl-CoA carboxylase (T478_1175). Relative abundance of all proteins identified in the global proteome is provided as an SI Appendix, Dataset S1.
Table 1.
Genome sizes and coding densities of select oligotrophic marine bacteria and previously sequenced Thaumarchaeota
| Characteristics | Oligotrophic pelagic marine bacteria | Thaumarchaeota | |||||
| Methylophilales sp. HTCC2181 (OM43) | Pelagibacter ubique HTCC1062 | Prochloroccocus marinus AS9601 | N. gargensis | Ca. N. limnia SFB1 | N. maritimus | Ca. N. brevis | |
| Size, Mbp | 1.304 | 1.309 | 1.670 | 2.834 | 1.743 | 1.645 | 1.232 |
| ORFs | 1,377 | 1,394 | 1,988 | 3,599 | 2,088 | 1,842 | 1,501 |
| Percentage coding | 95.0 | 96.1 | 91.2 | 81.5 | 84.8 | 90.8 | 94.6 |
| Percentage GC | 38 | 30 | 31 | 48 | 32 | 34 | 33 |
The translated ORFs, predicted from the assembled genome, were used as a reference to identify proteins in a global proteome of early stationary phase cells (SI Appendix and Fig. 1). The proteome recovered peptides mapping to 1,012 unique proteins, or roughly 70% of the total predicted proteins (SI Appendix, Dataset S1). Relative to previously investigated microbes, Ca. N. brevis translates a large fraction of its proteome under oligotrophic conditions (SI Appendix, Table S2).
Energy Metabolism.
The Ca. N. brevis genome encodes genes for all three subunits of ammonia monooxygenase (AMO) with the same order and orientation (amoACB; T478_0302, _0300, _0298) found in other marine Thaumarchaeota, and all three subunits were detected in the proteome (Fig. 1 and SI Appendix, Dataset S1). Although not among the top 15 most abundant proteins in terms of spectral counts, AmoB was highly abundant (top 5% of expressed proteins), as it is in the proteome of the ammonia-oxidizing bacterium Nitrosomonas europaea (27). The Ca. N. brevis genome also encodes for the 120-amino acid hypothetical protein previously termed AmoX [(28); T478_0301], located between amoA and amoC, and the proteome confirmed expression of this protein. As with all previously sequenced Thaumarchaeota, no hydroxylamine oxidoreductase homologs were identified. Five of the 15 most abundant proteins in the proteome were involved in energy production and conversion (Fig. 1 and SI Appendix, Dataset S1), and energy production proteins are abundant in the proteomes of other chemolithoautotrophic organisms (29); thus, highly abundant hypothetical proteins are promising candidates for additional proteins involved in energy generation.
Metalloenzyme-specific analyses conducted for Ca. N. brevis suggest that, similar to N. maritimus, there is a reliance on copper-containing electron transport proteins (SI Appendix, Dataset S2). The Ca. N. brevis genome encodes for 12 cupredoxin domain-containing proteins (Structural Classification of Proteins family 49550), which bind copper in a redox active fashion, compared with 27 proteins for N. maritimus. Many of the single-domain cupredoxins contain long N-terminal extensions lacking annotation, whereas two contain C-terminal PEFG sequences that likely target them to the cell membrane. Multicopper oxidases (Pfam07732) are not typically found in archaeal genomes outside the Thaumarchaeota and have been suggested as potential alternatives to the “missing” hydroxylamine oxidoreductase enzyme (24); Ca. N. brevis contains three multicopper oxidases, whereas N. maritimus contains six. Of the Ca. N. brevis multicopper oxidases, two were detected in the proteome (T478_0212, T478_1026), including the putative copper-containing nitrite (NO2−) reductase (nirK; T478_1026). nirK transcripts are abundant in some marine metatranscriptomes (7) and were abundant in the proteome (SI Appendix, Dataset S1).
Reductive N2O production from NO2− has been demonstrated in enrichment cultures of Ca. N. brevis (10) and in N. maritimus (30, 31), although it is unclear whether reductive N2O production originates from enzymatic or abiotic reactions. The Ca. N. brevis assembly encodes for two putative nitric oxide reductase accessory proteins (norQ, T478_0286, and norD, T478_0285), both of which were detected in the proteome. NorQ is essential for the activation of NorB, which catalyzes the reduction of NO to N2O in both nitrifying (32) and denitrifying (33) bacteria. However, no homologs of norB were identified in Ca. N. brevis or in any other thaumarchaeal genome. Although implicated in reductive N2O production, norB and norQ mutants of the bacterial nitrifier N. europaea still produce N2O but have a greatly diminished capability to degrade NO (32). Thus, the genomic data leave the mechanism of reductive N2O production in Ca. N. brevis unresolved.
Central Carbon Metabolism.
Candidate genes encoding for a partial 3-hydroxypropionate/4-hydroxybutyrate pathway were identified, suggesting the potential for carbon fixation in Ca. N. brevis (SI Appendix, Dataset S2). We identified proteins from all eleven enzymes, with a subunit of the acetyl-/propionyl-CoA carboxylase enzyme complex (T478_1175) among the most abundant proteins (Fig. 1), suggestive of active carbon fixation during growth. The thaumarchaeal 3-hydroxypropionate/4-hydroxybutyrate pathway was recently demonstrated to be the most efficient pathway for carbon fixation (23), which is likely an important adaptation for chemolithoautotrophic growth in the oligotrophic ocean.
Putative genes for a complete tricarboxylic acid cycle were also identified, and all were detected in the Ca. N. brevis proteome, with malate dehydrogenase among the most abundant proteins (Fig. 2). Glycolysis is apparently incomplete (genes encoding a pyruvate kinase and phosphofructokinase were absent), but a complete gluconeogenic pathway was identified (SI Appendix, Dataset S2). However, Ca. N. brevis may benefit from the presence of organic compounds. For example, putative transport proteins for the import of lipoproteins, glycerol, and glycine betaine were all identified in the genome, with several present in the proteome, suggestive of potential alternative substrate use. Similarly, the persistence of a small percentage (<10% of total cells) of putatively heterotrophic bacterial cells in the enrichment culture and reports of reliance on organic compounds in other marine Thaumarchaeota (13) leave open the potential that Ca. N. brevis may benefit from organic compounds produced by the bacteria or in the natural seawater medium for growth. We found, however, no effect of organic carbon addition on the growth rate or cell yield of Ca. N. brevis cultures in tests with 20 different organic compounds (SI Appendix, Table S3).
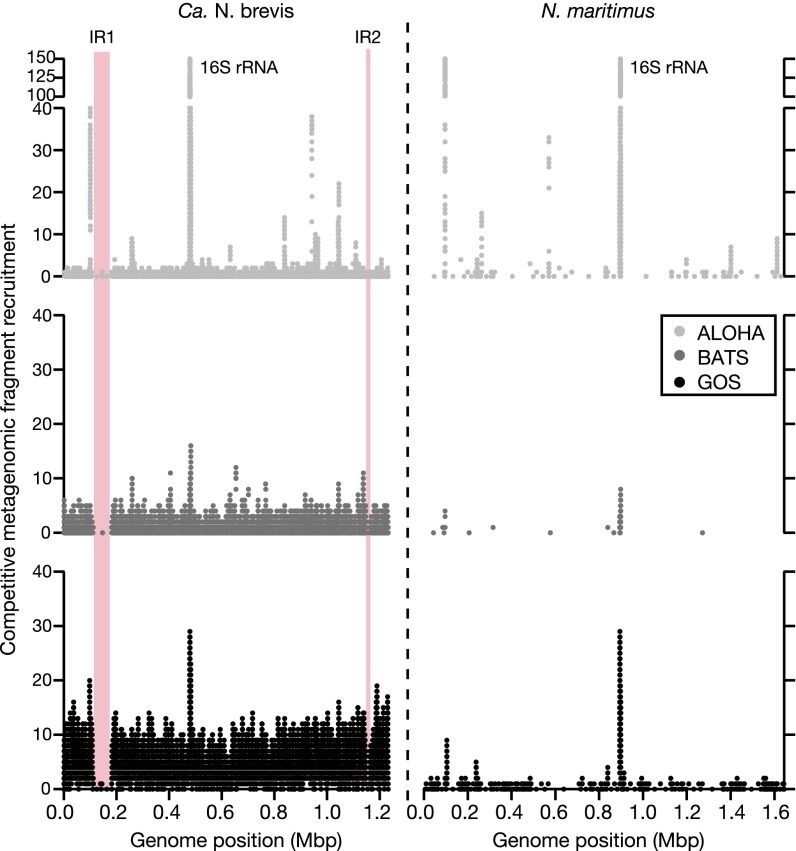
Fig. 2.
Sequences highly similar to Ca. N. brevis dominate marine metagenomes. Competitive metagenomic fragment recruitment between the Ca. N. brevis genome assembly (Left) and N. maritimus (Right) at >90% nucleotide identity in marine metagenomic datasets from the Hawai’i Ocean Time-series (ALOHA), Bermuda Atlantic Time-series Station (BATS), and the Global Ocean Sampling Expedition (GOS). Regions highlighted in red indicate genomic IR in Ca. N. brevis.
Vitamin and Amino Acid Biosynthesis.
Complete biosynthetic pathways for the B vitamin cofactors thiamin (B1), riboflavin (B2), pantothenate (B5), pyridoxine (B6), and biotin (B7) are present in the Ca. N. brevis genome (SI Appendix, Dataset S2). A near-complete pathway for cobalamin (B12) synthesis was also identified in the genome, missing only precorrin-6X reductase (cbiJ-cobK), which is also lacking in nearly all known cobalamin-producing archaea (34) except Methanococcus (35). Proteins within each of these pathways were detected in the proteome. Distributions of these vitamins in seawater have been suggested to explain the success of various phytoplankton lineages (36), although little is known about the source of them in seawater, particularly below the euphotic zone.
Consistent with the genetic capacity for B12 biosynthesis, Ca. N. brevis encodes for three major B12-requiring enzymes: methylmalonyl-CoA mutase (T478_0628), methionine synthase (T478_1032), and ribonucleoside reductase (T478_1341). The genome also encodes for the archaeal-specific cobalt chelatase (cbiX) and cobY-cobU from the oxygen independent B12 biosynthetic pathway, which does not require oxygen to produce the cobalt-binding corrin ring center of the vitamin (34). Because of its small genome size, Ca. N. brevis has a relatively large genetic investment in B12 synthesis, with 1.7% of the genome encoding B12-related genes compared with 0.7% in Salmonella (37). Our findings also support those of a recent metagenomic analysis showing the widespread distribution of thaumarchaeal B12 biosynthesis genes in the ocean (38). Six of the seven proteins for B1 biosynthesis and two of the three proteins in the B7 pathway were detected in the proteome. Vitamin B1 is required for several central carbon metabolism enzymes including transketolase (T478_1212, T478_1213) and acetolactate synthase (T478_0886, T478_0887), and vitamin B7 is a required coenzyme for the acetyl-/propionyl-CoA carboxylase enzyme complex (T478_1174, T478_1175, T478_1176). Other minimal genomes, such as Pelagibacter spp., lack the capability for complete B vitamin synthesis, uptake, and use (39, 40). The genomic and proteomic data presented here, together with the abundance of archaea in the mesopelagic (1), suggest Thaumarchaeota such as Ca. N. brevis are a potential source of multiple B vitamins required by microorganisms in the upper mesopelagic.
We identified complete or near-complete pathways for the synthesis of 18 amino acids, plus a near-complete pathway for methionine synthesis (SI Appendix, Dataset S2). We interpret apparent deficiencies in these pathways as gaps in our understanding of archaeal amino acid biosynthesis, rather than evidence of auxotrophy, as genes for all “missing” enzymes in the Ca. N. brevis genome are also absent in N. maritimus, which grows in minimal medium without added amino acids. Proteins in all amino acid biosynthesis pathways except asparagine were detected in the proteome. Although genes coding for known mechanisms of proline biosynthesis were not annotated, the absence of a canonical proline biosynthesis pathway was previously noted in other archaea and may be substituted by synthesis from l-ornithine (41). Again, the presence of several putative amino acid and oligopeptidetransporters lends support to the possibility that amino acids may be acquired exogenously, despite having genomic inventory for their biosynthesis.
Comparative Genomic Analyses Suggest Adaptations to the Surface Ocean.
Phylogenetic analysis of an alignment of concatenated ribosomal protein genes unambiguously associates Ca. N. brevis within the Thaumarchaeota (SI Appendix, Fig. S5), yet a comparative whole-genome analysis highlights the distinction between Ca. N. brevis and previously sequenced Thaumarchaeota. The average amino acid identity of aligned proteins between Ca. N. brevis and other thaumarchaeal genomes ranged from 34% (against Candidatus Nitrosphaera gargensis) to 75% (against N. maritimus) (SI Appendix, Table S4). Protein sequences from Ca. N. brevis and eight other thaumarchaeal genomes were clustered at a range of amino acid identities (SI Appendix, Fig. S6). Consistent with each new ammonia-oxidizing archaeal genome sequenced to date (18), Ca. N. brevis contains a large number of proteins that are either unique or highly divergent, relative to other thaumarchaea. Using a 50% amino acid identity threshold to define orthologs, the Ca. N. brevis genome contains 331 predicted proteins not present in any other thaumarchaeal predicted proteome (SI Appendix, Dataset S3).
We investigated the Ca. N. brevis proteins with <50% identity to other thaumarchaeal proteins as potentially adaptive to the pelagic environment from which it was enriched; specifically, the lower euphotic zone. UV radiation and reactive oxygen species are two potential physiological stresses present in sunlit waters. We identified two genes encoding putative deoxyribodipyrimidine photolyases (T478_0069 and T478_0078; (SI Appendix, Dataset S2), associated with DNA repair resulting from UV damage. Both of these proteins were detected in the dark-grown proteome, suggesting either that these proteins have an alternative function in Ca. N. brevis or that they are coregulated as part of a universal stress response, as they are in Escherichia coli (42). A unique putative alkyl hydroxy peroxidase (ahpC) associated with reactive oxygen and nitrogen stress response was also identified (T478_0940), although homologous sequences were also identified in several deep (4,000 m) ocean fosmids, suggesting this is not a surface ocean-specific gene. In addition to the two “unique” ahpC-like genes, five other genes encoding predicted proteins in the same family (Pfam00578) were identified in the Ca. N. brevis genome (SI Appendix, Dataset S2). Low trace metal concentrations in the surface ocean may also play a role in microbial adaptation to the oligotrophic surface ocean, including marine archaea (43). Consistent with this, several of the “unique” Ca. N. brevis genes encode putative metal transport proteins, including a ferrous iron transporter (T478_0963), a putative CorA-like Mg2+ or Co2+ transporter (T478_0228), and a Zn-binding protein (T478_0238).
The Ca. N. brevis genome is also distinguished by the lack of identifiable genes for several features reported in previously sequenced Thaumarchaeota. No genes encoding for flagellar synthesis or chemotaxis proteins were detected, suggesting a nonmotile lifestyle. Ca. N. brevis has no apparent capacity for biosynthesis of the osmolyte hydroxyectoine, as is present in the three sequenced Nitrosopumilus strains. The Ca. N. brevis genome does not encode for the Pst-type high-affinity phosphate transport system present in the N. maritimus genome, but it does encode for the transcriptional regulator PhoU (T478_0950) in a putative operon with a low-affinity phosphate transporter (Pit, T478_0951). We speculate that because subtropical North Pacific surface waters often contain residual phosphate, the genetic investment in a high-affinity phosphate transport system may not be necessary (44). Metabolism of methylphosphonic acid (MPn) by phosphate-starved microbes has recently been scrutinized as a possible explanation for the observed methane oversaturation in marine surface waters (45). N. maritimus was recently shown to synthesize MPn de novo, suggesting that planktonic marine archaea might be a natural source of MPn, and thus linked to marine methane dynamics (46). Surprisingly, the Ca. N. brevis genome does not encode for a complete MPn biosynthesis pathway. In particular, Ca. N. brevis does not encode for the key enzyme MpnS (46), suggesting it does not synthesize MPn. It remains to be seen whether other open ocean Thaumarchaeota also lack the capacity to synthesize MPn, but these findings show that MPn synthesis may not be universally conserved in planktonic Thaumarchaeota, and that changes in thaumarchaeal population structure may influence marine methane dynamics.
Evidence for Genome Streamlining in Marine Thaumarchaeota.
The genome streamlining hypothesis argues that species with large effective population sizes are under selective pressures that favor small genomes, reducing the material or energetic cost of cellular replication in nutrient-poor environments (47, 48). Streamlined genomes are found in diverse, uncultivated bacteria in the oligotrophic ocean (49, 50), and it has been suggested that evolution of the Archaea, in particular, has been dominated by reductive selection (51). As exemplified by Prochlorococcus (52), the uncultivated bacterial clade SAR86 (50), and Pelagibacter (39), reductive selection can result in a loss of metabolic versatility or nutritional dependencies (53), such as the loss of pathways for assimilation of oxidized forms of nitrogen or essential vitamin cofactors. The Ca. N. brevis genome has no apparent loss of cofactor or amino acid metabolism and the concomitant inclusion of a complete pathway for carbon fixation. The genome has the highest coding density of any Thaumarchaeota (94.6%; Table 1), although the coding density is lower than for streamlined bacterial genomes such as Pelagibacter (Table 1). We did not find evidence of selection for shorter proteins, as average protein length is not correlated with genome size within the Archaea, according to an analysis of all finished archaeal genomes in the Integrated Microbial Genomes (IMG) database (R2 < 0.01; n = 164).
Consistent with other streamlined genomes (39), the abundance of paralogous proteins is small, even when normalizing for genome size (SI Appendix, Table S5). In particular, the Ca. N. brevis genome contains a reduced number of genes involved in environmental sensing and regulation relative to other Thaumarchaeota. Transcriptional regulation in archaea is controlled by two families of basal transcription factors: transcription factor B (TFB) and TATA-binding proteins (54), with orthologous proteins present in eukaryotes. TFBs and TATA-binding proteins combine as TFB-TATA-binding protein pairs, with different regulons according to the pairing, allowing for a complex regulatory scheme with few proteins (54, 55). It has been hypothesized that organisms containing more TFBs may be better suited to changing environmental conditions (56). The N. maritimus genome has eight annotated TFBs, which is among the highest in the archaeal domain (56), suggesting a large network of potential regulatory complexes to respond to a changing environment. The Ca. N. brevis genome contains only four TFB, in contrast to eight to twelve for other sequenced Thaumarchaeota. Whether transcriptional regulation by factor swapping analogous to sigma factor switching in bacteria occurs within the Thaumarchaeota remains to be demonstrated (55).
Somewhat surprisingly for an oligotrophic microbe, there are fewer predicted transport proteins (77 predicted in IMG and 50 predicted using TransAAP; SI Appendix, Table S6 and SI Appendix, Dataset S2) compared with other aquatic Thaumarchaeota. This reduction in transport proteins is particularly manifest for ATP-binding cassette (ABC)-type transporters (there are 18 in the Ca. N. brevis genome vs. 31 in N. maritimus) and holds even when these estimates are normalized to genome size (14.6 vs. 18.9 ABC transporters per millions of base pairs genome). The identification and expression of two Amt-type ammonium transporters (T478_1378, T478_1350) gives further support for the hypothesis that ammonia-oxidizing archaea actively transport ammonium (NH4+) into the cell (13, 20) and is consistent with detection of these genes in environmental metatranscriptomes (8, 57).
Comparison with Marine Metagenomic and Metatranscriptomic Data.
We used competitive fragment recruitment (SI Appendix, Materials and Methods) to determine the relative recruitment to the Ca. N. brevis and N. maritimus genomes in more than 360 metagenomic and metatranscriptomic datasets from marine environments, including the Global Ocean Sampling (GOS) Expedition data (58) (SI Appendix, Dataset S4). At 90% nucleotide identity or higher within the GOS data, read recruitment to Ca. N. brevis was 30 times greater than to N. maritimus (Fig. 2), and Ca. N. brevis had higher recruitment in nearly twice as many samples (SI Appendix, Dataset S4). Two regions of the Ca. N. brevis genome were rarely observed in the metagenomic datasets, a characteristic associated with genomic islands, regions that are highly variable within a population of otherwise identical organisms (Figs. 1 and 2). Although N. maritimus also contains several genomic islands (5, 8), Ca. N. brevis’s island gene contents are distinct from those in N. maritimus. The first Ca. N. brevis island, associated with a negative deviation in GC content (Fig. 1), encodes for 75 predicted proteins, of which nearly all appear to be involved in cell surface modifications through glycosylation (SI Appendix, Table S7). These enzymes may act to modify the cell surface, changing the palatability to grazers (59) or reducing susceptibility to phage infection. The second, much smaller, island contains mostly genes with unknown function. Although nothing is known about thaumarchaeal phage or thaumarchaeal defenses against them, genomic islands in many marine microbes are dominated by genes involved in cell wall and polysaccharide biosynthesis and modification, as they are in Ca. N. brevis, suggesting an important role of phage in thaumarchaeal population dynamics (60). To this end, we did not identify lysogenic phage or phage integrases in the Ca. N. brevis genome. Further, we found no clustered regularly interspaced short palindromic repeats (CRISPR) or CRISPR-associated protein systems of phage defense, although a putative abortive infection phage resistance protein was identified (T478_0343).
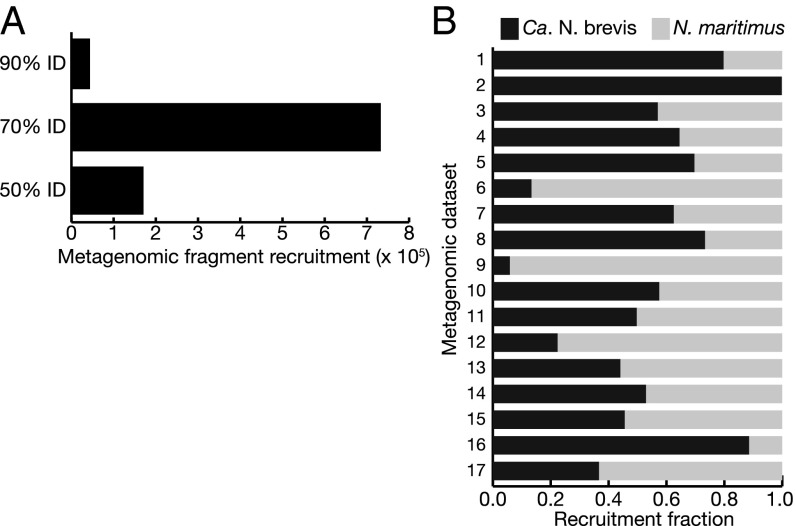
Two final observations from the competitive fragment recruitment analysis are the ubiquitous presence of thaumarchaeal genomes in the marine environment and the extent to which present cultivated strains do not represent this diversity. Even when recruitment to ribosomal RNA genes is excluded, genome fragments with >50% identity to either Ca. N. brevis or N. maritimus were found in all but one of the 366 datasets examined. Sequences >90% identity to Ca. N. brevis were abundant in the oligotrophic surface ocean (the GOS data), and the two oligotrophic time series datasets (Hawai'i Ocean Time-series and Bermuda Atlantic Time-series Station), implicating Ca. N. brevis as a globally abundant contributor to nitrification in ocean surface waters. The majority of the competitive fragment recruitment, however, was at nucleotide identities less than 90% (Fig. 3 and SI Appendix, Table S8). This indicates that a vast diversity of Thaumarchaeota distinct from Ca. N. brevis and N. maritimus exists in marine environments, and that further genomic and metabolic capability within this group remains to be explored.
Fig. 3.
(A) Combined metagenomic fragment recruitment to the Ca. N. brevis and N. maritimus genomes at three different nucleotide identity cutoffs. Bins are exclusive; that is, once a read is recruited at 90% identity, it is removed from the analysis and is thus not counted twice. Recruitment to ribosomal RNA genes has been excluded. (B) Detailed results of competitive fragment recruitment to Ca. N. brevis and N. maritimus in the 70–89% identity band from A indicating the fraction of total reads recruited to each genome. Metagenomic dataset numbers refer to the following accession numbers (preceeded by CAM_) in the CAMERA database: 1, PROJ_AntarcticaAquatic; 2, PROJ_BATS; 3, PROJ_Bacterioplankton; 4, PROJ_BotanyBay; 5, PROJ_HOT; 6, PROJ_LineIsland; 7, PROJ_MontereyBay; 8, PROJ_PML; 9, PROJ_PeruMarginSediment; 10, PROJ_SapeloIsland; 11, PROJ_SargassoSea; 12, PROJ_WesternChannelOMM; 13, P0000712; 14, P0000715; 15, P0000719; 16, P0000828; and 17, P0001028. Details of each metagenomic dataset are provided in SI Appendix, Table S8.
Multiple cultures and corresponding reference genomes from marine bacterial clades such as the cyanobacterium Prochlorococcus and heterotrophic bacterium Pelagibacter have provided important insights into the forces driving genome evolution and diversification in the oligotrophic ocean (61, 62). Hindered by a lack of relevant cultures, we know far less about the open ocean Thaumarchaeota, although they play similarly important roles in marine biogeochemical cycling. The genome and proteome presented here for Ca. N. brevis, originating from the largest contiguous biome on Earth (63), are the first step to uncovering similar ecological and evolutionary insights into a significant component of the microbial community in the expanding oligotrophic ocean (64).
Materials and Methods
Cultivation and Genome Sequencing.
The enrichment culture CN25 was grown under ammonia-oxidizing conditions in natural seawater-based oligotrophic north Pacific (ONP) medium (26) with 100 µM added NH4Cl and harvested onto 0.2-µm pore size filters. DNA was extracted using a modified phenol-chloroform extraction. Metagenomic sequencing was done on the Illumina HiSeq platform after paired-end library construction with a 2-Kbp insert size at the University of Maryland Institute for Genome Sciences Genomics Resource Center. Complete details can be found in the SI Appendix, Materials and Methods. The Ca. N. brevis CN25 genome has been deposited in the National Center for Biotechnology Information’s GenBank repository under accession number CP007026.
Scanning Electron Microscopy.
One hundred microliters CN25 culture was prefiltered through a 0.45-µm pore size syringe filter and then gently vacuum filtered onto 25 mm, 0.2 µm polycarbonate membrane filters, sequentially dehydrated, sputter coated, and prepared for observation with a Zeiss Supra 40VP scanning electron microscopy. Complete details can be found in the SI Appendix, Materials and Methods.
Genome Annotation and Metabolic Reconstruction.
Gene prediction and annotation were done using both the J. Craig Venter Institute’s microbial genome automated annotation pipeline and the Joint Genome Institute’s Integrated Microbial Genomes pipeline with subsequent manual investigation. Complete details can be found in the SI Appendix, Materials and Methods.
Global Proteome.
Early stationary phase CN25 cells grown under ammonia-oxidizing conditions in ONP medium were harvested by vacuum filtration onto single 0.2-µm pore size filters and frozen at −80 °C. Proteins were extracted using SDS extraction buffer, trypsin digested, purified, and concentrated. Proteins were identified by LC-MS of protein extracts using both one-dimensional and two-dimensional fractional chromatography. Mass spectral libraries were searched using SEQUEST HT (v 1.4). Complete details can be found in the SI Appendix, Materials and Methods.
Comparative Genomics and Phylogenetic Analysis.
Ortholog clustering was conducted using CD-Hit at the indicated alignment cutoffs with subsequent pairwise BLASTP alignments. Phylogenetic analysis was done using a concatenated alignment of 43 ribosomal proteins, and a tree was generated as described in the SI Appendix, Materials and Methods.
Metagenomic Fragment Recruitment.
Details of the competitive fragment recruitment analysis can be found in the SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank Jason Smith and Marguerite Blum for obtaining seawater for cultivation and John McCutcheon for helpful comments on a previous version of the manuscript. This work was funded by startup funds from the University of Maryland Center for Environmental Science (to A.E.S.); National Science Foundation awards OCE-1260006 (to A.E.S.), OCE-1259994 (to C.L.D.), OCE-1031271, and OCE-1233261 (to M.A.S.); the Life Technologies Foundation and Beyster Fund of the San Diego Foundation to the J. Craig Venter Institute; and support from the Gordon and Betty Moore Foundation under awards GBMF2724, GBMF3782, and GBMF3934 (to M.A.S.) and GBMF3307 (to A.E.S.). A.E.S. is an associate in the Integrated Microbial Biodiversity program of the Canadian Institute for Advanced Research. This is University of Maryland Center for Environmental Science contribution number 4949.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. CP007026).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1416223112/-/DCSupplemental.
References
- 1.Karner MB, DeLong EF, Karl DM. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature. 2001;409(6819):507–510. doi: 10.1038/35054051. [DOI] [PubMed] [Google Scholar]
- 2.Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P. Mesophilic Crenarchaeota: Proposal for a third archaeal phylum, the Thaumarchaeota. Nat Rev Microbiol. 2008;6(3):245–252. doi: 10.1038/nrmicro1852. [DOI] [PubMed] [Google Scholar]
- 3.Spang A, et al. Distinct gene set in two different lineages of ammonia-oxidizing archaea supports the phylum Thaumarchaeota. Trends Microbiol. 2010;18(8):331–340. doi: 10.1016/j.tim.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 4.DeLong EF. Archaea in coastal marine environments. Proc Natl Acad Sci USA. 1992;89(12):5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tully BJ, Nelson WC, Heidelberg JF. Metagenomic analysis of a complex marine planktonic thaumarchaeal community from the Gulf of Maine. Environ Microbiol. 2012;14(1):254–267. doi: 10.1111/j.1462-2920.2011.02628.x. [DOI] [PubMed] [Google Scholar]
- 6.Stewart FJ, Ulloa O, DeLong EF. Microbial metatranscriptomics in a permanent marine oxygen minimum zone. Environ Microbiol. 2012;14(1):23–40. doi: 10.1111/j.1462-2920.2010.02400.x. [DOI] [PubMed] [Google Scholar]
- 7.Hollibaugh JT, Gifford S, Sharma S, Bano N, Moran MA. Metatranscriptomic analysis of ammonia-oxidizing organisms in an estuarine bacterioplankton assemblage. ISME J. 2011;5(5):866–878. doi: 10.1038/ismej.2010.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baker BJ, Lesniewski RA, Dick GJ. Genome-enabled transcriptomics reveals archaeal populations that drive nitrification in a deep-sea hydrothermal plume. ISME J. 2012;6(12):2269–2279. doi: 10.1038/ismej.2012.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ingalls AE, et al. Quantifying archaeal community autotrophy in the mesopelagic ocean using natural radiocarbon. Proc Natl Acad Sci USA. 2006;103(17):6442–6447. doi: 10.1073/pnas.0510157103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Santoro AE, Buchwald C, McIlvin MR, Casciotti KL. Isotopic signature of N(2)O produced by marine ammonia-oxidizing archaea. Science. 2011;333(6047):1282–1285. doi: 10.1126/science.1208239. [DOI] [PubMed] [Google Scholar]
- 11.Könneke M, et al. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature. 2005;437(7058):543–546. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- 12.Martens-Habbena W, Berube PM, Urakawa H, de la Torre JR, Stahl DA. Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature. 2009;461(7266):976–979. doi: 10.1038/nature08465. [DOI] [PubMed] [Google Scholar]
- 13.Qin W, et al. Marine ammonia-oxidizing archaeal isolates displate obligate mixotrophy and wide ecotypic variation. Proc Natl Acad Sci USA. 2014;111(34):12504–12509. doi: 10.1073/pnas.1324115111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tourna M, et al. Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc Natl Acad Sci USA. 2011;108(20):8420–8425. doi: 10.1073/pnas.1013488108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lehtovirta-Morley LE, et al. Characterisation of terrestrial acidophilic archaeal ammonia oxidisers and their inhibition and stimulation by organic compounds. FEMS Microbiol Ecol. 2014;89(3):542–552. doi: 10.1111/1574-6941.12353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hallam SJ, et al. Genomic analysis of the uncultivated marine crenarchaeote Cenarchaeum symbiosum. Proc Natl Acad Sci USA. 2006;103(48):18296–18301. doi: 10.1073/pnas.0608549103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blainey PC, Mosier AC, Potanina A, Francis CA, Quake SR. Genome of a low-salinity ammonia-oxidizing archaeon determined by single-cell and metagenomic analysis. PLoS ONE. 2011;6(2):e16626. doi: 10.1371/journal.pone.0016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spang A, et al. The genome of the ammonia-oxidizing Candidatus Nitrososphaera gargensis: Insights into metabolic versatility and environmental adaptations. Environ Microbiol. 2012;14(12):3122–3145. doi: 10.1111/j.1462-2920.2012.02893.x. [DOI] [PubMed] [Google Scholar]
- 19.Hatzenpichler R, et al. A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc Natl Acad Sci USA. 2008;105(6):2134–2139. doi: 10.1073/pnas.0708857105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lehtovirta-Morley LE, Stoecker K, Vilcinskas A, Prosser JI, Nicol GW. Cultivation of an obligate acidophilic ammonia oxidizer from a nitrifying acid soil. Proc Natl Acad Sci USA. 2011;108(38):15892–15897. doi: 10.1073/pnas.1107196108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swan BK, et al. Genomic and metabolic diversity of Marine Group I Thaumarchaeota in the mesopelagic of two subtropical gyres. PLoS ONE. 2014;9(4):e95380. doi: 10.1371/journal.pone.0095380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo H, et al. Single-cell genomics shedding light on marine Thaumarchaeota diversification. ISME J. 2014;8(3):732–736. doi: 10.1038/ismej.2013.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Könneke M, et al. Ammonia-oxidizing archaea use the most energy-efficient aerobic pathway for CO2 fixation. Proc Natl Acad Sci USA. 2014;111(22):8239–8244. doi: 10.1073/pnas.1402028111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walker CB, et al. Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc Natl Acad Sci USA. 2010;107(19):8818–8823. doi: 10.1073/pnas.0913533107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arp DJ, Chain PSG, Klotz MG. The impact of genome analyses on our understanding of ammonia-oxidizing bacteria. Annu Rev Microbiol. 2007;61(1):503–528. doi: 10.1146/annurev.micro.61.080706.093449. [DOI] [PubMed] [Google Scholar]
- 26.Santoro AE, Casciotti KL. Enrichment and characterization of ammonia-oxidizing archaea from the open ocean: Phylogeny, physiology and stable isotope fractionation. ISME J. 2011;5(11):1796–1808. doi: 10.1038/ismej.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pellitteri-Hahn MC, Halligan BD, Scalf M, Smith L, Hickey WJ. Quantitative proteomic analysis of the chemolithoautotrophic bacterium Nitrosomonas europaea: Comparison of growing- and energy-starved cells. J Proteomics. 2011;74(4):411–419. doi: 10.1016/j.jprot.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 28.Schleper C, Jurgens G, Jonuscheit M. Genomic studies of uncultivated archaea. Nat Rev Microbiol. 2005;3(6):479–488. doi: 10.1038/nrmicro1159. [DOI] [PubMed] [Google Scholar]
- 29.Markert S, et al. Status quo in physiological proteomics of the uncultured Riftia pachyptila endosymbiont. Proteomics. 2011;11(15):3106–3117. doi: 10.1002/pmic.201100059. [DOI] [PubMed] [Google Scholar]
- 30.Löscher C, et al. Production of oceanic nitrous oxide by ammonia-oxidizing archaea. Biogeosciences. 2012;9:2419–2429. [Google Scholar]
- 31.Stieglmeier M, et al. Aerobic nitrous oxide production through N-nitrosating hybrid formation in ammonia-oxidizing archaea. ISME J. 2014;8(5):1135–1146. doi: 10.1038/ismej.2013.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beaumont HJE, Lens SI, Reijnders WNM, Westerhoff HV, van Spanning RJM. Expression of nitrite reductase in Nitrosomonas europaea involves NsrR, a novel nitrite-sensitive transcription repressor. Mol Microbiol. 2004;54(1):148–158. doi: 10.1111/j.1365-2958.2004.04248.x. [DOI] [PubMed] [Google Scholar]
- 33.Zumft WG. Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev. 1997;61(4):533–616. doi: 10.1128/mmbr.61.4.533-616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. Comparative genomics of the vitamin B12 metabolism and regulation in prokaryotes. J Biol Chem. 2003;278(42):41148–41159. doi: 10.1074/jbc.M305837200. [DOI] [PubMed] [Google Scholar]
- 35.Kim W, Major TA, Whitman WB. Role of the precorrin 6-X reductase gene in cobamide biosynthesis in Methanococcus maripaludis. Archaea. 2005;1(6):375–384. doi: 10.1155/2005/903614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sañudo-Wilhelmy SA, et al. Multiple B-vitamin depletion in large areas of the coastal ocean. Proc Natl Acad Sci USA. 2012;109(35):14041–14045. doi: 10.1073/pnas.1208755109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roth JR, Lawrence JG, Rubenfield M, Kieffer-Higgins S, Church GM. Characterization of the cobalamin (vitamin B12) biosynthetic genes of Salmonella typhimurium. J Bacteriol. 1993;175(11):3303–3316. doi: 10.1128/jb.175.11.3303-3316.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doxey AC, Kurtz DA, Lynch MDJ, Sauder LA, Neufeld JD. Aquatic metagenomes implicate Thaumarchaeota in global cobalamin production. ISME J. 2014 doi: 10.1038/ismej.2014.1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giovannoni SJ, et al. Genome streamlining in a cosmopolitan oceanic bacterium. Science. 2005;309(5738):1242–1245. doi: 10.1126/science.1114057. [DOI] [PubMed] [Google Scholar]
- 40.Carini P, et al. Discovery of a SAR11 growth requirement for thiamin’s pyrimidine precursor and its distribution in the Sargasso Sea. ISME J. 2014;8(8):1727–1738. doi: 10.1038/ismej.2014.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Graupner M, White RH. Methanococcus jannaschii generates L-proline by cyclization of L-ornithine. J Bacteriol. 2001;183(17):5203–5205. doi: 10.1128/JB.183.17.5203-5205.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rozen Y, Dyk TK, LaRossa RA, Belkin S. Seawater activation of Escherichia coli gene promoter elements: Dominance of rpoS control. Microb Ecol. 2001;42(4):635–643. doi: 10.1007/s00248-001-1012-x. [DOI] [PubMed] [Google Scholar]
- 43.Amin SA, et al. Copper requirements of the ammonia-oxidizing archaeon Nitrosopumilus maritimus SCM1 and implications for nitrification in the marine environment. Limnol Oceanogr. 2013;58(6):2037–2045. [Google Scholar]
- 44.Martiny AC, Huang Y, Li W. Occurrence of phosphate acquisition genes in Prochlorococcus cells from different ocean regions. Environ Microbiol. 2009;11(6):1340–1347. doi: 10.1111/j.1462-2920.2009.01860.x. [DOI] [PubMed] [Google Scholar]
- 45.Karl DM, et al. Aerobic production of methane in the sea. Nat Geosci. 2008;1(7):473–478. [Google Scholar]
- 46.Metcalf WW, et al. Synthesis of methylphosphonic acid by marine microbes: A source for methane in the aerobic ocean. Science. 2012;337(6098):1104–1107. doi: 10.1126/science.1219875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mira A, Ochman H, Moran NA. Deletional bias and the evolution of bacterial genomes. Trends Genet. 2001;17(10):589–596. doi: 10.1016/s0168-9525(01)02447-7. [DOI] [PubMed] [Google Scholar]
- 48.Lynch M. Streamlining and simplification of microbial genome architecture. Annu Rev Microbiol. 2006;60:327–349. doi: 10.1146/annurev.micro.60.080805.142300. [DOI] [PubMed] [Google Scholar]
- 49.Swan BK, et al. Prevalent genome streamlining and latitudinal divergence of planktonic bacteria in the surface ocean. Proc Natl Acad Sci USA. 2013;110(28):11463–11468. doi: 10.1073/pnas.1304246110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dupont CL, et al. Genomic insights to SAR86, an abundant and uncultivated marine bacterial lineage. ISME J. 2012;6(6):1186–1199. doi: 10.1038/ismej.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wolf YI, Koonin EV. Genome reduction as the dominant mode of evolution. BioEssays. 2013;35(9):829–837. doi: 10.1002/bies.201300037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dufresne A, et al. Genome sequence of the cyanobacterium Prochlorococcus marinus SS120, a nearly minimal oxyphototrophic genome. Proc Natl Acad Sci USA. 2003;100(17):10020–10025. doi: 10.1073/pnas.1733211100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morris JJ, Lenski RE, Zinser ER. The Black Queen Hypothesis: Evolution of dependencies through adaptive gene loss. MBio. 2012;3(2):e00036-12. doi: 10.1128/mBio.00036-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Facciotti MT, et al. General transcription factor specified global gene regulation in archaea. Proc Natl Acad Sci USA. 2007;104(11):4630–4635. doi: 10.1073/pnas.0611663104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Decker KB, Hinton DM. Transcription regulation at the core: Similarities among bacterial, archaeal, and eukaryotic RNA polymerases. Annu Rev Microbiol. 2013;67(67):113–139. doi: 10.1146/annurev-micro-092412-155756. [DOI] [PubMed] [Google Scholar]
- 56.Turkarslan S, et al. Niche adaptation by expansion and reprogramming of general transcription factors. Mol Syst Biol. 2011;7:554. doi: 10.1038/msb.2011.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gifford SM, Sharma S, Rinta-Kanto JM, Moran MA. Quantitative analysis of a deeply sequenced marine microbial metatranscriptome. ISME J. 2011;5(3):461–472. doi: 10.1038/ismej.2010.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rusch DB, et al. The Sorcerer II Global Ocean Sampling expedition: Northwest Atlantic through eastern tropical Pacific. PLoS Biol. 2007;5(3):e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Palenik B, et al. The genome of a motile marine Synechococcus. Nature. 2003;424(6952):1037–1042. doi: 10.1038/nature01943. [DOI] [PubMed] [Google Scholar]
- 60.Rodriguez-Valera F, et al. Explaining microbial population genomics through phage predation. Nat Rev Microbiol. 2009;7(11):828–836. doi: 10.1038/nrmicro2235. [DOI] [PubMed] [Google Scholar]
- 61.Coleman ML, et al. Genomic islands and the ecology and evolution of Prochlorococcus. Science. 2006;311(5768):1768–1770. doi: 10.1126/science.1122050. [DOI] [PubMed] [Google Scholar]
- 62.Grote J, et al. Streamlining and core genome conservation among highly divergent members of the SAR11 clade. MBio. 2012;3(5):e00252-12. doi: 10.1128/mBio.00252-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Karl DM. A sea of change: Biogeochemical variability in the North Pacific Subtropical Gyre. Ecosystems (N Y) 1999;2(3):181–214. [Google Scholar]
- 64.Polovina JJ, Howell EA, Abecassis M. Ocean's least productive waters are expanding. Geophys Res Lett. 2008;35(3):L03618. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.