Significance
T box riboswitches generally regulate gene expression at the level of transcription attenuation by utilizing a specific tRNA as the regulatory ligand. Conserved features of these elements are under development as antibiotic targets. T box sequences in the phylum Actinobacteria, including pathogens like Mycobacterium tuberculosis, lack conserved motifs essential for function in other T box RNAs and are predicted to regulate gene expression at the level of translation initiation. Demonstration that these RNAs function as T box riboswitches is crucial to determine whether T box-targeting antibiotics will be effective against these organisms. This study provides the first evidence, to our knowledge, for T box regulation of translation and new information about novel modes of tRNA recognition.
Keywords: regulation, Gram-positive, tRNA, riboswitch, translational control
Abstract
The T box riboswitch regulates many amino acid-related genes in Gram-positive bacteria. T box riboswitch-mediated gene regulation was shown previously to occur at the level of transcription attenuation via structural rearrangements in the 5′ untranslated (leader) region of the mRNA in response to binding of a specific uncharged tRNA. In this study, a novel group of isoleucyl-tRNA synthetase gene (ileS) T box leader sequences found in organisms of the phylum Actinobacteria was investigated. The Stem I domains of these RNAs lack several highly conserved elements that are essential for interaction with the tRNA ligand in other T box RNAs. Many of these RNAs were predicted to regulate gene expression at the level of translation initiation through tRNA-dependent stabilization of a helix that sequesters a sequence complementary to the Shine–Dalgarno (SD) sequence, thus freeing the SD sequence for ribosome binding and translation initiation. We demonstrated specific binding to the cognate tRNAIle and tRNAIle-dependent structural rearrangements consistent with regulation at the level of translation initiation, providing the first biochemical demonstration, to our knowledge, of translational regulation in a T box riboswitch.
Regulation of aminoacyl-tRNA synthetase (aaRS) gene expression is essential for bacterial survival because misregulation of these genes can result in tRNA misacylation or induction of the stringent response due to accumulation of uncharged tRNA (1, 2). Many aaRS genes in Gram-positive bacteria are regulated by T box riboswitches (3), which are cis-acting RNA regulatory elements that control gene expression through structural rearrangements in the 5′ untranslated (leader) region of the transcript in response to the aminoacylation state of the cognate tRNA (4–6). T box riboswitch control of essential genes has led to targeting of conserved elements of T box RNA for development of antibiotic agents (7–9).
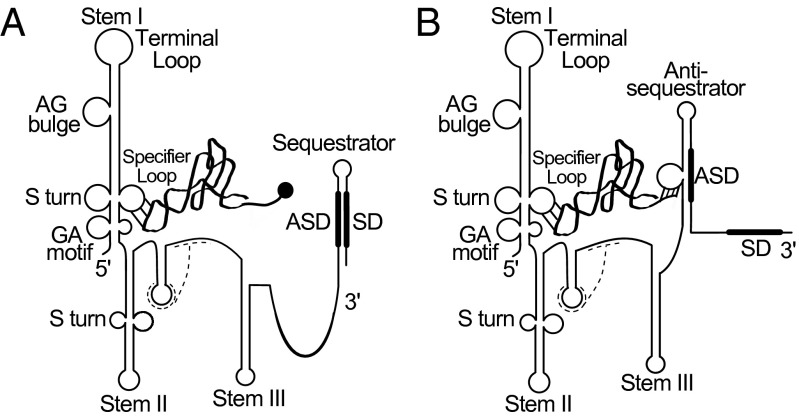
Conserved elements within T box riboswitches include three large stem–loop structures (Stems I, II, and III), a pseudoknot element (Stem IIA/B) between Stems II and III, and mutually exclusive terminator and antiterminator structures (10–13). The stem–loop structures are shown in Fig. 1, but mutually exclusive terminator and antiterminator structures are substituted for analogous sequestrator and antisequestrator structures, respectively, to illustrate translational regulation (described below). The most important feature in Stem I is the Specifier Sequence, a 3-nt sequence corresponding to a codon that matches the amino acid specificity of the downstream gene; this sequence binds to the anticodon of the cognate tRNA and is the primary element that defines specific tRNA binding (14–16). The Specifier Sequence is located in an internal loop (the Specifier Loop) of Stem I, adjacent to a conserved S-turn motif that [in conjunction with the GA motif, which forms a kink-turn below the Specifier Loop (17–19)] is responsible for presentation of the Specifier Sequence to the tRNA anticodon (18, 20). The apical region of Stem I contains the conserved AG bulge and terminal loop (6), which interdigitate to create a platform that interacts with the tRNA elbow in the glycyl-tRNA synthetase gene (glyQS) T box RNA (18, 21). However, it is unclear whether glyQS structural information is generally applicable to other T box RNAs because of the absence of the Stem II and IIA/B conserved domains (15) and an unusual arrangement of sequences in the apical portion of Stem I that differs from the consensus T box arrangement (22).
Fig. 1.
Proposed T box riboswitch regulation at the level of translation initiation. (A) Charged tRNA interacts with the Specifier Sequence of the leader RNA through the anticodon but is unable to stabilize the antisequestrator element, resulting in formation of the sequestrator helix and inhibition of translation initiation. (B) Uncharged tRNA can interact with both the Specifier Sequence and the antisequestrator helix, which frees the SD sequence to allow translation initiation. The dashed lines represent Stem IIA/B pseudoknot elements, the ribbon represents tRNA, the circle represents an amino acid charged onto tRNA, and the thick lines represent SD and ASD regions as labeled.
The Stem II domain usually contains a second S-turn motif (6) and is immediately followed by the Stem IIA/B pseudoknot (12). The roles of these domains are unclear, because some T box riboswitches (e.g., glyQS, as noted above) function without them (23), whereas others (e.g., tyrS found in tyrosyl-tRNA synthetase gene) lose expression when these domains are mutated (12). Nearly all T box RNAs contain a Stem III domain, but features of this element vary widely (6). Stem III is followed by mutually exclusive terminator and antiterminator structures. The highly conserved 14-nt T box sequence forms the 5′ side of the antiterminator element, which is composed of two short helices separated by a 7-nt bulge (24–26). Pairing of sequences from the 3′ side of the antiterminator helix with sequences further downstream results in formation of the terminator helix, which results in reduced expression of the downstream coding sequence.
All T box riboswitches studied to date regulate expression at the level of transcription attenuation. Formation of the terminator helix is prevented by stabilization of the antiterminator element through binding of the acceptor end of the cognate uncharged tRNA to complementary residues in the antiterminator bulge (11). In contrast, many ileS genes in the phylum Actinobacteria lack apparent terminator and antiterminator structures; instead, they are predicted to be regulated at the level of translation initiation via mutually exclusive structures that hide the Shine–Dalgarno (SD) sequence of the downstream gene by pairing with a complementary anti-SD (ASD) sequence (sequestrator helix) or release the SD by pairing the ASD with a complementary sequence (antisequestrator helix) (27, 28). By analogy with attenuation control, the sequestrator helix is predicted to form when the majority of the cognate tRNAIle is aminoacylated, resulting in low expression of the downstream ileS coding sequence; uncharged tRNAIle is predicted to stabilize the antisequestrator helix (which resembles the antiterminator in genes regulated by transcription attenuation), releasing the SD to allow efficient ileS translation (Fig. 1).
We recognized that many ileS genes in Actinobacteria exhibit other unusual leader RNA features. Many of these sequences, including the sequences from Mycobacterium tuberculosis and other pathogens, lack the apical portion of Stem I, including the conserved AG bulge, terminal loop, and S-turn motif, which indicates that the interaction with the tRNA elbow and presentation of the Specifier Sequence to the anticodon differ from other T box RNAs. Analysis of the Actinobacteria ileS genes is important to determine whether the T box riboswitch is functional in this important group of organisms, and the effect of the unusual features on the T box regulatory mechanism. In this study, we demonstrate that genes in this group exhibit specific tRNAIle binding and tRNA-dependent modulation of RNA structure and ribosome binding.
Results
Unique Features of ileS Leader RNAs in the Phylum Actinobacteria.
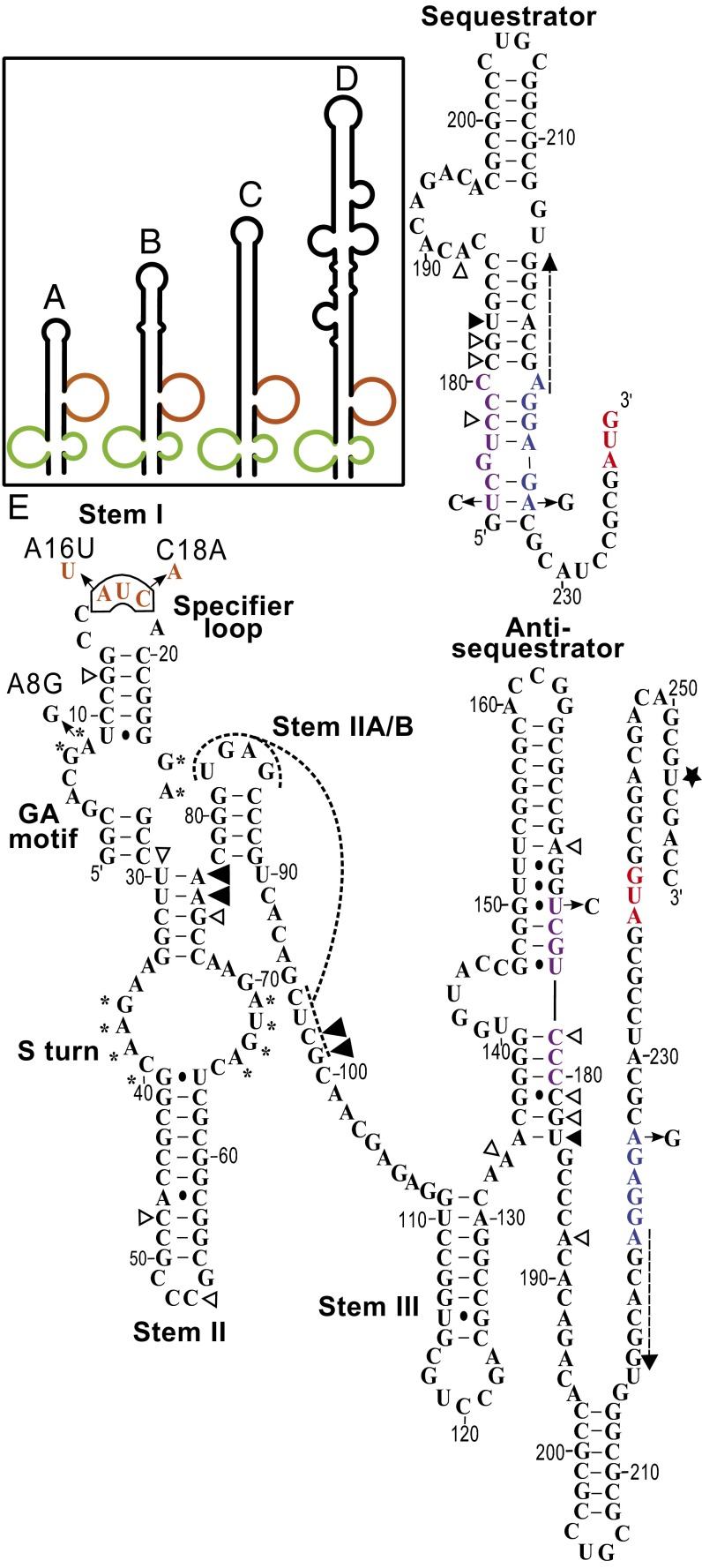
The ileS gene sequences from Actinobacteria genomes were examined for the presence of primary sequence and secondary structure elements conserved in T box riboswitches. Genomes from 15 genera have sequences consistent with features of the previously described T box genes and were designated as “canonical” (Fig. 1, Fig. S1, and Table S1). No recognizable T box riboswitches were found upstream of the ileS genes in 14 genera (Table S1). Genomes from the remaining genera have ileS gene sequences that differ from the canonical arrangement, mainly in the predicted structure of the Stem I domain. The ileS leader sequences from the genus Mobiluncus and some from Actinomyces contain unusually structured Stem I regions (USSRs) that are shorter than the canonical Stem I structures (Fig. 2 A–D, Fig. S1, and Table S1). The GA motif and the Specifier Loop are present at the base of the USSR, but the sequences above the Specifier Loop are not predicted to form the conserved AG bulge and terminal loop. The S-turn adjacent to the Specifier Sequence (12) is also absent, which positions the Specifier Sequence in a bulge instead of an internal loop. The ileS genes from most other genera contain shortened Stem I structures, designated the ultrashort Stem I (US) class, and retain only the bottom portion of Stem I that includes the GA motif and the Specifier Loop, with the Specifier Sequence located in a terminal loop that is typically 6 nt in length (Fig. 2E, Fig. S1, and Table S1). This shortening corresponds to the absence of ∼60 nt above the Specifier Loop, including the highly conserved residues of the AG bulge and terminal loop.
Fig. 2.
T box RNA secondary structures found in Actinobacteria. (A–D) USSR class Stem I structures (refer to Fig. S1 for nucleotide sequence) in Actinomyces sp. oral taxon 178 str. F0338, Actinomyces sp. oral taxon 448 str. F0400, Mobiluncus curtisii, and Mobiluncus mulieris, respectively. The GA motif is shown in green, and the Specifier Loop is shown in brown. (E) Secondary structure of US class RNA from N. farcinica ileS gene (nucleotides 1–251; nucleotides 252–258 are the vector sequence for primer binding). Sequestrator structure corresponds to nucleotides 153–239. Arrows show mutations made in the study, white triangles represent locations of tRNAIle-independent primer extension inhibition stops, large black triangles represent tRNAIle-dependent stops independent of the antisequestrator helix–tRNA acceptor end interaction, small black triangles represent tRNAIle-dependent stops dependent on the antisequestrator helix–RNA acceptor end interaction, the black star represents the position of the toeprint, the dashed arrow represents the annealing position of the antisense oligonucleotide used in the RNase H cleavage assays, the asterisk (*) represents conserved residues that participate in the GA or S-turn structural elements, dashed lines represent the pseudoknot structure, blue residues represent the SD sequence, purple residues indicate the ASD sequence, red residues represent the AUG start codon, and brown residues represent the AUC Specifier Sequence.
Stem II domains of the ileS leader RNAs vary in size and predicted secondary structure, but in all cases, they retain an S-turn motif (Fig. 2E and Fig. S1). The 5′ GAAC and 3′ AGUA S-turn sequences are positioned 2 nt and 3 nt, respectively, from the upper helix, rather than immediately adjacent to the helix, as found in most T box leader RNAs (6). The Stem IIA/B pseudoknot structure is present in most of the leader RNAs, except those leader RNAs in the Propionibacterieae family. Stem III domains are variable in size and are difficult to detect in a few of the ileS genes in this group.
Some Actinobacteria species (primarily in the Rubrobacter and Coriobacteriia classes) are predicted to regulate ileS gene expression at the level of transcription attenuation, as previously described for the majority of T box riboswitches, based on easily recognizable terminator/antiterminator structures (Fig. S1). However, most ileS genes in the phylum Actinobacteria are predicted to be regulated at the level of translation initiation (6, 26, 27) (Fig. S1).
Specific Binding of tRNAIle to ileS Leader RNAs.
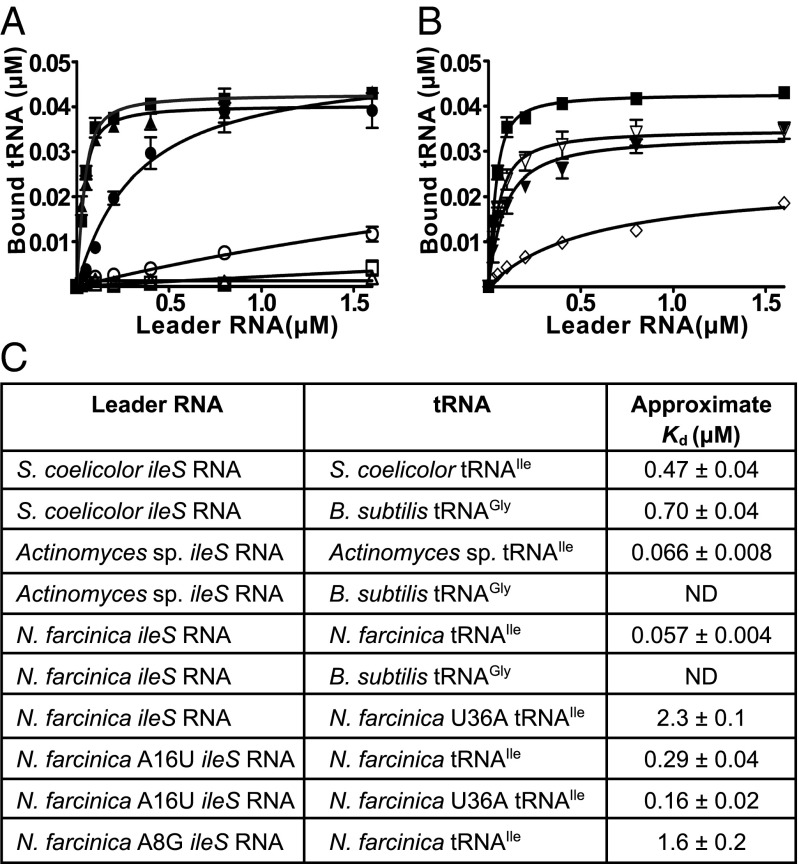
The ileS leader RNAs from three structural subclasses were tested for specific binding to tRNAIle (Fig. 3 A and C). The approximate Kd values for the interaction with tRNAIle are comparable to the Kd of the glyQS RNA for its cognate tRNAGly (0.2 μM) (18). The observed binding is specific, because binding to the noncognate tRNAGly was not detected for Nocardia farcinica and Actinomyces sp. ileS RNAs. For Streptomyces coelicolor ileS RNA, the approximate Kd was approximately twofold increased and the maximum amount of bound tRNA was fourfold reduced for noncognate tRNAGly (Fig. 3 A and C); the lower specificity may be due, in part, to the requirement for different buffer conditions for efficient binding of the S. coelicolor ileS RNA to the cognate tRNAIle, possibly because of different folding requirements for this longer RNA.
Fig. 3.
tRNA-binding assays. The [α-32P]-labeled tRNA was incubated with increasing concentrations of leader RNA. Bound and unbound tRNA species were separated by size exclusion filtration, and binding was expressed as the concentration of bound tRNA. (A) ■, N. farcinica ileS leader RNA binding to tRNAIle; □, N. farcinica ileS leader RNA binding to B. subtilis tRNAGly; ▲, Actinomyces sp. ileS leader RNA binding to tRNAIle; △, Actinomyces sp. ileS leader RNA binding to B. subtilis tRNAGly; ●, S. coelicolor ileS leader RNA binding to tRNAIle; ○, S. coelicolor ileS leader RNA binding to B. subtilis tRNAGly. (B) ▼, A16U N. farcinica ileS leader RNA binding to tRNAIle; ▽, A16U N. farcinica ileS leader RNA binding to U36A tRNAIle; ◇, N. farcinica ileS leader RNA binding to U36A tRNAIle. (C) Approximate Kd values for binding of tRNA to ileS leader RNA. n ≥ 3 ± SEM. ND, no detectable binding.
Specificity was tested further by introduction of an A16U mutation in the N. farcinica ileS leader RNA that changes the Specifier Sequence from AUC (Ile codon) to UUC (Phe codon; Fig. 2E); this mutation resulted in a fivefold reduction of affinity for tRNAIle (Fig. 3 B and C). N. farcinica tRNAIle with a compensatory U36A mutation (which changes the Ile anticodon to a Phe anticodon) showed 40-fold reduced affinity for the WT ileS leader RNA. Binding of tRNAIleU36A to the A16U ileS leader RNA resulted in partial restoration of affinity, as demonstrated by a 14-fold decrease in the Kd relative to the affinity of the WT ileS leader RNA for tRNAIleU36A. These results are consistent with binding of glyQS RNA to tRNAGly that was disrupted by the introduction of codon-anticodon mismatches (15).
An A8G mutation was introduced into the GA motif of N. farcinica ileS leader RNA (Fig. 2E) to authenticate ileS genes further as members of the T box family. In the context of other T box leader RNAs, this mutation disrupts the kink-turn and results in loss of tRNA-dependent antitermination (17). The A8G mutation decreased affinity for tRNAIle ∼28-fold (Fig. 3C), consistent with the effect of analogous mutations in other T box leader RNAs, and indicating the importance of the kink-turn structural element in the context of a US T box RNA.
tRNAIle Binding Induces Structural Changes in the N. farcinica ileS Leader RNA Consistent with Regulation at the Level of Translation Initiation.
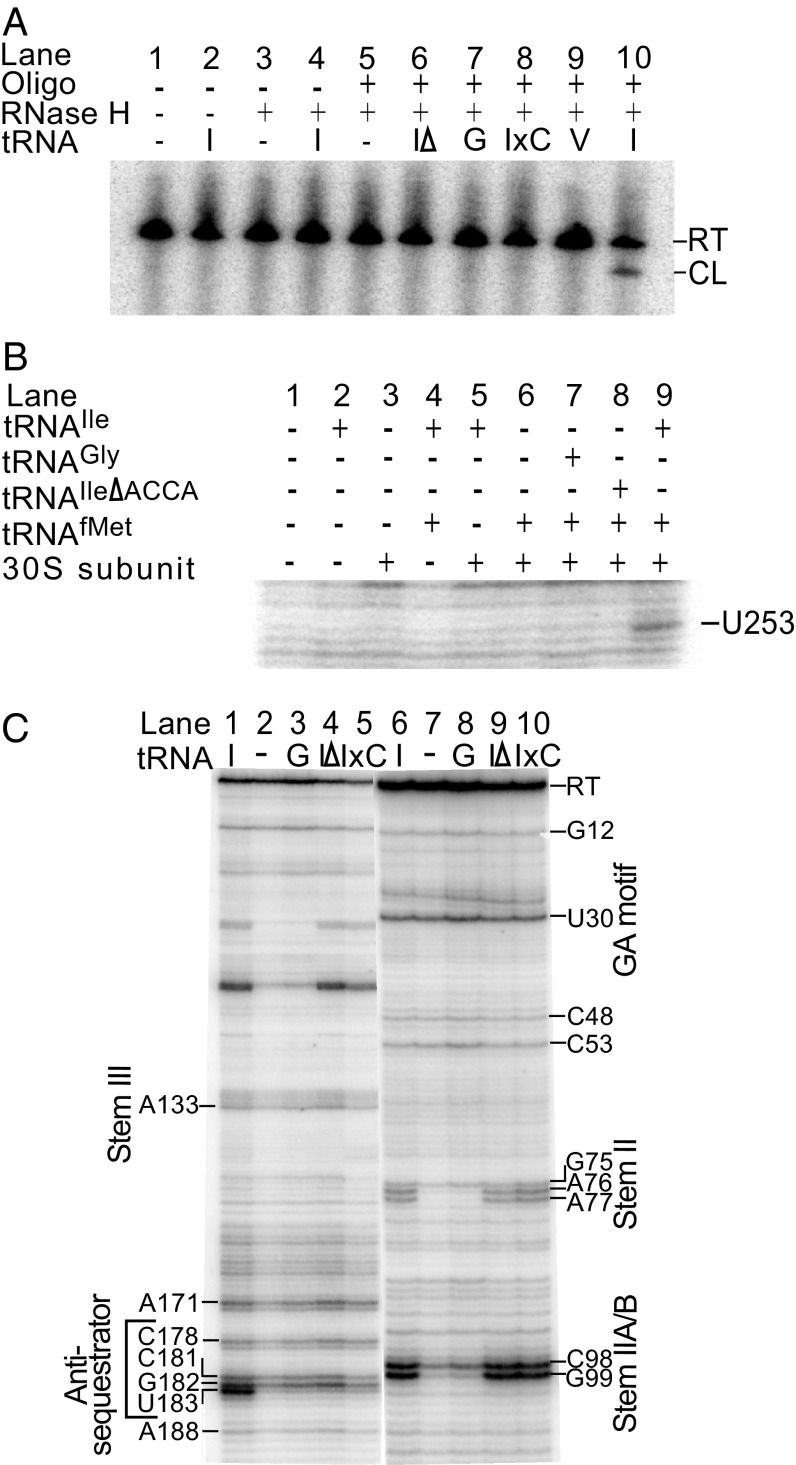
The translational regulation model predicts that the SD sequence is more available for 30S subunit binding in the presence of tRNAIle due to stabilization of an antisequestrator element that includes the ASD sequence that otherwise pairs with the SD sequence (Fig. 1). Binding of a DNA oligonucleotide complementary to the region immediately adjacent to the predicted SD sequence of the N. farcinica ileS leader RNA (Fig. 2E), followed by digestion with RNase H (which cleaves RNA/DNA duplexes), was used to monitor accessibility of this region in the presence and absence of tRNAIle. An RNase H cleavage product, which indicates increased SD region availability, was observed in the reaction containing tRNAIle (Fig. 4A, lane 10). No cleavage products were detected in the absence of tRNA or in the presence of noncognate tRNAGly or tRNAVal, or variants of tRNAIle that are unable to stabilize the antisequestrator helix (tRNAIleΔACCA, a deletion variant missing 4 nt at the tRNAIle acceptor end that are predicted to pair with residues in the bulge of the antisequestrator element, and tRNAIleEx1C, a charged tRNAIle mimic with an extra cytosine at the 3′ end of tRNA; Fig. 4A, lanes 6–9). These results indicate that the SD sequence becomes more accessible in the presence of tRNAIle and that accessibility is dependent on both the Specifier Sequence–anticodon and antisequestrator–tRNA acceptor end interactions.
Fig. 4.
US RNA structural changes. (A) N. farcinica ileS leader region was transcribed in the presence and absence of tRNA, annealed to an oligonucleotide complementary to the SD region, and incubated with RNase H. Primer extension inhibition analysis of N. farcinica ileS leader RNA is shown in the presence and absence of tRNA using [γ-32P]-labeled oligonucleotides complementary to positions 310–330 (B) and 267–289 (C) of the transcript. In C, lanes 1–5 show the 3′ regions of the transcript and lanes 6–10 show the 5′ region of the transcript. CL, 218-base RNase H cleavage product; G, tRNAGly; I, tRNAIle; IΔ, tRNAIleΔACCA; IxC, tRNAIleEx1C; RT, 289-base full-length readthrough product; V, tRNAVal.
tRNAIle-Dependent 30S Ribosomal Subunit Binding to N. farcinica ileS Leader RNA.
Primer extension inhibition (“toeprint”) assays were used to identify the ribosome-binding site on the N. farcinica ileS RNA and the dependence of ribosomal binding on the presence of tRNAIle. A reverse transcriptase stop, caused by a bound 30S ribosomal subunit when the initiator tRNAfMet is positioned in the P site, usually occurs 16–17 nt downstream of the adenosine (A) of the start codon (29–31). Incubation of N. farcinica ileS RNA with 30S subunits, initiator tRNAfMet, and tRNAIle resulted in a reverse transcriptase stop at U253, 17 nt downstream of the A of the predicted start codon (Fig. 4B, lane 9). Noncognate tRNAGly or tRNAIleΔACCA (which lacks residues predicted to pair with the antisequestrator element) failed to yield a toeprint product, indicating dependence of ribosome binding on the cognate tRNA and the antisequestrator–tRNA acceptor end interaction (Fig. 4B, lanes 7 and 8).
RNA secondary and tertiary structure can also cause inhibition of reverse transcriptase activity, and identification of primer extension inhibition products was used to investigate the ileS leader RNA structure. A reverse transcriptase stop observed at position U183, the 3′ terminal nucleotides of the antisequestrator helix, was more abundant in reactions containing tRNAIle (Fig. 4C, lane 1), consistent with the hypothesis that the antisequestrator helix is stabilized upon interaction with tRNAIle.
Products at G99 and G98 correspond to the 3′ end of the predicted Stem IIA/B pseudoknot. The G99 product was present only in reactions with tRNAIle, tRNAIle∆ACCA, or tRNAIleEx1C (Fig. 4C, lanes 6, 9, and 10), and the C98 product was present in all reactions but was more abundant in the presence of tRNAIle or tRNAIle acceptor end variants. Similarly, for products corresponding to the Stem II domain, the G75 product was tRNA-independent but the A76 and A77 products appeared only in reactions with tRNAIle or tRNAIle acceptor end variants. These results indicate that Stem II and Stem IIA/B can form in the absence of tRNA but are stabilized in the presence of cognate tRNAIle, independent of the antisequestrator–acceptor end interaction; this finding suggests that these elements are stabilized upon binding of the cognate tRNA, regardless of its aminoacylation status. The reverse transcriptase products corresponding to the terminal loop of Stem II (C53 and C48), GA motif (U30), and top of Stem I (G12) were present in all of the reactions, independent of the cognate tRNA, which indicates that these elements form before interaction with the cognate tRNA.
Discussion
In this study, the ileS T box sequences found in the phylum Actinobacteria were classified into three groups: canonical, US, and USSR, based on the structure of their Stem I domain. Members of each group were demonstrated to bind the cognate tRNAIle specifically, despite the absence (in the US and USSR RNAs) of conserved elements previously shown to be crucial for tRNA binding. tRNAIle was shown to promote ileS RNA structural rearrangements, including stabilization of the antisequestrator helix and an increase in availability of the SD sequence for binding of the 30S subunit consistent with regulation at the level of translation initiation. Although phylogenetic studies have predicted this level of regulation for T box genes (27, 28), this study provides the first biochemical evidence, to our knowledge, for T box gene regulation at a level other than transcription attenuation.
To date, T box regulation at the level of translation initiation has been identified only in organisms of the phylum Actinobacteria. Selective pressures that led to the evolution of this T box regulatory mechanism are not clear but do not appear to be linked to G + C content or environmental niche, both of which vary widely in Actinobacteria (32). Translational regulation may provide these organisms with a more rapid response to environmental stimuli compared with regulation at the level of transcription attenuation, because the full-length gene transcript is available for immediate initiation of translation. However, translational regulation may have higher metabolic costs, because resources are consumed in generation and maintenance of the untranslated transcript.
Transcriptional T box regulation has been predicted for organisms in Rubrobacter and Coriobacteriia classes; however, these organisms were suggested to be assigned incorrectly to the Actinobacteria phylum (32). Removal of these classes from the phylum would make translational T box regulation a unique molecular feature of Actinobacteria. Molecular features found in a conserved protein sequences in closely related species are referred to as conserved signature intels (CSIs). Mechanistic variation of T box regulation could serve as an RNA CSI for the Actinobacteria. Structures found within Stem I could also serve as RNA molecular markers, because Actinobacteria species cluster in the phylogenetic tree based on their ileS Stem I structure (Fig. S2). We also recognized an association between unusual T box features and genes that encode proteins with known CSIs. For example, ileS genes in Actinomyces and Mobiluncus encode a protein with a 3-aa insert that provides a molecular signature of these genera (32) and also contain the USSR form of Stem I. Phylogenetic relationships may allow identification of evolutionary advantages provided by the unusual T box RNA features.
Attempts to target T box riboswitches for development of antibiotic agents are in progress, but testing has focused on T box systems that regulate gene expression at the level of transcription attenuation (7, 9). T box-targeted antibiotic compounds bind to the helix–bulge–helix structure of the antiterminator and prevent tRNA-dependent stabilization of the antiterminator, thereby leading to loss of downstream gene expression (7). A similar helix–bulge–helix structure is found in the Actinobacteria antisequestrator element, which would allow binding of the antibiotic compound and inhibition of translation. Demonstration of T box riboswitch functionality in Actinobacteria is crucial for their validation as antibiotic targets in organisms in this phylum, and the assays described here will be useful in these efforts.
Specifier Sequence presentation for the interaction with the anticodon in the US and USSR variants significantly differs from the well-characterized tyrS and glyQS T box RNA due to the absence of the S-turn motif in Stem I. Both the US and USSR Stem I lack the interaction with the tRNA elbow found in the glyQS structure, either because the corresponding region is completely absent (US) or because the domain above the Specifier Loop lacks the elements that participate in this interaction in glyQS (USSR). The importance of this interaction in RNAs that have these domains suggests that the ileS RNAs have evolved to bind their tRNA without it or to replace it with other interactions.
The discovery of the US and USSR variants adds to the growing list of T box riboswitches with unusual structural arrangements. No T box sequences that lack both the apical region of Stem I and the Stem II and Stem IIA/B pseudoknot domains have been found, which suggests that the system can tolerate the absence of either, but not both, of the domains. Identification of T box variants with noncanonical Stem I structures also suggests that T box gene regulation might be more widespread than previously thought. Bioinformatic approaches to T box RNA identification might have missed some T box RNAs that lack canonical features, and it is necessary to use methods that do not rely on the canonical Stem I features. Future analyses of T box riboswitch variants are likely to reveal additional noncanonical examples of this regulatory system, and may provide new insight into how these RNAs can recognize their tRNA ligands.
Materials and Methods
Bacterial Species, DNA Constructs, and RNA Synthesis.
The ileS gene and tRNAIle sequences were derived from the genomes of N. farcinica [National Center for Biotechnology Information (NCBI) AP006618.1], Actinomyces sp. oral taxon 178 str. F0338 (NCBI AEUH01000170.1), and S. coelicolor A32 (NCBI AL939111.1). tRNAGly and tRNAVal sequences were derived from the genome of Bacillus subtilis (BGSC 1A40).
DNA constructs were generated by ligating complementary pairs of oligonucleotides (Integrated DNA Technologies) (23) or, for B. subtilis tRNAVal template, amplifying the sequence of interest by PCR using chromosomal DNA and complementary oligonucleotides (33) (Table S2). Most constructs were inserted into plasmid vectors, and templates for T7 RNA polymerase (RNAP) transcription reactions were generated by PCR of the inserts from the plasmid, by PCR of the ligation reactions, or by BsrDI digestions of the plasmid for generation of B. subtilis tRNAVal and Actinomyces sp. tRNAIle templates (Table S2). Mutations were introduced by using pairs of oligonucleotides containing the mutation in a ligation reaction, by primers containing the mutation in PCR, or by oligonucleotide-directed mutagenesis of the plasmid as in a study by Wilson-Mitchell et al. (34). The N. farcinica SD and ASD sequences were changed to the sequences of the Escherichia coli consensus (by an A226G mutation in combination with a compensatory U174C ASD mutation) to facilitate primer extension inhibition assays with E. coli 30S subunits. Positions 65–83 of the Actinomyces sp. ileS leader sequence were substituted with 5′-GCCGGCCACGGC-3′ to shorten the Stem II secondary structure.
RNA was synthesized using laboratory-prepared T7 RNAP and gel-purified (15). For tRNA-binding assays, the tRNA was uniformly labeled by including [α-32P]-UTP (800 Ci/mmol; 1 Ci = 37 GBq) to a final concentration of 0.85 μM in the transcription reaction. RNA concentration was determined using an ND-1000 spectrophotometer (NanoDrop Technologies, Inc.) or by analysis in a Packard Tri-Carb 2100TR liquid scintillation counter.
tRNA-Binding Assays and Approximate Kd Determination.
Uniformly labeled tRNA at 0.05 μM was combined with leader RNA at the indicated concentrations in 1× transcription buffer (15) for N. farcinica and Actinomyces sp. leader RNAs, and in 1× HiFi buffer (35) for S. coelicolor leader RNA. The reactions were heated to 65 °C for 5 min, slow-cooled to 40 °C, and passed through a Nanosep 30K Omega filter microconcentrator (Life Sciences). The fluid retained on the filter was combined with Packard BioScience Ultima Gold scintillation fluid and counted in a Packard Tri-Carb 2100TR liquid scintillation counter. Approximate Kd was determined using GraphPad Prism 4 software nonlinear regression analysis.
RNase H Cleavage Assays.
Labeled N. farcinica ileS leader RNA was generated in single-round transcription reactions as previously described (33), except that transcription was initiated by addition of 150 μM ApC (Sigma), CTP was omitted from transcription initiation reactions to form halted complexes at position +11, and transcription elongation was carried out in the presence of tRNA. RNA products were incubated with an antisense DNA oligonucleotide complementary to sequences immediately upstream of the SD sequence (GGCACGA, positions 215–221) at 37 °C for 5 min and digested with RNase H (10 U/μL; Ambion) at 37 °C for 10 min. The reactions were terminated by phenol-chloroform extraction, and products (full-length readthrough product = 289 bases, cleavage product = 218 bases) were resolved by denaturing 6% (wt/vol) PAGE and visualized by PhosphorImager analysis (Molecular Dynamics).
Primer Extension Inhibition Assays.
DNA oligonucleotide 310–330 complementary to the 3′ region of N. farcinica ileS leader RNA (Table S3) was 5′-labeled with [γ-32P]-ATP (6,000 Ci/mmol, 1 Ci = 37 GBq) (36). Labeled oligonucleotide at 0.05 μM was annealed to N. farcinica ileS leader RNA at 10 nM in 1× extension buffer (36) by heating to 65 °C for 5 min and slow-cooling to 40 °C. tRNA was added at 1.7 μM to the reaction, and mixtures were incubated for 10 min at 37 °C. The 30S ribosomal subunits (37) at 1.5 μM, E. coli tRNAfMet (Sigma–Aldrich) at 0.9 μM, and/or water was added, and mixtures were incubated at 45 ° for 10 min. Avian myeloblastosis virus reverse transcriptase (1 unit per reaction, Thermoscript RT-PCR; Invitrogen) and dNTPs at 40 μM were added to the reaction, and mixtures were incubated at 45 °C for 15 min. The reactions were terminated by phenol-chloroform extraction, followed by precipitation in precipitation/inactivation buffer [1 M guanidinium thiocyanate, 0.167% N-lauryl sarcosine, 10 mM DTT, 83% (vol/vol) isopropanol]. The products were resolved by denaturing 6% (wt/vol) PAGE and visualized by PhosphorImager analysis (38). A DNA sequencing ladder was generated using a DNA Sequenase 2.0 kit (USB Corporation), a DNA template that contained nucleotides 1–251 of the N. farcinica ileS gene followed by 91 nt of the vector, and the same downstream primer as in the primer extension inhibition assays. For resolution of changes in the 5′ region of the leader RNA, primer 267–289 (Table S3) complementary to sequences closer to the 5′ end of the leader RNA was used as described above, and 30S ribosomal subunits and tRNAfMet were omitted.
Supplementary Material
Acknowledgments
This work was supported by NIH Institute of General Medical Sciences Grant R01 GM047823.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1424175112/-/DCSupplemental.
References
- 1.Bedouelle H, Guez V, Vidal-Cros A, Hermann M. Overproduction of tyrosyl-tRNA synthetase is toxic to Escherichia coli: A genetic analysis. J Bacteriol. 1990;172(7):3940–3945. doi: 10.1128/jb.172.7.3940-3945.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baick J-W, et al. Growth inhibition of Escherichia coli during heterologous expression of Bacillus subtilis glutamyl-tRNA synthetase that catalyzes the formation of mischarged glutamyl-tRNA1 Gln. J Microbiol. 2004;42(2):111–116. [PubMed] [Google Scholar]
- 3.Henkin TM, Grundy FJ. Sensing metabolic signals with nascent RNA transcripts: The T box and S box riboswitches as paradigms. Cold Spring Harb Symp Quant Biol. 2006;71:231–237. doi: 10.1101/sqb.2006.71.020. [DOI] [PubMed] [Google Scholar]
- 4.Henkin TM. Riboswitch RNAs: Using RNA to sense cellular metabolism. Genes Dev. 2008;22(24):3383–3390. doi: 10.1101/gad.1747308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith AM, Fuchs RT, Grundy FJ, Henkin TM. Riboswitch RNAs: Regulation of gene expression by direct monitoring of a physiological signal. RNA Biol. 2010;7(1):104–110. doi: 10.4161/rna.7.1.10757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gutiérrez-Preciado A, Henkin TM, Grundy FJ, Yanofsky C, Merino E. Biochemical features and functional implications of the RNA-based T-box regulatory mechanism. Microbiol Mol Biol Rev. 2009;73(1):36–61. doi: 10.1128/MMBR.00026-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orac CM, et al. Synthesis and stereospecificity of 4,5-disubstituted oxazolidinone ligands binding to T-box riboswitch RNA. J Med Chem. 2011;54(19):6786–6795. doi: 10.1021/jm2006904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou S, Acquaah-Harrison G, Jack KD, Bergmeier SC, Hines JV. Ligand-induced changes in T box antiterminator RNA stability. Chem Biol Drug Des. 2012;79(2):202–208. doi: 10.1111/j.1747-0285.2011.01274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jentzsch F, Hines J. Interfacing medicinal chemistry with structural bioinformatics: Implications for T box riboswitch RNA drug discovery. BMC Bioinformatics. 2012;13(Suppl 2):1–5. doi: 10.1186/1471-2105-13-S2-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henkin TM, Glass BL, Grundy FJ. Analysis of the Bacillus subtilis tyrS gene: Conservation of a regulatory sequence in multiple tRNA synthetase genes. J Bacteriol. 1992;174(4):1299–1306. doi: 10.1128/jb.174.4.1299-1306.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Green NJ, Grundy FJ, Henkin TM. The T box mechanism: tRNA as a regulatory molecule. FEBS Lett. 2010;584(2):318–324. doi: 10.1016/j.febslet.2009.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rollins SM, Grundy FJ, Henkin TM. Analysis of cis-acting sequence and structural elements required for antitermination of the Bacillus subtilis tyrS gene. Mol Microbiol. 1997;25(2):411–421. doi: 10.1046/j.1365-2958.1997.4851839.x. [DOI] [PubMed] [Google Scholar]
- 13.Grundy FJ, Henkin TM. Conservation of a transcription antitermination mechanism in aminoacyl-tRNA synthetase and amino acid biosynthesis genes in gram-positive bacteria. J Mol Biol. 1994;235(2):798–804. doi: 10.1006/jmbi.1994.1038. [DOI] [PubMed] [Google Scholar]
- 14.Grundy FJ, Yousef MR, Henkin TM. Monitoring uncharged tRNA during transcription of the Bacillus subtilis glyQS gene. J Mol Biol. 2005;346(1):73–81. doi: 10.1016/j.jmb.2004.11.051. [DOI] [PubMed] [Google Scholar]
- 15.Yousef MR, Grundy FJ, Henkin TM. Structural transitions induced by the interaction between tRNA(Gly) and the Bacillus subtilis glyQS T box leader RNA. J Mol Biol. 2005;349(2):273–287. doi: 10.1016/j.jmb.2005.03.061. [DOI] [PubMed] [Google Scholar]
- 16.Grundy FJ, Henkin TM. tRNA as a positive regulator of transcription antitermination in B. subtilis. Cell. 1993;74(3):475–482. doi: 10.1016/0092-8674(93)80049-k. [DOI] [PubMed] [Google Scholar]
- 17.Winkler WC, Grundy FJ, Murphy BA, Henkin TM. The GA motif: An RNA element common to bacterial antitermination systems, rRNA, and eukaryotic RNAs. RNA. 2001;7(8):1165–1172. doi: 10.1017/s1355838201002370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang J, Ferré-D’Amaré AR. Co-crystal structure of a T-box riboswitch stem I domain in complex with its cognate tRNA. Nature. 2013;500(7462):363–366. doi: 10.1038/nature12440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J, Nikonowicz EP. Solution structure of the K-turn and Specifier Loop domains from the Bacillus subtilis tyrS T-box leader RNA. J Mol Biol. 2011;408(1):99–117. doi: 10.1016/j.jmb.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J, Henkin TM, Nikonowicz EP. NMR structure and dynamics of the Specifier Loop domain from the Bacillus subtilis tyrS T box leader RNA. Nucleic Acids Res. 2010;38(10):3388–3398. doi: 10.1093/nar/gkq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grigg JC, et al. T box RNA decodes both the information content and geometry of tRNA to affect gene expression. Proc Natl Acad Sci USA. 2013;110(18):7240–7245. doi: 10.1073/pnas.1222214110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henkin TM. The T box riboswitch: A novel regulatory RNA that utilizes tRNA as its ligand. Biochim Biophys Acta. 2014;1839(10):959–963. doi: 10.1016/j.bbagrm.2014.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yousef MR, Grundy FJ, Henkin TM. tRNA requirements for glyQS antitermination: A new twist on tRNA. RNA. 2003;9(9):1148–1156. doi: 10.1261/rna.5540203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grundy FJ, Rollins SM, Henkin TM. Interaction between the acceptor end of tRNA and the T box stimulates antitermination in the Bacillus subtilis tyrS gene: A new role for the discriminator base. J Bacteriol. 1994;176(15):4518–4526. doi: 10.1128/jb.176.15.4518-4526.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gerdeman MS, Henkin TM, Hines JV. Solution structure of the Bacillus subtilis T-box antiterminator RNA: Seven nucleotide bulge characterized by stacking and flexibility. J Mol Biol. 2003;326(1):189–201. doi: 10.1016/s0022-2836(02)01339-6. [DOI] [PubMed] [Google Scholar]
- 26.Grundy FJ, Moir TR, Haldeman MT, Henkin TM. Sequence requirements for terminators and antiterminators in the T box transcription antitermination system: Disparity between conservation and functional requirements. Nucleic Acids Res. 2002;30(7):1646–1655. doi: 10.1093/nar/30.7.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seliverstov AV, Putzer H, Gelfand MS, Lyubetsky VA. Comparative analysis of RNA regulatory elements of amino acid metabolism genes in Actinobacteria. BMC Microbiol. 2005;5(1):54. doi: 10.1186/1471-2180-5-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vitreschak AG, Mironov AA, Lyubetsky VA, Gelfand MS. Comparative genomic analysis of T-box regulatory systems in bacteria. RNA. 2008;14(4):717–735. doi: 10.1261/rna.819308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schaefer EM, Hartz D, Gold L, Simoni RD. Ribosome-binding sites and RNA-processing sites in the transcript of the Escherichia coli unc operon. J Bacteriol. 1989;171(7):3901–3908. doi: 10.1128/jb.171.7.3901-3908.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fredrick K, Noller HF. Accurate translocation of mRNA by the ribosome requires a peptidyl group or its analog on the tRNA moving into the 30S P site. Mol Cell. 2002;9(5):1125–1131. doi: 10.1016/s1097-2765(02)00523-3. [DOI] [PubMed] [Google Scholar]
- 31.Ringquist S, Gold L. Toeprinting assays. Mapping by blocks to reverse transcriptase primer extension. Methods Mol Biol. 1998;77:283–295. doi: 10.1385/0-89603-397-X:283. [DOI] [PubMed] [Google Scholar]
- 32.Gao B, Gupta RS. Phylogenetic framework and molecular signatures for the main clades of the phylum Actinobacteria. Microbiol Mol Biol Rev. 2012;76(1):66–112. doi: 10.1128/MMBR.05011-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grundy FJ, Winkler WC, Henkin TM. tRNA-mediated transcription antitermination in vitro: Codon-anticodon pairing independent of the ribosome. Proc Natl Acad Sci USA. 2002;99(17):11121–11126. doi: 10.1073/pnas.162366799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilson-Mitchell SN, Grundy FJ, Henkin TM. Analysis of lysine recognition and specificity of the Bacillus subtilis L box riboswitch. Nucleic Acids Res. 2012;40(12):5706–5717. doi: 10.1093/nar/gks212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huggins W, Shapkina T, Wollenzien P. Conformational energy and structure in canonical and noncanonical forms of tRNA determined by temperature analysis of the rate of s(4)U8-C13 photocrosslinking. RNA. 2007;13(11):2000–2011. doi: 10.1261/rna.656907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fuchs RT, Grundy FJ, Henkin TM. S-adenosylmethionine directly inhibits binding of 30S ribosomal subunits to the SMK box translational riboswitch RNA. Proc Natl Acad Sci USA. 2007;104(12):4876–4880. doi: 10.1073/pnas.0609956104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Powers T, Noller HF. A functional pseudoknot in 16S ribosomal RNA. EMBO J. 1991;10(8):2203–2214. doi: 10.1002/j.1460-2075.1991.tb07756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Artsimovitch I, Henkin TM. In vitro approaches to analysis of transcription termination. Methods. 2009;47(1):37–43. doi: 10.1016/j.ymeth.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.