Significance
Brain cancer is one of the deadliest human tumors and is characterized by several genetic changes leading to impairment of tumor suppressive pathways and oncogene activation. These genetic alterations promote subsequent molecular changes, including modifications of cellular metabolism, which are believed to contribute to cancer pathogenesis. Conversely, the role of metabolic changes in regulation of genomic stability in brain cancer has not been investigated. Our work shows that alterations of mitochondrial metabolism promote genetic loss of the p53 tumor suppressor and transformation via a mechanism involving reactive oxygen species. Overall, our findings suggest a causative link between metabolic alterations and loss of tumor suppressive control in the central nervous system, with implications for our understanding of brain cancer pathogenesis.
Keywords: mitochondrial metabolism, brain cancer, p53
Abstract
Alterations of mitochondrial metabolism and genomic instability have been implicated in tumorigenesis in multiple tissues. High-grade glioma (HGG), one of the most lethal human neoplasms, displays genetic modifications of Krebs cycle components as well as electron transport chain (ETC) alterations. Furthermore, the p53 tumor suppressor, which has emerged as a key regulator of mitochondrial respiration at the expense of glycolysis, is genetically inactivated in a large proportion of HGG cases. Therefore, it is becoming evident that genetic modifications can affect cell metabolism in HGG; however, it is currently unclear whether mitochondrial metabolism alterations could vice versa promote genomic instability as a mechanism for neoplastic transformation. Here, we show that, in neural progenitor/stem cells (NPCs), which can act as HGG cell of origin, inhibition of mitochondrial metabolism leads to p53 genetic inactivation. Impairment of respiration via inhibition of complex I or decreased mitochondrial DNA copy number leads to p53 genetic loss and a glycolytic switch. p53 genetic inactivation in ETC-impaired neural stem cells is caused by increased reactive oxygen species and associated oxidative DNA damage. ETC-impaired cells display a marked growth advantage in the presence or absence of oncogenic RAS, and form undifferentiated tumors when transplanted into the mouse brain. Finally, p53 mutations correlated with alterations in ETC subunit composition and activity in primary glioma-initiating neural stem cells. Together, these findings provide previously unidentified insights into the relationship between mitochondria, genomic stability, and tumor suppressive control, with implications for our understanding of brain cancer pathogenesis.
Alterations of mitochondrial metabolism are found in several cancers (1). This can occur through inactivation of components of the tricarboxylic acid (TCA) cycle and electron transport chain (ETC) (1–5). In particular, high-grade gliomas (HGGs) display mutations in the TCA enzymes isocitrate dehydrogenase IDH1 and IDH2 (5). Notably, gliomas also present mutations in mitochondrial DNA (mtDNA) and alterations of the ETC, but whether these are early or late events in cancer pathogenesis remains to be determined (6–14). Finally, p53, which has emerged as an important regulator of mitochondrial metabolism and cellular redox control (15–17), is often found mutated or functionally inactivated in HGG. Its inactivation in neural progenitor/stem cells (NPCs), which act as HGG cells of origin, contributes to gliomagenesis (18–22). In particular, deletion of a significant portion of the p53 DNA binding domain induces the accumulation of cooperative oncogenic events, thus leading to HGG (21). However, it remains to be determined whether p53 metabolic functions contribute to suppression of neoplastic transformation in the nervous system. Although these studies suggest an involvement of altered mitochondria metabolism in brain tumorigenesis, direct evidence of its role as a driver or contributing factor in pathogenesis of HGG and other human cancers is missing. More generally, the role of mitochondrial dysfunction in regulation of tumor suppressive control remains only partially investigated.
Here, we studied the effect of oxidative metabolism inhibition in normal NPCs. Our findings show that inhibition of respiration via knockdown (KD) of the complex I subunit NDUFA10 or by reducing mtDNA copy number results in p53 genetic loss, via a mechanism involving generation of reactive oxygen species (ROS) and ROS-mediated oxidative damage. In turn, this causes a glycolytic switch, a marked growth advantage, and tumor formation upon transplantation in the mouse brain. Overall, this study reveals that, in NPCs, the relationship between p53 and mitochondrial metabolism is bidirectional, with p53 being activator of mitochondrial metabolism as well as target for genetic inactivation upon inhibition of respiratory chain activity.
Results
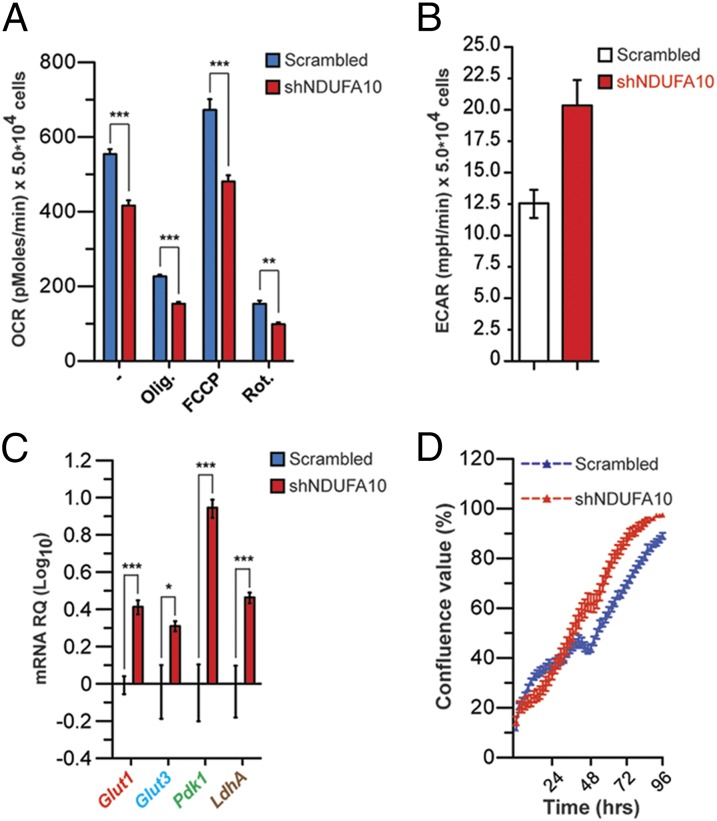
We studied the effect of oxidative metabolism inhibition in NPCs derived from the subventricular zone (SVZ), one of the two main postnatal neurogenic niches involved in brain tumorigenesis (20, 23). To this end, we inhibited the ETC by knocking down the ETC complex I component NADH dehydrogenase (ubiquinone) 1α subcomplex, 10 of complex I (NDUFA10) (Fig. S1A). Short hairpin RNA (shRNA)-mediated NDUFA10 KD resulted in decreased oxygen consumption rate (OCR) both at steady state and upon treatment with the complex I inhibitor rotenone and complex V inhibitor oligomycin (Fig. 1A). Maximal respiration was also reduced in shNDUFA10 cells (Fig. 1A). Diminished ETC activity was associated with increased extracellular acidification rate (ECAR) and lactate levels (Fig. 1B and Fig. S1B). In agreement with their glycolytic metabolism, shNDUFA10 cells displayed increased expression of genes involved in glucose uptake and metabolism Glut1, Glut3, Pdk1, and LdhA (Fig. 1C). Finally, acquisition of glycolytic metabolism was associated with growth advantage (Fig. 1D). Thus, NPCs appears to activate glycolytic metabolism upon inhibition of ETC activity.
Fig. 1.
Silencing of a complex I component results in metabolic shift and growth advantage in NPCs. (A) OCR in shNDUFA10 NPCs under basal conditions, following addition of oligomycin (0.1 μg⋅mL−1), FCCP (0.4 μM), or rotenone (0.2 μM) (data are represented as mean ± SEM for n = 3; **P < 0.01, ***P < 0.001). (B) ECAR in scramble and shNDUFA10-transfected NPCs. Lactate production was measured under basal conditions or after oligomycin treatment. (C) Quantitative RT-PCR (QPCR) expression analysis of genes involved in glucose uptake and metabolism in scrambled and shNDUFA10 NPCs. Results are normalized to β-actin expression levels (n = 3; *P < 0.05, **P < 0.01, ***P < 0.001). (D) Cell growth of scrambled and shNDUFA10-transduced NPCs, as assessed by INCUCYTE Live-Cell Imaging System.
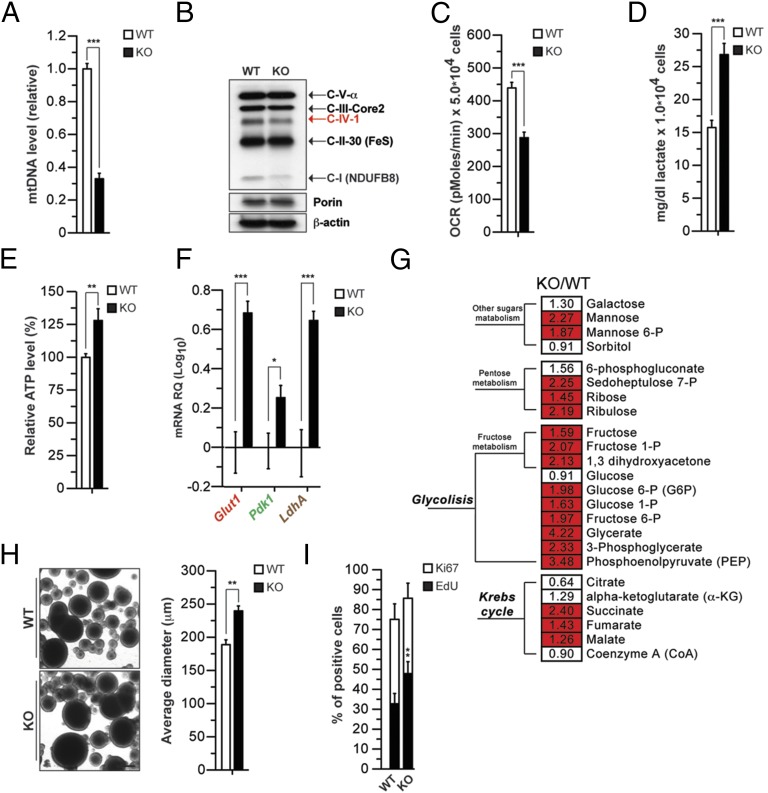
To determine whether these changes could occur also in another model of mitochondrial dysfunction, we used genetically modified NPCs, where oxidative phosphorylation is decreased due to loss of thymidine kinase 2 (TK2), a key component of the salvage pathway for nucleotide biosynthesis within mitochondria (24–26). TK2 knockout (KO) animals are ataxic and die by postnatal day 15 due to defects in multiple tissues, including brain (24–26). TK2 deficiency in postmitotic cells results in decreased mtDNA synthesis, in turn leading to diminished expression of mtDNA-encoded ETC components and impaired ETC (24–26). We isolated NPCs from the SVZ of WT and KO mice (preparation WT1 and KO1; Dataset S1). TK2 KO NPCs showed decreased mtDNA levels (Fig. 2A) and reduced expression of the mtDNA-encoded complex IV subunit I (C-IV-I) and the nuclear DNA-encoded C-I NDUFB8 (Fig. 2B). This in turn led to impaired mitochondrial oxidative capacity, as measured by detection of OCR (Fig. 2C). Defects in ETC function were accompanied by increased lactate and ATP levels (Fig. 2 D and E). Notably, TK2 KO cells displayed increased expression of Glut1, Pdk1, and LdhA (Fig. 2F). shRNA-mediated KD of TK2 (Fig. S1C) led to similar metabolic alterations (Fig. S1 D and E). Analysis of the TK2 KO metabolome revealed clear changes in glycolysis as well as sugar metabolism and Krebs cycle (Fig. 2G and Dataset S1). Overall, these data together with NDUFA10 KD experiments show that NPCs, unlike postmitotic neurons (25), are able to activate glycolysis upon inhibition of oxidative metabolism. These metabolic changes correlated with increased growth properties, as, when plated at clonal density in nonadherent conditions, KO NPCs formed larger neurospheres (Fig. 2H). Furthermore, in adherent conditions, TK2 KO NPC cultures displayed an increased number of cells in S phase (Fig. 2I). This phenotype was not associated with changes in basal apoptosis (Fig. S1F). When switched to differentiating conditions, TK2 KO cultures displayed an increased number of Nestin-positive undifferentiated cells and generated more TuJ1-positive neurons (Fig. S2 A–C). Finally, when switched back to proliferation media, KO cells proliferated more readily and formed larger neurospheres, suggesting an increased number of cells resistant to differentiation (Fig. S2 D and E). These data indicate that ETC impairment leads to increased proliferation and altered differentiation properties in NPCs.
Fig. 2.
Reduction in mitochondrial copy number leads to ETC defects, glycolysis induction, and growth advantage. (A) Relative quantification of mtDNA levels in wild-type (WT) and TK2 KO (KO) NPCs using quantitative RT-PCR (QPCR) (n = 3; ***P < 0.001). (B) Levels of ETC mtDNA-encoded (red) and nuclear-encoded (black) proteins at steady state using an antibody mix against OXPHOS complexes. Porin and β-actin are shown as loading controls for mitochondria and total protein extracts, respectively. (C) OCR in NPCs under basal conditions. Data are average of three independent experiments as mean ± SEM. NPCs were obtained from n = 3 animals for each genotype; ***P < 0.001; lactate (D) and relative ATP levels (E) in NPCs. Measurements were made in triplicate (data are represented as mean ± SEM for n = 3; **P < 0.01, ***P < 0.001). (F) Expression of the genes Glut1, Pdk1, and LdhA in WT and TK2 KO NPCs (expressed as levels over WT cells). (G) Heat map showing the ratio of the metabolite levels between KO and WT NPCs, and their statistical significance of the difference (Welch’s two-samples t test). Cells shaded in red indicate higher metabolite levels in KO NPCs with P < 0.001. Cells not shaded indicate no significant difference (P > 0.05). The number in each cell indicates fold changes over WT. (H) Proliferation of NPCs measured using the neurosphere assay. Neurosphere diameter is increased in restimulated KO NPCs (n = 3; **P < 0.001, Student’s t test). (Scale bar: 100 µm.) (I) Proliferation of NPCs by EdU labeling (2 h) and immunodetection of Ki67. Graph shows quantification of EdU and Ki67.
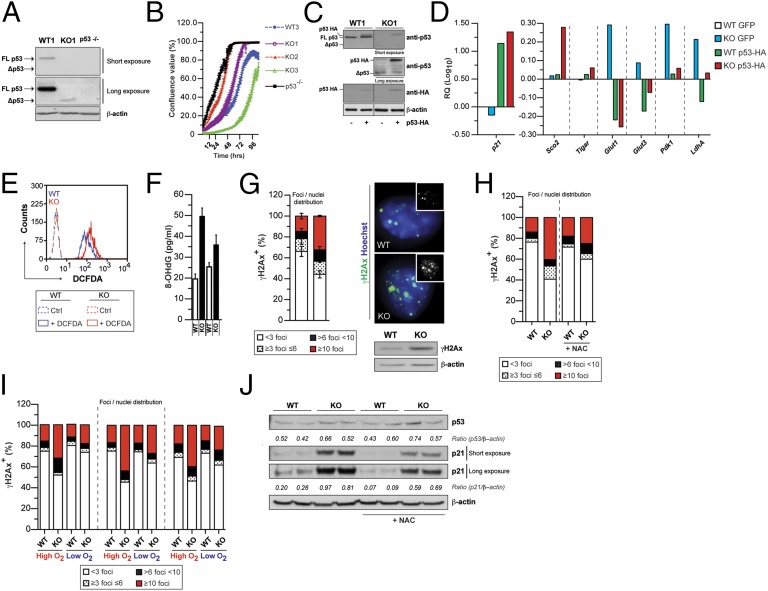
We next investigated the mechanisms underlying the metabolic switch to glycolysis in ETC-impaired NPCs. As these phenotypic changes are features of p53-deficient cells (17), we studied whether ETC-impaired cells displayed alterations of the p53 pathway. We observed a complete lack of full-length (FL) p53 expression and the presence of a shorter isoform (Δp53) along with reduced p21 expression (Fig. 3A, Fig. S3A, and Table S1) in TK2 KO cells. In agreement with the loss of FL p53 and decreased p21 levels, KO cells failed to arrest upon ionizing irradiation (IR) (Fig. S3B). We sequenced p53 cDNA in search of potential mutations in its coding region and found a p53 truncation, which was caused by a 44-nt deletion, creating a premature stop codon (Fig. S3C). Interestingly, C-terminal p53 truncations have been described in HGG cells (27). We next analyzed two additional TK2 KO NPC preparations (Table S1). KO2 cells carried a missense mutation in the p53 DNA binding domain (Fig. S3C), which corresponded to codon-281 hot-spot mutation found in human cancers including glioma (www-p53.iarc.fr/). In contrast, KO3 cells carried a silent mutation in codon 75 (Fig. S3C). Interestingly, KO2 and KO3 cells displayed a marked down-regulation of p16INK4a expression at both mRNA and protein levels (Fig. S3D). p53 mutations were also found in KO4 and KO5 cell preparations (Table S1). shNDUFA10 NPCs displayed a missense mutation in the p53 DNA binding domain (hot-spot codon 135 in human p53; Fig. S3E). In all cases, KO cell preparations and shNDUFA10 cells were analyzed at passages 5–6 from isolation/infection (Table S1). Notably, we were unable to detect p53 mutations/deletions in WT cells from four individual cell preparations up to passages 12–14 (Table S1). Overall, our data suggest that inhibition of oxidative metabolism leads to p53 genetic inactivation in NPCs.
Fig. 3.
ETC impairment leads to p53 genetic inactivation via a ROS-dependent mechanism. (A) Western blot showing expression of full-length (FL) p53 as well as the delete Δp53 form in WT and TK2 KO (KO) NPCs. p53−/− NPCs were used as negative control. (B) Increased growth properties of KO1, KO2, and p53 KO NPCs as assessed by a High Definition (HD) imaging system; growth curves representative of two independent experiments; (C) Western blot analysis of WT (WT1) and KO (KO1) NPCs infected with WT-p53-HA GFP or GFP alone (Left). (D) Representative expression analysis of p21WAF1, Sco2, Tigar, Glut1, Glut3, Pdk1, and LdhA levels (Right) upon retroviral expression of exogenous WT-p53. Results are normalized to β-actin expression level. (E) Representative plot of intracellular ROS levels, as assessed by FACS analysis using the dichlorofluorescein (DCFDA) dye. (F) Quantification of oxidized DNA marker 8-hydroxy-2′-deoxyguanosine (8-OHdG) in WT and KO NPCs cultured in normoxic condition. (G) Percentage of cells displaying defined number of γH2Ax foci/nuclei (Left). Immunofluorescence analysis of phosphorylated histone γH2Ax foci (Top Right) in NPCs. Western blot analysis of γH2Ax levels (Bottom Right). (H) γH2Ax foci quantification in WT and TK2 KO NPCs with or without the antioxidant N-acetyl cysteine (NAC) (0.1 mM). (I) γH2Ax in three independent preparations (Table S2) of KO and matched WT NPCs in high and low O2. The graph (Left) shows the percentage of cells displaying defined number of γH2Ax foci/nuclei. (J) Western blot analysis of p53 and p21WAF1 (p21) levels at steady state in the presence or absence of NAC. Cells were treated for 7 d with NAC before any measurement.
p53 mutational status, not p16INK4a down-regulation, correlated with enhanced proliferative capacity, as KO1 and KO2 cells displayed a growth advantage over KO3 and WT cells (Fig. 3B and Fig. S3D). KO3 cells grew even slower than WT cells (Fig. 3B). p53 loss-of-function mutations found in ETC-impaired cells correlated with increased ATP and lactate levels (Fig. S3F), a phenotype observed in NPCs derived from the p53 germline KO (Fig. S3F), as well as in Cre-infected p53flox/flox cells (Fig. S3 G and H). Furthermore, KO1 and KO2 cells displayed an impaired G1/S checkpoint upon IR, whereas KO3 cells behaved like wild-type cells (Fig. S4). These data suggest that p53 loss is responsible for the growth and metabolic phenotypes observed upon ETC inhibition in NPCs. Indeed, when we reintroduced WT p53 in KO1 cells, we observed a rescue in p21 expression and down-regulation of Glut1, Glut3, LdhA, and Pdk1 (Fig. 3 C and D).
We next investigated the mechanisms underlying p53 loss upon inhibition of oxidative metabolism. As ETC dysfunction is known to promote alterations of cellular redox (28, 29) and ROS can directly cause DNA damage (30, 31), we hypothesized that ROS could be involved in causing p53 genetic inactivation in ETC-impaired NPCs. To test this, we analyzed KO cells at isolation from the SVZ. Indeed, we found that ROS are increased in KO cells (Fig. 3E), and this is associated with augmented 8-hydroxy-2′-deoxyguanosine (8-OHdG) levels, a marker of DNA oxidative damage (Fig. 3F). 8-OHdG elevation correlated with increased γH2AX foci and levels, suggesting induction of double-strand breaks and ensuing DNA damage response activation (Fig. 3G). KO NPCs displayed aberrant nuclear morphology with multilobated nuclei and increased number of micronuclei (Fig. S5A). FACS analysis of KO cells also revealed increased hyper-diploid DNA content, an index of chromosomal abnormalities (Fig. S5B). Similar changes in nuclear morphology were obtained when cells were cultured for 6 d in the presence of oligomycin to induced ETC dysfunction (Fig. S5C). To confirm the involvement of ROS in promotion of genomic instability, we treated WT and KO NPCs with the ROS scavenger N-acetyl cysteine (NAC). Indeed, NAC normalized ROS levels, γH2AX foci number, and nuclear morphology (Fig. S5D and Fig. 3H). Similarly, normalization of γH2AX was achieved by culturing KO NPCs in low oxygen (Fig. 3I). Notably, freshly isolated KO cells displayed increased p53 activation, which was ROS dependent and was associated with reduced growth (Fig. 3J and Fig. S5E). These findings suggest that increased selective pressure to overcome p53-mediated cell cycle arrest in the presence of mitogenic signals along with increased ROS-mediated DNA damage represents a potential mechanism for p53 loss in NPCs. To test this, we cultured KO cells in low and high O2 from isolation and analyzed oxidative damage and the p53 status through passaging. Although KO NPC preparations acquired p53 mutations in high O2, we failed to detect any mutation in low O2 conditions up to passage 8 (Fig. S5F and Table S1).
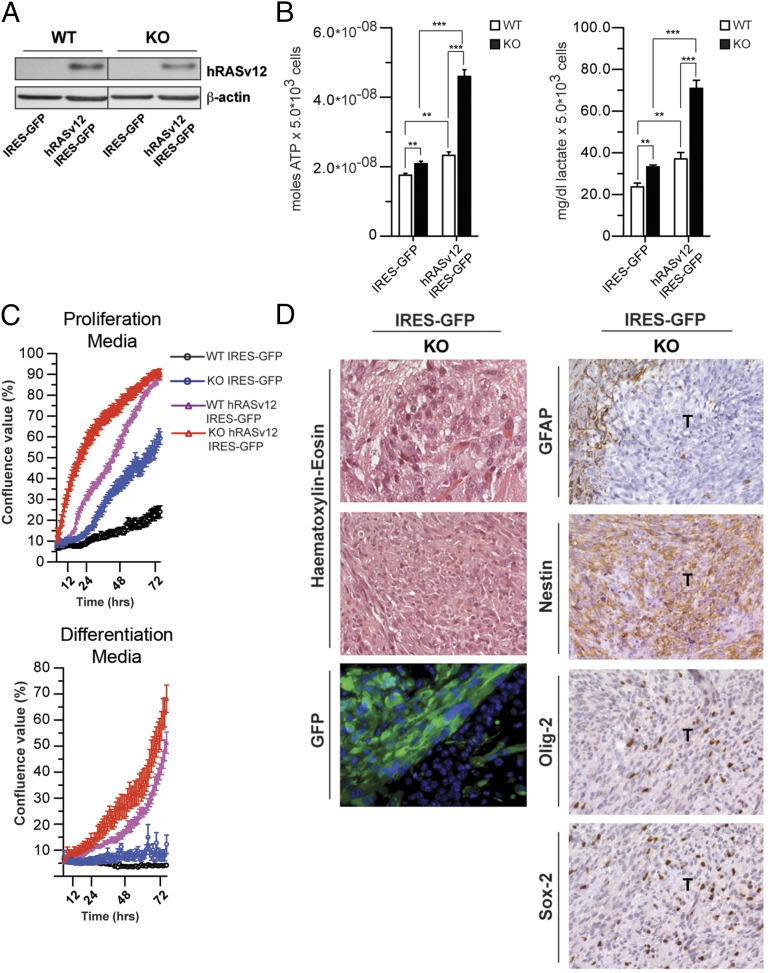
p53 inactivation is predicted to contribute to overcoming the growth-suppressive response to oncogenic activation. Hence, we analyzed the growth properties of WT and KO cells transduced with hRASV12 and control vector viral particles (Fig. 4A). hRASV12 KO cells displayed higher lactate and ATP levels (Fig. 4B). Although oncogenic hRAS did not induce growth arrest in WT NPCs unlike in fibroblasts, hRASV12 KO cells grew faster than control cells in either the presence or absence of growth factors (Fig. 4C).
Fig. 4.
ETC-impaired cells are susceptible to neoplastic transformation. (A) Western blot analysis of WT and KO (KO1 preparation) NPCs infected with hRASv12 IRES-GFP or IRES-GFP alone. (B) ATP and lactate production under basal conditions in WT and KO NPCs transduced with either IRES-GFP or hRASv12 IRES-GFP retrovirus. (**P < 0.01, ***P < 0.0001, Student’s t test; error bars are SD). (C) Growth curves analysis and representative images of WT and KO cells transduced with control (IRES-GFP) or hRASv12 IRES-GFP retroviral particles in proliferation (Top) and differentiation (Bottom) culture conditions. (D, Left) Histological and fluorescence microscopy analysis of allografts, derived from KO IRES-GFP cells. WT and KO cells were cultured as neurospheres, transduced with IRES-GFP, and orthotopically allografted into the caudoputamen of NOD/SCID immunosuppressed mice. No tumors were obtained in mice in which WT cells were xenografted. H&E histology shows large, cytoplasm-rich cells in KO tumor cells. Anti-GFP immunostaining was used to identify the tumor-initiating cells (bottom row). (Right) Immunohistochemical staining for the astrocyte marker GFAP shows that all tumors are negative with positive cells only in the adjacent host tissue. Immunohistochemical analysis of the neural stem cell marker Nestin shows a strong, diffuse labeling of tumor cells. The neural progenitor marker Olig-2 shows that many, but not all, of tumor cells present strong nuclear expression. The progenitor cell marker Sox-2 is expressed in a smaller subset of tumor cells. [Scale bar: 80 µm (first row), 320 µm (second row), and 160 µm (remaining panels).]
As p53 loss has been shown to promote HGG development in mouse via increased genomic instability (21), we reasoned that ETC-impaired, p53-deficient cells could become tumorigenic. To this end, we orthotopically transplanted WT and KO1 cells into the brain of recipient mice. To allow for identification of transplanted cells, NPCs were first transduced with IRES-GFP retroviral particles. None of the mice injected with WT cells developed brain tumors (0 of 13). In contrast, KO IRES-GFP cells were able to form poorly differentiated tumors in transplanted immune-compromised (one of four) as well as immune-competent (one of five) mice, which diffusely infiltrated the host brain (Fig. 4D and Table S2). Overall, these findings indicate that impairment of mitochondrial respiration in neural stem cells can result in inactivation of the p53 pathway and favors tumor transformation.
Finally, we investigated whether respiratory chain alterations correlated with p53 mutations in primary HGG cells. To this end, we took advantage of a panel of glioma-initiating neural stem (GNS) cells derived from resected HGG (G1, G2, G3, G4, G144, and G166). These cells represent a subpopulation within the bulk of established tumors bearing neural stem-like features, which can initiate glioma when transplanted in recipient animals (32). We noticed that a number of lines displayed enhanced growth properties (G3, G4, G144, and G166; Fig. S6A). Among them, only G3, G144, and G166 also showed elevated lactate levels and ECAR (Fig. S6B). These cells carried hot-spot mutations in the p53 DNA binding domain (Fig. S6 C and D). We then explored the status of the respiratory chain by using the MitoProfile Total OXPHOS antibody mixture and individual antibodies for different ETC components (Fig. S6E). Interestingly, p53-mutated cells showed reduced expression of the mtDNA-encoded C-IV subunit I (C-IV-1) and C-IV-2, as well as the nuclear DNA-encoded C-IV-4 (Fig. S6E). We then measured whether these changes correlated with impaired OCR. Uncoupling the mitochondria with an optimal concentration of carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone (FCCP) (3 μM) rapidly increased respiration, yielding an OCR value for maximal O2 consumption in p53-proficient cells, whereas it was significantly lower in p53-mutated GNS cells (Fig. S6 F and G). Consistent with the decreased expression of C-IV components, we observed significant impairment of C-IV activity (Fig. S6H) using treatment with antimycin A, which blocks electron flux to complex III, and a complex IV direct agonist [N,N,N′,N′-tetramethyl-p-phenylenediamine (TMPD)/ascorbate]. Changes in ETC subunit levels were not recapitulated by p53 loss alone or concomitant with expression of p53 mutants in NPCs, suggesting that alterations in ETC subunit composition are not downstream to p53 inactivation (Fig. S6 I and J). Altogether, these findings indicate that ETC alterations are associated with p53 mutations and glycolytic metabolism in GNS cells.
Discussion
This work suggests a role for mitochondrial metabolism in the regulation of tumor suppressive mechanisms and transformation in the CNS. In particular, we showed that respiratory chain dysfunction can lead to p53 genetic inactivation and transformation in NPCs (Fig. S7). Furthermore, in accordance with the reported role of p53 in suppressing transformation of NPCs (18–22), ETC-impaired/p53-deficient NPCs grow faster upon oncogenic activation and are capable of forming brain tumors in a subset of orthotopically transplanted animals. The incomplete penetrance observed in these experiments may be due to the fact that other cooperative oncogenic events may have to be acquired, as previously suggested (21), and indeed these might be favored by ROS increase as well as by p53 loss. Genome stability could also be affected via metabolic stress-dependent inhibition of metabolism-sensitive DNA repair enzymes, such as poly(ADP-ribose) polymerase (33), or indirectly via iron/sulfur (Fe/S) cluster formation (34–37) as part of a mitochondrial retrograde signaling.
Our data indicate that selective pressure to overcome ROS-mediated p53 activation along with increased ROS-mediated DNA damage contribute to p53 genetic loss in NPCs. Loss of p53 in turn leads to a metabolic switch and potentially favors acquisition of other oncogenic mutations that are yet to be identified. The importance of the redox state in the mechanism leading to p53 mutation is clearly shown by the fact that, by reducing oxygen levels, we were able to block the appearance of p53 mutations in ETC-impaired NPCs. It is conceivable that ROS originating from dysfunctional mitochondria synergize with ROS produced by growth factor signaling, as NPCs are cultured in the presence of highly mitogenic growth factors. In vivo, elevated ROS levels within the highly vascular SVZ niche have been proposed to fuel NPC expansion via growth factor signaling (38, 39). However, ROS can also lead to respiratory chain dysfunction via ROS-mediated damage to ETC components and mtDNA (30, 40–42). Respiratory chain inhibition would further augment ROS generation, thus promoting a vicious circle of oxidative stress (28, 29, 43). Finally, we observed an association between altered ETC composition, dysfunctional respiratory chain function, and p53 mutations in primary HGG cells. These findings suggest a twist in the relationship between oxidative metabolism, the p53 tumor suppressive pathway, and cellular redox status in somatic stem cells (Fig. S7): although in normal cells p53 positively regulates oxidative metabolism and antioxidant defenses, inhibition of the respiratory chain can lead to p53 genetic inactivation via a ROS-dependent mechanism, eventually contributing to malignant transformation.
Based on the classical model of tumorigenesis, an initial mutation of an oncogene or a tumor suppressor leads to subsequent molecular changes ultimately resulting in malignant transformation, including alterations of mitochondrial metabolism. We propose an additional model, whereby an initial alteration of oxidative metabolism can lead to increased mutation rate that eventually results in oncogenic mutations and tumor transformation. In highly proliferative epithelial tissues, which are more exposed to environmental factors, the first model would apply more frequently. In this respect, UV irradiation has recently been demonstrated to induce accumulation of p53 mutations at high frequency, thus accelerating BRAF(V600E)-driven melanomagenesis (44). In contrast, in progenitor/stem cells within the adult brain, which are less exposed to environmental insults, the initial event might be a metabolic defect, such as alterations of mitochondrial respiration. In fact, with the exception of exposure to ionizing radiations, a strong correlation between brain tumors and exposure to environmental carcinogens was never clearly demonstrated (45, 46). It is important to note that there is lack of evidence that hereditary diseases carrying mutations in nuclear DNA and mtDNA-encoded mitochondrial factors, such as TK2, display increased cancer susceptibility. However, the severity of phenotypes affecting cells in theses conditions may not provide an incontrovertible answer to this question. In particular, neurodegeneration phenotypes observed in these hereditary diseases could have an earlier onset compared with more indolent tumor phenotypes, thus limiting the value of an epidemiological approach.
Experimental Procedures
Animals.
Germline TK2 KO mice that harbor a progressive lost of mtDNA were developed in the laboratory of A.K. (Karolinska Institute, Stockholm, Sweden) (26). All animal work described in this study had the approvals from the University of Leicester and University College London review boards and was part of project licences granted by the Home Office.
Mouse Neural Stem Cell Culture.
Isolation of adult mouse NPCs was performed as previously described. Brains were dissected to remove the olfactory bulbs, cerebellum, and brainstem. An area encompassing the SVZ surrounding the lateral wall of the forebrain ventricle was dissected. Tissue was dissociated with accutase for 15 min at 37 °C and mechanically dissociated. Cells were plated onto laminin-precoated culture dish in expansion media [RHA-B media supplemented with 10 ng/mL of both basic FGF (bFGF) and EGF]. For neurosphere cultures, cells were plated at clonal density (20 × 103 cells per mL) and cultured for 7 d in vitro (DIV). For differentiation analysis, single cells were plated at 2.5 × 105 cells per mL on laminin-coated glass coverslips (Ø13 mm) in expansion media, before the subsequent withdrawal of bFGF and EGF growth factors to facilitate differentiation over 8 DIV (3 d without EGF followed by 8 d in the absence of growth factors).
ATP Levels and Lactate Production.
Total ATP levels were determined using the CellTiter-Glo Luminescent assay as per the manufacturer’s instruction (Promega). Lactate was measured using the Trinity Biotech lactate assay. Data are expressed as moles of ATP/relative ATP level (percentage) and milligrams per deciliter lactate, respectively, and all of the values were normalized to cells number.
Metabolic Profiling.
Metabolic profiles were obtained for each individual genotype using the Metabolon Platform (Metabolon), as described in ref. 47. Processing of each sample (six KO and five WT samples) was conducted using a proprietary series of organic and aqueous extractions to remove the protein fraction while allowing maximum recovery of small molecules. The resulting extract was divided into two fractions: one for analysis by liquid chromatography (LC) and one for analysis by GC. Samples were placed briefly on a TurboVap (Zymark) to remove the organic solvent. Samples were then frozen, vacuum dried, and then prepared for either LC/MS or GC/MS. Compounds above the detection threshold were identified by comparison with library entries of purified standards or recurrent unknown entities. Identification of known chemical entities was based on comparison with metabolomic library entries of purified standards.
Supplementary Material
Acknowledgments
We thank Salvador Moncada [University College London (UCL)], Doug Turnbull, Robert Taylor (University of Newcastle), Andrea Cossarizza (University of Modena and Reggio Emilia), Doug Green (St. Jude Childrens Hospital), Gerry Melino (MRC Toxicology Unit, Leicester), Sarah Ann-Martin (Barts Cancer Institute), Steven Pollard (Edinburgh University), and Pablo Rodriguez-Viciana and Asim Khwaja (UCL) for reagents, support, and critical discussion. Finally, we thank the UCL Scientific Services, the Cancer Genome Engineering facility, and the UCL Biological Services Unit. In the laboratory of P.S., this work was supported by Medical Research Council, by The Brain Tumour Charity (through a generous donation from the Brian Cross family), and by a donation from David Hunter and Wendy Tansey in memory of Peter Clark. S. Brandner and N.V.H. received support from University College London Hospitals Comprehensive Biomedical Research Centre and the Brain Tumour Charity. S. Brandner (Institute of Neurology, UCL) acknowledges the Neurosurgical Team at the National Hospital for their continued support of the brain tumor bank. V.G. was recipient of travel fellowships from Boehringer Ingelheim (2010) and European Molecular Biology Organization (2011). This work was also in part supported by Associazione Italiana per la Ricerca sul Cancro Investigator Grant 11450 and Ministero Sanità Ricerca Finalizzata (2009) (to V.D.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.R.G. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1413165112/-/DCSupplemental.
References
- 1.Frezza C, Gottlieb E. Mitochondria in cancer: Not just innocent bystanders. Semin Cancer Biol. 2009;19(1):4–11. doi: 10.1016/j.semcancer.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 2.Amary MF, et al. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J Pathol. 2011;224(3):334–343. doi: 10.1002/path.2913. [DOI] [PubMed] [Google Scholar]
- 3.Prensner JR, Chinnaiyan AM. Metabolism unhinged: IDH mutations in cancer. Nat Med. 2011;17(3):291–293. doi: 10.1038/nm0311-291. [DOI] [PubMed] [Google Scholar]
- 4.Reitman ZJ, et al. Profiling the effects of isocitrate dehydrogenase 1 and 2 mutations on the cellular metabolome. Proc Natl Acad Sci USA. 2011;108(8):3270–3275. doi: 10.1073/pnas.1019393108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cairns RA, Mak TW. Oncogenic isocitrate dehydrogenase mutations: Mechanisms, models, and clinical opportunities. Cancer Discov. 2013;3(7):730–741. doi: 10.1158/2159-8290.CD-13-0083. [DOI] [PubMed] [Google Scholar]
- 6.DeHaan C, et al. Mutation in mitochondrial complex I ND6 subunit is associated with defective response to hypoxia in human glioma cells. Mol Cancer. 2004;3:19. doi: 10.1186/1476-4598-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kirches E, et al. High frequency of mitochondrial DNA mutations in glioblastoma multiforme identified by direct sequence comparison to blood samples. Int J Cancer. 2001;93(4):534–538. doi: 10.1002/ijc.1375. [DOI] [PubMed] [Google Scholar]
- 8.Larman TC, et al. Cancer Genome Atlas Research Network Spectrum of somatic mitochondrial mutations in five cancers. Proc Natl Acad Sci USA. 2012;109(35):14087–14091. doi: 10.1073/pnas.1211502109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lueth M, et al. Somatic mitochondrial mutations in pilocytic astrocytoma. Cancer Genet Cytogenet. 2009;192(1):30–35. doi: 10.1016/j.cancergencyto.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 10.Marin-Valencia I, et al. Analysis of tumor metabolism reveals mitochondrial glucose oxidation in genetically diverse human glioblastomas in the mouse brain in vivo. Cell Metab. 2012;15(6):827–837. doi: 10.1016/j.cmet.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oliva CR, et al. Acquisition of temozolomide chemoresistance in gliomas leads to remodeling of mitochondrial electron transport chain. J Biol Chem. 2010;285(51):39759–39767. doi: 10.1074/jbc.M110.147504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vega A, et al. mtDNA mutations in tumors of the central nervous system reflect the neutral evolution of mtDNA in populations. Oncogene. 2004;23(6):1314–1320. doi: 10.1038/sj.onc.1207214. [DOI] [PubMed] [Google Scholar]
- 13.Kiebish MA, Han X, Cheng H, Seyfried TN. In vitro growth environment produces lipidomic and electron transport chain abnormalities in mitochondria from non-tumorigenic astrocytes and brain tumours. ASN Neuro. 2009;1(3):pii: e00011. doi: 10.1042/AN20090011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou Y, et al. Metabolic alterations in highly tumorigenic glioblastoma cells: Preference for hypoxia and high dependency on glycolysis. J Biol Chem. 2011;286(37):32843–32853. doi: 10.1074/jbc.M111.260935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones RG, Thompson CB. Tumor suppressors and cell metabolism: A recipe for cancer growth. Genes Dev. 2009;23(5):537–548. doi: 10.1101/gad.1756509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lane D, Levine A. p53 Research: The past thirty years and the next thirty years. Cold Spring Harb Perspect Biol. 2010;2(12):a000893. doi: 10.1101/cshperspect.a000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muller PA, Vousden KH. p53 mutations in cancer. Nat Cell Biol. 2013;15(1):2–8. doi: 10.1038/ncb2641. [DOI] [PubMed] [Google Scholar]
- 18.Alcantara Llaguno S, et al. Malignant astrocytomas originate from neural stem/progenitor cells in a somatic tumor suppressor mouse model. Cancer Cell. 2009;15(1):45–56. doi: 10.1016/j.ccr.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chow LM, et al. Cooperativity within and among Pten, p53, and Rb pathways induces high-grade astrocytoma in adult brain. Cancer Cell. 2011;19(3):305–316. doi: 10.1016/j.ccr.2011.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacques TS, et al. Combinations of genetic mutations in the adult neural stem cell compartment determine brain tumour phenotypes. EMBO J. 2010;29(1):222–235. doi: 10.1038/emboj.2009.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y, et al. Expression of mutant p53 proteins implicates a lineage relationship between neural stem cells and malignant astrocytic glioma in a murine model. Cancer Cell. 2009;15(6):514–526. doi: 10.1016/j.ccr.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng H, et al. p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature. 2008;455(7216):1129–1133. doi: 10.1038/nature07443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Llaguno SA, Chen J, Kwon CH, Parada LF. Neural and cancer stem cells in tumor suppressor mouse models of malignant astrocytoma. Cold Spring Harb Symp Quant Biol. 2008;73:421–426. doi: 10.1101/sqb.2008.73.005. [DOI] [PubMed] [Google Scholar]
- 24.Akman HO, et al. Thymidine kinase 2 (H126N) knockin mice show the essential role of balanced deoxynucleotide pools for mitochondrial DNA maintenance. Hum Mol Genet. 2008;17(16):2433–2440. doi: 10.1093/hmg/ddn143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bartesaghi S, et al. Loss of thymidine kinase 2 alters neuronal bioenergetics and leads to neurodegeneration. Hum Mol Genet. 2010;19(9):1669–1677. doi: 10.1093/hmg/ddq043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou X, et al. Progressive loss of mitochondrial DNA in thymidine kinase 2-deficient mice. Hum Mol Genet. 2008;17(15):2329–2335. doi: 10.1093/hmg/ddn133. [DOI] [PubMed] [Google Scholar]
- 27.Kim EL, et al. Comparative assessment of the functional p53 status in glioma cells. Anticancer Res. 2005;25(1A):213–224. [PubMed] [Google Scholar]
- 28.Li N, et al. Mitochondrial complex I inhibitor rotenone induces apoptosis through enhancing mitochondrial reactive oxygen species production. J Biol Chem. 2003;278(10):8516–8525. doi: 10.1074/jbc.M210432200. [DOI] [PubMed] [Google Scholar]
- 29.Muller FL, Liu Y, Van Remmen H. Complex III releases superoxide to both sides of the inner mitochondrial membrane. J Biol Chem. 2004;279(47):49064–49073. doi: 10.1074/jbc.M407715200. [DOI] [PubMed] [Google Scholar]
- 30.Ziech D, Franco R, Pappa A, Panayiotidis MI. Reactive oxygen species (ROS)—induced genetic and epigenetic alterations in human carcinogenesis. Mutat Res. 2011;711(1-2):167–173. doi: 10.1016/j.mrfmmm.2011.02.015. [DOI] [PubMed] [Google Scholar]
- 31.Sedelnikova OA, et al. Role of oxidatively induced DNA lesions in human pathogenesis. Mutat Res. 2010;704(1-3):152–159. doi: 10.1016/j.mrrev.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pollard SM, et al. Glioma stem cell lines expanded in adherent culture have tumor-specific phenotypes and are suitable for chemical and genetic screens. Cell Stem Cell. 2009;4(6):568–580. doi: 10.1016/j.stem.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 33.Luo X, Kraus WL. On PAR with PARP: Cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes Dev. 2012;26(5):417–432. doi: 10.1101/gad.183509.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stehling O, Elsässer HP, Brückel B, Mühlenhoff U, Lill R. Iron-sulfur protein maturation in human cells: Evidence for a function of frataxin. Hum Mol Genet. 2004;13(23):3007–3015. doi: 10.1093/hmg/ddh324. [DOI] [PubMed] [Google Scholar]
- 35.Thierbach R, et al. Specific alterations of carbohydrate metabolism are associated with hepatocarcinogenesis in mitochondrially impaired mice. Hum Mol Genet. 2012;21(3):656–663. doi: 10.1093/hmg/ddr499. [DOI] [PubMed] [Google Scholar]
- 36.Thierbach R, et al. Targeted disruption of hepatic frataxin expression causes impaired mitochondrial function, decreased life span and tumor growth in mice. Hum Mol Genet. 2005;14(24):3857–3864. doi: 10.1093/hmg/ddi410. [DOI] [PubMed] [Google Scholar]
- 37.Veatch JR, McMurray MA, Nelson ZW, Gottschling DE. Mitochondrial dysfunction leads to nuclear genome instability via an iron-sulfur cluster defect. Cell. 2009;137(7):1247–1258. doi: 10.1016/j.cell.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Le Belle JE, et al. Proliferative neural stem cells have high endogenous ROS levels that regulate self-renewal and neurogenesis in a PI3K/Akt-dependant manner. Cell Stem Cell. 2011;8(1):59–71. doi: 10.1016/j.stem.2010.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ottone C, et al. Direct cell-cell contact with the vascular niche maintains quiescent neural stem cells. Nat Cell Biol. 2014;16(11):1045–1056. doi: 10.1038/ncb3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kowaltowski AJ, Vercesi AE. Mitochondrial damage induced by conditions of oxidative stress. Free Radic Biol Med. 1999;26(3-4):463–471. doi: 10.1016/s0891-5849(98)00216-0. [DOI] [PubMed] [Google Scholar]
- 41.Moncada S, Erusalimsky JD. Does nitric oxide modulate mitochondrial energy generation and apoptosis? Nat Rev Mol Cell Biol. 2002;3(3):214–220. doi: 10.1038/nrm762. [DOI] [PubMed] [Google Scholar]
- 42.Choksi KB, Boylston WH, Rabek JP, Widger WR, Papaconstantinou J. Oxidatively damaged proteins of heart mitochondrial electron transport complexes. Biochim Biophys Acta. 2004;1688(2):95–101. doi: 10.1016/j.bbadis.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 43.Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: A dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Viros A, et al. Ultraviolet radiation accelerates BRAF-driven melanomagenesis by targeting TP53. Nature. 2014;511(7510):478–482. doi: 10.1038/nature13298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwartzbaum JA, Fisher JL, Aldape KD, Wrensch M. Epidemiology and molecular pathology of glioma. Nat Clin Pract Neurol. 2006;2(9):494–503; quiz 1 p following 516. doi: 10.1038/ncpneuro0289. [DOI] [PubMed] [Google Scholar]
- 46.Ostrom QT, et al. The epidemiology of glioma in adults: A “state of the science” review. Neuro Oncol. 2014;16(7):896–913. doi: 10.1093/neuonc/nou087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tufi R, et al. Enhancing nucleotide metabolism protects against mitochondrial dysfunction and neurodegeneration in a PINK1 model of Parkinson’s disease. Nat Cell Biol. 2014;16(2):157–166. doi: 10.1038/ncb2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.