Abstract
The key material for bioethanol production is cellulose, one of the main components of the plant cell wall. Enzymatic depolymerization of cellulose, an essential step in bioethanol production, can be accomplished by fungal and bacterial cellulases. Most of the biochemically characterized bacterial cellulases come from only a few of cellulose degrading bacteria thus limiting our knowledge of a range of cellulolytic activities that exist in nature. The recent explosion of genomic data offers a unique opportunity to search for novel cellulolytic activities; however, the absence of clear understanding of structural and functional features that are important for reliable computational identification of cellulases precludes their exploration in the genomic datasets. Here we explore the diversity of cellulases and propose a genomic approach to overcome this bottleneck.
Cellulose and cellulases
The dramatic rate of fossil fuels depletion and the resulting global economic and environmental consequences have spurred the search for alternative renewable energy sources such as biofuels. One of the promising materials for biofuel production is plant biomass [1], which contains large amounts of the sugar polymers cellulose (a polymer of β-1,4 linked glucose) and hemicelluloses (polymers composed of xylose, mannose, galactose, rhamnose, arabinose and other sugars [2]. These can be broken down by a mixture of enzymes into simple sugars that are fermentable to produce ethanol [3]. Although cellulose is largely present as crystalline fibers that are highly recalcitrant to hydrolysis, its biomass content is typically larger than that of hemicellulose [3] and consequently, cellulases are the key enzymes for bioethanol production. Individual cellulose polymers form rigid microfiber structures stabilized by inter- and intramolecular hydrogen bonds and van der Waals interactions between glucose residues in the fibers, which significantly contributes to its recalcitrance [4]. This network of bonds leads to a mostly uniform arrangement of fibers and the resulting crystalline cellulose lacks enzyme-accessible surface morphologies, further enhancing recalcitrance to hydrolysis [5].
All cellulases are glycoside hydrolase (GH) enzymes that utilize the same catalytic mechanism of acid-base catalysis with inversion or retention of glucose anomeric configuration [6]. There are two common types of the cellulase active sites. Glycoside hydrolyses with open (groove, cleft) active sites typically exhibit endocellulolytic activity (endocellulases), binding anywhere along the length of the cellulose molecule and hydrolyzing the β-1,4 glycosidic linkage, while those with tunnel-like active site exhibit exocellulolytic activity (cellobiohydrolases), binding at the ends of the cellulose molecule [7] and producing unit-length oligosaccharide products. Typically, exocellulases are processive enzymes, i.e. they are attached to the cellulose chain until it is completely hydrolyzed [7, 8], while endocellualses can be both processive and non-processive [7]. Efficiency of processive cellulases may greatly contribute to the rate-limiting step of cellulose hydrolysis [8]. Cellualses with endo- mode of action appear to be represented by a larger number of protein folds (Table 1). This indicates that endocellulases are either more evolutionary diverse or many novel exocellulases are yet to be found [9]. Many cellulases are multi-domain proteins and in addition to the catalytic domain have accessory domains such as carbohydrate binding modules (CBM) connected by a flexible linker [10]. The main role of CBM is to help cellulases bind cellulose, although it may also participate in initial disruption of cellulose fibers [11]. Cellulases preferentially bind to the amorphous or somewhat disordered (e.g. through acid pre-treatment) regions on the surface of the crystalline cellulose fiber [12]. Endocellulases (sometimes along with CBMs) help to disrupt the cellulose fibers and create accessible ends, while cellobiohydrolases continue the degradation by removing di- and oligosaccharides (usually 2–4 residues) from the ends of the disrupted cellulose fibers [13].
Table 1.
Examples of cellulases with endo- and exo- mode of action
| CAZy family | Accession Number | Fold | Mode of action | Reference |
|---|---|---|---|---|
| GH5 | Q47916 | (β/α) 8 | Endo | [19] |
| GH5 | CAB76938.1 | (β/α) 8 | Exo | [51] |
| GH6 | Q53488 | atypical β/α barrel | Endo | [52] |
| GH6 | AAA62211.1 | atypical β/α barrel | Exo | [53] |
| GH7 | P56680 | β-jelly roll | Endo | [54] |
| GH7 | A7LN91 | β-jelly roll | Exo | [27] |
| GH8* | AAA73867.1 | (α/α) 6 | Endo | [55] |
| GH9 | Q02934 | (α/α) 6 | Endo | [56] |
| GH9 | Q6RSN8 | (α/α) 6 | Exo | [57] |
| GH12* | O33897 | β-jelly roll | Endo | [25] |
| GH23* | 2XQO_A | α8 superhelical | Endo | [33] |
| GH44* | Q934F9 | (β/α) 8 | Endo | [58] |
| GH45* | Q9P868 | β6-barrel | Endo | [59] |
| GH48 | P37698 | (α/α) 6 | Endo | [60] |
| GH48 | Q8KKF7 | (α/α) 6 | Exo | [61] |
| GH51* | P77865 | (β/α) 8 | Endo | [26] |
| GH61* | O14405 | β-sandwich with an Ig-like topology. β9α5 | Endo | [62] |
| GH74* | Q9WYE1 | 7-fold β-propeller | Endo | [63] |
does not have confirmed cellualses with exo- mode of action in CAZy [30]
Lack of standards in cellulase enzymology
Several biochemical methods are commonly used to determine the substrate-specificity of cellulases and the endo-/exo- mode of action. The reducing sugar assays involve incubating purified enzyme with cellulose-containing substrates along with a binding reagent (i.e. dinitrosalicylic acid), which reacts with glucose, released during the incubation, to create fluorescent compounds, which are then detected spectrophotometrically [14]. In the halo assay, a gene coding for a putative cellulase is introduced into a non-cellulolytic microorganism, such as Escherichia coli, which is then grown on cellulose substrates stained with Congo Red. Colonies carrying cellulase genes are screened by formation of halo plaques resulting from degradation of the stained cellulose by the bacterial colony [15]. Viscosimetry and thin layer chromatography (TLC) assays are commonly used to determine exo- versus endo- mode of action, for example, exocellulases reduce viscosisty of solution of CMC much slower than endocellualses, while running incubation products of a cellulase on a gel will show whether shorter, such as glucose, cellobiose (exo- mode of action) or longer oligosaccharides, such as cellotriose, cellotetraose (endo- mode of action) are present [16, 17].
Adding to the challenge of biochemical characterization of cellulases is the multi-substrate specificity. Many of the biochemically confirmed cellulases are active on variety of substrates in addition to cellulose, such as xylan, lichenan, mannan. For example, cellulase Cel5E from Pseudomonas fluorescens was active on caboxymethyl cellulose (CMC), lichenan, Avicel (or microcrystalline cellulose) and ASC (acid-swollen cellulose) but completely inactive on xylan [18]. Cellulase CelG form Fibrobacter succinogenes belongs to the same GH family 5 but showed high activity on CMC and xylan and was completely inactive on Avicel and lichenan [19]. By contrast, some cellulases are active only on cellulose derivatives. For example, a GH family 5 cellulase cel5B from Termobifida fusca is able to degrade only cellulose-containing substrates (CMC, Avicel and MN300 (native fibrous cellulose), but is completely inactive on other substrates [20]. The vast majority of researchers use CMC degradation as indication of the cellulolytic activity. Therefore, here we consider documented CMC hydrolysis as the minimum requirement for an enzyme to be annotated as a biochemically confirmed cellulase. The multi-substrate specificity of cellulases and the persistent lack of data about activity on substrates other than CMC emphasize the need for adoption of a universal methodology for cellulase validation and characterization (Box 1).
Box 1. Current problems in cellulase studies and proposed solutions.
1. Experimental
-
1.1
Lack of standardization in the use of certain assays and substrates for experimental cellulase determination:
Devise a standard assay or a set of assays for unambiguous and reliable identification of cellulases
-
1.2
Poor taxonomic representation among experimentally studied organisms:
Obtain genome sequences and biochemically characterize potential cellulases from taxonomically diverse organisms
2. Computational
-
2.1
Cellulases are found in 12 unrelated protein families
Develop a natural classification system for each cellulase-containing protein family
-
2.2
There are multiple substrate specificities other than cellulose in each of the cellulase-containing families. There are no known genomic markers for cellulases. Current models for genomic identification of cellulases are not specific:
Identify class-specific genomic markers for cellulases
Develop sensitive, cellulase-specific models
Validate models via iterative experiment-computation approach
Known cellulolytic bacteria: a few of the many
Bacteria that are either known to be or potentially could be cellulolytic are widely distributed in nature. However, the best studied cellulose-degraders, such as Clostridium thermocellum, Clostridium cellulolyticum, Caldicellulosiruptor bescii (previously known as Anaerocellum thermophilum) belong to the same phylum, Firmicutes. Despite numerous studies of microbial cellulolytic apparatus [21–24], only about 20 genomes of known cellulose degraders have been fully sequenced so far. Recent genomic studies have identified many bacteria that contain arrays of various glycosyl hydrolases (many of which could be cellulases [21, 22]). Therefore it is likely that only a small fraction of the cellulolytic world has been annotated and studied to date and more experimental and genomic investigation of potential cellulase degraders from diverse taxa and habitats is needed.
CAZy database: a bridge from enzymology to genomics
The CAZy (Carbohydrate-Active Enzymes) database provides classification of enzymes (e.g. glycoside hydrolases, glycosyl transferases) and substrate-binding modules involved in various types of carbohydrate metabolism based on sequence comparison. All known cellulases are found within twelve GH families of the CAZy database and can be described with two enzyme commission numbers: EC 3.2.1.4 (endoglucanase) and EC 3.2.1.91 (cellobiohydorlase). Families GH5 and GH9 appear to have the largest number of biochemically characterized cellulases. This could be partly due to the fact that cellulases from these families are abundant in the model cellulolytic bacteria. Yet, many enzymes that effectively hydrolyse cellulose belong to other, smaller CAZy families, for example the Cel12A cellulase from Rhodothermus marinus (GH12) [25], endoglucanase F from F. succinogenes S85 (GH51) [26], and CbhI from Fusicoccum sp. (GH7) [27]. This indicates that the search for potential efficient cellulases should be substantially broadened.
Although the collection of the carbohydrate enzyme data in CAZy provides a very useful resource for enzymologists, annotations could be significantly improved. For example, the term ‘characterized’ in CAZy is applied equally to proteins that have been characterized biochemically and to those for which the functions have been predicted computationally. As we show, computational predictions for cellulases are currently unreliable; therefore knowing the source of information for annotation would be helpful. Nevertheless, CAZy provides a much needed connection between enzymology and genomics and can be considerably enhanced with improved computational models.
Challenges of genomic identification of cellulases
In order to search for cellulases in the ever increasing genomic and metagenomic data reliable sequence-based methods for their identification must be available. Current computational methodologies require that proteins should be conserved enough in sequence to carry out full-length sequence similarity searches (e.g. BLAST) or they should have specific markers, such as distinctive protein domains and domain combinations, motifs, accessory proteins, etc (see [28] for details) to yield reliable predictions. To illustrate the problems of genomic identification of cellulases, we compare their relevant features to those of another common enzyme involved in carbohydrate metabolism, hexokinase (the first enzyme of the glycolysis pathway). BLAST searches with a hexokinase seem quite reliable, whereas those with confirmed cellulases produce much more ambiguous results, where similar sequences could be annotated with a variety of definitions other than cellulase. Automated annotation of new genomes depends heavily on the identification of similar proteins by BLAST, so this ambiguity greatly complicates identification of potential novel cellulases.
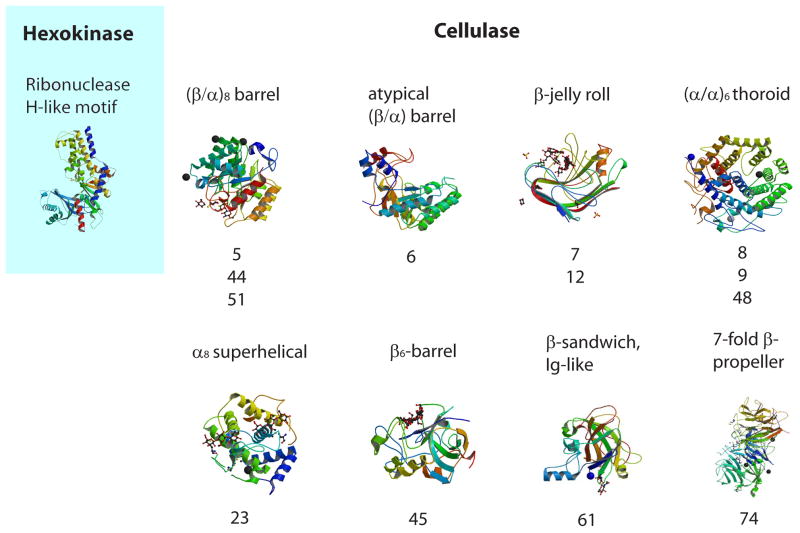
From a structural perspective, hexokinases belong to a single protein fold (Figure 1). All proteins that catalyze the ATP-dependent conversion of aldo- and keto-hexose sugars to the hexose-6-phosphate [29] have the same Ribonuclease H-like motif fold and belong to the same protein family ‘hexokinase’. On the other hand, proteins that catalyze the hydrolysis of the β-1,4 glucoside bond using the same mechanism of acid-base catalysis (cellulases) belong to at least 8 unrelated protein folds (Fig. 1) further differentiating into even more protein families [30]. For example, cellulase Cel5E from P. fluorescens has an (β/α)8 fold and belongs to GH family 5 [18] (family classification according to the CAZy database [30]), cellulase Egl-257 from Bacillus circulans has an (α/α)6 barrel fold and belongs to GH family 8 [31], cellulase cel44a from C. thermocellum has TIM-like barrel and β-sandwich domain fold and belongs to GH family 44 [32]. Recent biochemical and genomics studies identified cellulases in 11 or 13 CAZy families [9, 30, 33]. Cellulases therefore are representatives of a large class of non-homologous isofunctional enzymes [34], i.e. proteins catalyzing the same biochemical reaction that have evolved independently and are unrelated in sequence and structure. Therefore, in contrast to hexokinase, cellulases from each protein family must be treated as independent cases in any type of genomic analysis. This is a potential problem, which is easily solvable although it dramatically increases the amount of data analysis.
Figure 1.
Hexokinase and cellulases: structural conservation and diversity. Corresponding CAZy families are listed below structures. (images are taken from the RCSB PDB (www.pdb.org). The following labels correspond to PDB accession numbers: ‘Hexokinase’ - 1ig8[43]; ‘GH5, GH44, GH51’ - 1e5j[44]; ‘GH6’ - 2boe[45]; ‘GH7, GH12’ - 2jen[46]; ‘GH8, GH9, GH48’ - 1ia6[47]; ‘GH23’ – 2xqo[33]; ‘GH45’ - 4eng[48]; ‘GH61’ - 2vtc[49]; ‘GH74’ - 2cn2[50].
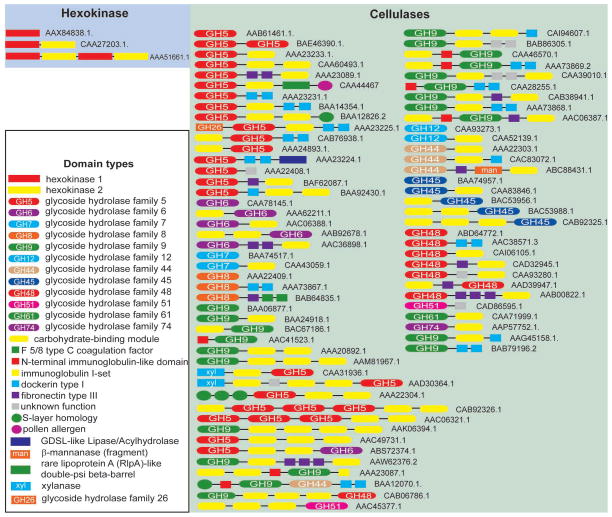
In addition to pairwise sequence similarity searches, the second powerful tool used in automated annotation is protein domain architecture identified using domain specific profile Hidden Markov Models (HMMs). HMMs are built from multiple sequence alignments and represent probabilities of certain amino acids to be located at certain position in a domain. Again, hexokinases can be easily distinguished from other enzymes based on their domain architecture (Figure 2). Nearly all hexokinases display a conserved combination of two protein domains termed “hexokinase_1” (Pfam accession PF00349) in the N-terminus and “hexokinase_2” (PF03727) in the C-terminus. Detection of these domains in any protein sequence unambiguously identifies it as a hexokinase. There is essentially no diversity in the domain architecture of hexokinases: less than 10% of sequences exhibit a duplicated version of the dual domain protein (Figure 2) and less than 1% contain other unrelated domains.
Figure 2.
Hexokinase and cellulases: conservation and diversity of domain architectures. Accession numbers for sequences are shown.
By contrast, identification of cellulases by domain architecture is problematic due to two characteristics. First, cellulases display an extremely wide diversity of domain architectures even within the same protein family (Figure 2). Second, and more critical, the HMMs currently available to recognize cellulases are built from multiple alignments that include both cellulases and similar in sequence non-cellulases and thus are not able to differentiate between members of the same protein family that have different substrate specificities. To illustrate this problem, we compare known activities of enzymes that belong to GH5 family (Pfam PF00150, Cellulase) to that of GH19 family (Pfam 00182, Glyco_hydro_19). GH19 is a large family (more than 1000 sequences in current databases) in which all 165 experimentally studied enzymes exhibit a single activity - chitinase (EC 3.2.1.14). GH5 family is comparable in size (just over 2000 sequences), however, among 373 experimentally studied enzymes from this family, at least 12 different activities other than cellulase have been reported (data from the CAZy database). Therefore, datasets retrieved with the current Cellulase domain model [35] may contain primarily non-cellulases and therefore would not be helpful to experimentalists.
Metagenomes: gold mines that need sluicing rather than panning
Metagenomic exploration of environments where lignocellulose is being effectively decomposed is the most promising path toward the discovery of novel cellulases. Recent advances in metagenomics resulted in generating genomic datasets from diverse environments, including fresh-water [36], the ocean [37], guts of insects [38], ruminants [39] and even the human intestine [40]. Such datasets have a tremendous potential to reveal novel cellulolytic capabilities. For example, the recent metagenomic study of a cow rumen has uncovered tens of thousands of putative cellulases [41] thus truly becoming a gold mine for their future exploration. However, the very same computational problems that we have outlined above prevented unambiguous identification of true cellulases in this dataset; investigators had to narrow down their list of targets for experimental validation randomly and the reported success rate was around 50% [41]. Clearly, a more efficient and cost effective method of mining is urgently needed.
Proposed computational solutions
Natural classification systems based on evolutionary relationships between sequences are instrumental in dealing with complex biological systems [28]. Because cellulases are found in protein families with different evolutionary histories and belong to different protein folds, the evolutionary path of each cellulase-containing protein family must be evaluated independently. To build a natural classification system for cellulases, classes must be defined using a phylogenomic approach, where related sequences of enzymatic domains are collected, properly aligned using available structural information and then clustered (e.g. via phylogenetic tree construction). Independent genomic markers, such as specific combinations of enzymatic and accessory domains, genome neighborhoods, etc, must be then identified for each individual class. In order to link biochemical activities to genomically identified classes, all available information on substrate-specificity of individual sequences must be mapped onto individual classes.
An effective natural classification scheme will assist in searching for novel cellulolytic activities in genomic datasets by identifying markers for that can be used to differentiate cellulases from related enzymes with different substrate specificity. While it is difficult to discern a pattern of accessory domains when looking at all sequences of a given GH family, focusing on a class of related proteins within a family may reveal specific accessory domains associated with that class. Most of the biochemically confirmed cellulases have carbohydrate-binding module domains (Figure 2), and cellulases with the same catalytic domains tend to more efficiently degrade recalcitrant crystalline cellulose in case if they contain a larger number of CBMs [27, 42]. Thus, identifying CBMs that are class-specific should be productive for better classification of the catalytic domains. Similarly, analysis of genome neighborhoods may reveal certain types of genes consistently found in proximity to genes encoding biochemically confirmed cellulases. Then the presence of these genes in proximity to genes encoding unknown GHs would suggest that it can be a cellulase (a guilty-by-association approach). Lastly, analysis of the aligned sequences can identify class-specific patterns of conserved amino acids, whose potential role in substrate specificity could be revealed by mapping onto available 3D structures and homology models. Aligned sequences of specific classes can also be turned into specific and sensitive domain models (e.g. HMM) for each of the catalytic domains or, where appropriate, for their combinations with auxiliary domains. Such models will become an essential tool, to search specifically for cellulases in ever-increasing genomic and metagenomic datasets. With new, refined models it should be possible to reduce the search space for cellulases by orders of magnitude and to provide experimentalists with a short list of enzymes that are more likely to be a true cellulase. Newly developed cellulase-specific models should be deposited to relevant databases (Pfam, CAZy) to ensure their availability to the scientific community.
The need for specific cellulase models is pressing. We now have hundreds of environmental sequencing samples that contain more than 1 billion sequences including datasets from such “cellulolytic” environments as termite gut [38] and cow rumen [41]. Together with still largely unexplored complete genomes of cellulose-degraders, metagenomic data creates a great reservoir for finding novel cellulolytic activities. There is also a need for a much closer collaboration between experimentalists and computational scientists in this area. The existing biochemical characterization has been performed on a small subset of closely related organisms; therefore a substantial number of experiments will be needed to fill gaps on substrate-specificity within newly identified classes of cellulase-containing families. Better standardization of cellulase assays and more thorough assessment of activity on a variety of carbohydrate polymers will greatly improve our ability to link sequence classes to enzyme activities (Box 1).
Conclusion
In conclusion, we would like to point out several contentious areas in practical biotechnology that might be addressed using computational genomics in the near future. First, there is a clear difference between enzymes in their ability to hydrolyze cellulose substrates, such as non-treated, raw and pre-treated plant material (e.g. switchgrass, wood pulp). Such differences could be due to inherent enzymatic domain properties (e.g. Kcat, product release) or associations with accessory domains that enhance substrate binding (e.g. CBM). Thus, one of the targets for computational studies would be associating the experimentally determined characteristics of various cellulases with both catalytic site conservation and the accessory domain architecture. The more enzymes with known sequence, structure and biochemical activities are available, the more powerful associations and therefore predictions could be made. The resulting data would be applicable to enzyme engineering as well to searches for better catalysts within a reduced sequence and structure space.
Second, many challenges are posed by the engineering of cellulases to be robust under harsh industrial settings (e.g. temperature, solvents, ionic conditions). Hence, better understanding the cellulase active site and enzymatic functions at the sequence level could enable protein engineering that would maintain catalytic properties while enhancing protein robustness.
Finally, a better communication between leading world cellulase researchers must be established in effort of standardization of both experimental and computational approaches to studies of cellulases. One way of accomplishing this goal would be creation of a freely available Internet resource that would include internationally accepted methodologies for biochemical and computational cellulase studies and a curated and updatable database of confirmed cellulases. To improve accessibility of such a resource, we recommend merging it with already existing web-resources, such as the CAZy database mentioned above.
Acknowledgments
The BioEnergy Science Center (BESC) is a U.S. Department of Energy Bioenergy Research Center supported by the Office of Biological and Environmental Research in the DOE Office of Science. We thank numerous colleagues across BESC for stimulating discussions.
List of literature
- 1.Lynd LR, et al. How biotech can transform biofuels. Nat Biotechnol. 2008;26:169–172. doi: 10.1038/nbt0208-169. [DOI] [PubMed] [Google Scholar]
- 2.Popper Z, et al. Evolution and Diversity of Plant Cell Walls: From Algae to Flowering Plants. Annu Rev Plant Biol. 2011;62:8.1–8.24. doi: 10.1146/annurev-arplant-042110-103809. [DOI] [PubMed] [Google Scholar]
- 3.Ragauskas AJ, et al. The path forward for biofuels and biomaterials. Science. 2006;311:484–489. doi: 10.1126/science.1114736. [DOI] [PubMed] [Google Scholar]
- 4.Cheng G, et al. Transition of cellulose crystalline structure and surface morphology of biomass as a function of ionic liquid pretreatment and its relation to enzymatic hydrolysis. Biomacromolecules. 2011;12:933–941. doi: 10.1021/bm101240z. [DOI] [PubMed] [Google Scholar]
- 5.Zhou W, et al. Cellulose hydrolysis in evolving substrate morphologies I: a general modeling formalism. Biotechnol Bioeng. 2009;104:261–274. doi: 10.1002/bit.22389. [DOI] [PubMed] [Google Scholar]
- 6.Davies G, Henrissat B. Structures and mechanisms of glycosyl hydrolases. Structure. 1995;3:853–859. doi: 10.1016/S0969-2126(01)00220-9. [DOI] [PubMed] [Google Scholar]
- 7.Kurasin M, Väljamäe P. Processivity of cellobiohydrolases is limited by the substrate. J Biol Chem. 2011;286:169–177. doi: 10.1074/jbc.M110.161059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beckham GT, et al. Applications of computational science for understanding enzymatic deconstruction of cellulose. Curr Opin Biotechnol. 2011;22:231–238. doi: 10.1016/j.copbio.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 9.Gilbert HJ. The Biochemistry and Structural Biology of plant cell wall deconstruction. Plant Physiol. 2010;153:444–455. doi: 10.1104/pp.110.156646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fontes CM, Gilbert HJ. Cellulosomes: highly efficient nanomachines designed to deconstruct plant cell wall complex carbohydrates. Annu Rev Biochem. 2010;79:655–681. doi: 10.1146/annurev-biochem-091208-085603. [DOI] [PubMed] [Google Scholar]
- 11.Shoseyov O, et al. Carbohydrate binding modules: biochemical properties and novel applications. Microbiol Mol Biol Rev. 2006;70:283–295. doi: 10.1128/MMBR.00028-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Canilha L, et al. A study on the pretreatment of a sugarcane bagasse sample with dilute sulfuric acid. J Ind Microbiol Biotechnol. 2011 doi: 10.1007/s10295-10010-10931-10292. [DOI] [PubMed] [Google Scholar]
- 13.White AR, Brown RM., Jr Enzymatic hydrolysis of cellulose: Visual characterization of the process. Proc Natl Acad Sci USA. 1981;78:1047–1051. doi: 10.1073/pnas.78.2.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem. 1959;31:426–428. [Google Scholar]
- 15.Teather RM, Wood PJ. Use of Congo-Red polysaccharide interactions in enumeration and characterization of cellulolytic bacteria from bovine rumen. Appl Environ Microbiol. 1982;43:777–782. doi: 10.1128/aem.43.4.777-780.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Irwin DC, et al. Activity studies of eight purified cellulases: specificity, synergism, and binding domain effects. Biotechnol Bioeng. 1993;42:1002–1013. doi: 10.1002/bit.260420811. [DOI] [PubMed] [Google Scholar]
- 17.Cohen R, et al. Processive endoglucanase active in crystalline cellulose hydrolysis by the brown rot basidiomycete Gloeophyllum trabeum. Appl Environ Microbiol. 2005;71:2412–2417. doi: 10.1128/AEM.71.5.2412-2417.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hall J, et al. The non-catalytic cellulose-binding domain of a novel cellulase from Pseudomonas fluorescens subsp. cellulosa is important for the efficient hydrolysis of Avicel. Biochem J. 1995;309:749–756. doi: 10.1042/bj3090749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iyo AH, Forsberg CW. Endoglucanase G from Fibrobacter succinogenes S85 belongs to a class of enzymes characterized by a basic C-terminal domain. Can J Microbiol. 1996;42:934–943. doi: 10.1139/m96-120. [DOI] [PubMed] [Google Scholar]
- 20.Posta K, et al. Cloning, characterization and phylogenetic relationships of cel5B, a new endoglucanase encoding gene from Thermobifida fusca. J Basic Microbiol. 2004;44:383–399. doi: 10.1002/jobm.200410422. [DOI] [PubMed] [Google Scholar]
- 21.Weiner RM, et al. Complete genome sequence of the complex carbohydrate-degrading marine bacterium, Saccharophagus degradans strain 2-40 T. PLoS Genet. 2008;4:e1000087. doi: 10.1371/journal.pgen.1000087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barabote RD, et al. Complete genome of the cellulolytic thermophile Acidothermus cellulolyticus 11B provides insights into its ecophysiological and evolutionary adaptations. Genome Res. 2009;19:1033–1043. doi: 10.1101/gr.084848.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moraïs S, et al. Cellulase-xylanase synergy in designer cellulosomes for enhanced degradation of a complex cellulosic substrate. MBio. 2010;1:e00285–00210. doi: 10.1128/mBio.00285-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rincon MT, et al. Abundance and diversity of dockerin-containing proteins in the fiber-degrading rumen bacterium, Ruminococcus flavefaciens FD-1. PLoS One. 2010;5:e12476. doi: 10.1371/journal.pone.0012476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crennell SJ, et al. The structure of Rhodothermus marinus Cel12A, A highly thermostable family 12 endoglucanase, at 1.8 A° resolution. J Mol Biol. 2002;320:883–897. doi: 10.1016/s0022-2836(02)00446-1. [DOI] [PubMed] [Google Scholar]
- 26.Malburg SR, et al. Catalytic properties of the cellulose-binding endoglucanase F from Fibrobacter succinogenes S85. Appl Environ Microbiol. 1997;63:2449–2453. doi: 10.1128/aem.63.6.2449-2453.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanokratana P, et al. Identification and expression of cellobiohydrolase (CBHI) gene from an endophytic fungus, Fusicoccum sp. (BCC4124) in Pichia pastoris. Protein Expr Purif. 2008;58:148–153. doi: 10.1016/j.pep.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 28.Wuichet K, Zhulin IB. Origins and diversification of a complex signal transduction system in prokaryotes. Sci Signal. 2010;3:ra50. doi: 10.1126/scisignal.2000724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Finn RD, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cantarel BL, et al. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2009;37:D233–238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hakamada Y, et al. Enzymatic properties, crystallization, and deduced amino acid sequence of an alkaline endoglucanase from Bacillus circulans. Biochim Biophys Acta. 2002;1570:174–180. doi: 10.1016/s0304-4165(02)00194-0. [DOI] [PubMed] [Google Scholar]
- 32.Kitago Y, et al. Crystal structure of Cel44A, a glycoside hydrolase family 44 endoglucanase from Clostridium thermocellum. J Biol Chem. 2007;282:35703–35711. doi: 10.1074/jbc.M706835200. [DOI] [PubMed] [Google Scholar]
- 33.Bras JLA, et al. Structural insights into a unique cellulase fold and mechanism of cellulose hydrolysis. Proc Natl Acad Sci USA. 2011;108:5237–5242. doi: 10.1073/pnas.1015006108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Omelchenko MV, et al. Non-homologous isofunctional enzymes: a systematic analysis of alternative solutions in enzyme evolution. Biol Direct. 2010;5:31. doi: 10.1186/1745-6150-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou F, et al. Large-scale analyses of glycosylation in cellulases. Genomics Proteomics Bioinformatics. 2009;7:194–199. doi: 10.1016/S1672-0229(08)60049-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Debroas D, et al. Metagenomic approach studying the taxonomic and functional diversity of the bacterial community in a mesotrophic lake (Lac du Bourget--France) Environ Microbiol. 2009;9:2412–2424. doi: 10.1111/j.1462-2920.2009.01969.x. [DOI] [PubMed] [Google Scholar]
- 37.Yooseph S, et al. The Sorcerer II Global Ocean Sampling expedition: expanding the universe of protein families. PLoS Biol. 2007;5:e16. doi: 10.1371/journal.pbio.0050016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Warnecke F, et al. Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature. 2007;450:560–565. doi: 10.1038/nature06269. [DOI] [PubMed] [Google Scholar]
- 39.Brulc JM, et al. Gene-centric metagenomics of the fiber-adherent bovine rumen microbiome reveals forage specific glycoside hydrolases. Proc Natl Acad Sci USA. 2009;106:1948–1953. doi: 10.1073/pnas.0806191105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hess M, et al. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science. 2011;331:463–467. doi: 10.1126/science.1200387. [DOI] [PubMed] [Google Scholar]
- 42.Baba Y, et al. Alternative splicing produces two endoglucanases with one or two carbohydrate-binding modules in Mucor circinelloides. J Bacteriol. 2005;187:3045–3051. doi: 10.1128/JB.187.9.3045-3051.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuser PR, et al. The high resolution crystal structure of yeast hexokinase PII with the correct primary sequence provides new insights into its mechanism of action. J Biol Chem. 2000;275:20814–20821. doi: 10.1074/jbc.M910412199. [DOI] [PubMed] [Google Scholar]
- 44.Fort S, et al. Mixed-linkage cellooligosaccharides: a new class of glycoside hydrolase inhibitors. Chem biochem. 2001;2:319–325. doi: 10.1002/1439-7633(20010504)2:5<319::AID-CBIC319>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 45.Larsson AM, et al. Crystal structure of Thermobifida fusca endoglucanase Cel6A in complex with substrate and inhibitor: the role of tyrosine Y73 in substrate ring distortion. Biochemistry. 2005;44:12915–12922. doi: 10.1021/bi0506730. [DOI] [PubMed] [Google Scholar]
- 46.Gloster TM, et al. Characterization and three-dimensional structures of two distinct bacterial xyloglucanases from families Gh5 and Gh12. J Biol Chem. 2007;282:19177–19189. doi: 10.1074/jbc.M700224200. [DOI] [PubMed] [Google Scholar]
- 47.Parsiegla G, et al. Crystal structure of the cellulase Cel9M enlightens structure/function relationships of the variable catalytic modules in glycoside hydrolases. Biochemistry. 2002;41:11134–11142. doi: 10.1021/bi025816m. [DOI] [PubMed] [Google Scholar]
- 48.Davies GJ, et al. Structure determination and refinement of the Humicola insolens endoglucanase V at 1.5 A resolution. Acta Crystallogr D Biol Crystallogr. 1996;52:7–17. doi: 10.1107/S0907444995009280. [DOI] [PubMed] [Google Scholar]
- 49.Karkehabadi S, et al. The first structure of a glycoside hydrolase family 61 member, Cel61B from the Hypocrea jecorina, at 1.6 A resolution. J Mol Biol. 2008;383:144–154. doi: 10.1016/j.jmb.2008.08.016. [DOI] [PubMed] [Google Scholar]
- 50.Martinez-Fleites C, et al. Crystal structures of Clostridium thermocellum xyloglucanase, Xgh74A, reveal the structural basis for xyloglucan recognition and degradation. J Biol Chem. 2006;281:24922–24933. doi: 10.1074/jbc.M603583200. [DOI] [PubMed] [Google Scholar]
- 51.Zverlov VV, et al. A newly described cellulosomal cellobiohydrolase, CelO, from Clostridium thermocellum: investigation of the exo-mode of hydrolysis, and binding capacity to crystalline cellulose. Microbiology. 2002;148:247–255. doi: 10.1099/00221287-148-1-247. [DOI] [PubMed] [Google Scholar]
- 52.Lin F, et al. Cloning and sequencing of an endo-beta-1,4-glucanase gene mcenA from Micromonospora cellulolyticum 86W-16. J Ind Microbiol. 1994;13:344–350. doi: 10.1007/BF01577217. [DOI] [PubMed] [Google Scholar]
- 53.Zhang S, et al. Characterization of a Thermomonospora fusca exocellulase. Biochemistry. 1995;34:3386–3395. doi: 10.1021/bi00010a030. [DOI] [PubMed] [Google Scholar]
- 54.Davies GJ, et al. Oligosaccharide specificity of a family 7 endoglucanase: insertion of potential sugar-binding subsites. J Biotechnol. 1997;57:91–100. doi: 10.1016/s0168-1656(97)00092-8. [DOI] [PubMed] [Google Scholar]
- 55.Fierobe HP, et al. Purification and characterization of endoglucanase C from Clostridium cellulolyticum. Catalytic comparison with endoglucanase A. Eur J Biochem. 1993;217:557–565. doi: 10.1111/j.1432-1033.1993.tb18277.x. [DOI] [PubMed] [Google Scholar]
- 56.Hazlewood GP, et al. Gene sequence and properties of CelI, a family E endoglucanase from Clostridium thermocellum. J Gen Microbiol. 1993;139:307–316. doi: 10.1099/00221287-139-2-307. [DOI] [PubMed] [Google Scholar]
- 57.Schubot FD, et al. Structural basis for the exocellulase activity of the cellobiohydrolase CbhA from Clostridium thermocellum. Biochemistry. 2004;43:1163–1170. doi: 10.1021/bi030202i. [DOI] [PubMed] [Google Scholar]
- 58.Rincon MT, et al. EndB, a newly identified family 44 cellulase from the rumen cellulolytic bacterium Ruminococcus flavefaciens 17, binds to cellulose via a novel cellulose binding domain and to a 130kDa R. flavefaciens protein via a dockerin domain. Appl Environ Microbiol. 2001;67:4426–4431. doi: 10.1128/AEM.67.10.4426-4431.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eberhardt RY, et al. Primary sequence and enzymic properties of two modular endoglucanases, Cel5A and Cel45A, from the anaerobic fungus Piromyces equi. Microbiology. 2000;146:1999–2008. doi: 10.1099/00221287-146-8-1999. [DOI] [PubMed] [Google Scholar]
- 60.Reverbel-Leroy C, et al. Molecular study and overexpression of the Clostridium cellulolyticum celF cellulase gene in Escherichia coli. Microbiology. 1996;142:1013–1023. doi: 10.1099/00221287-142-4-1013. [DOI] [PubMed] [Google Scholar]
- 61.Sanchez MM, et al. Exo-mode of action of cellobiohydrolase Cel48C from Paenibacillus sp. BP-23. Eur J Biochem. 2003;270:2913–2919. doi: 10.1046/j.1432-1033.2003.03673.x. [DOI] [PubMed] [Google Scholar]
- 62.Karlsson J, et al. Homologous expression and characterization of Cel61A (EG IV) of Trichoderma reesei. Eur J Biochem. 2001;268:6498–6507. doi: 10.1046/j.0014-2956.2001.02605.x. [DOI] [PubMed] [Google Scholar]
- 63.Chhabra SR, et al. Regulation of endo-acting glycosyl hydrolases in the hyperthermophilic bacterium Thermotoga maritima grown on glucan- and mannan-based polysaccharides. Appl Environ Microbiol. 2001;68:545–554. doi: 10.1128/AEM.68.2.545-554.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]