Abstract
IMPORTANCE
Human papillomavirus type 16 (HPV-16) is a major causative factor in oropharyngeal squamous cell carcinoma (OPSCC). The detection of primary OPSCC is often delayed owing to the challenging anatomy of the oropharynx.
OBJECTIVE
To investigate the feasibility of HPV-16 DNA detection in pretreatment and posttreatment plasma and saliva and its potential role as a marker of prognosis.
DESIGN, SETTING, AND PARTICIPANTS
This is a retrospective analysis of a prospectively collected cohort. Among a cohort of patients with oropharyngeal and unknown primary squamous cell carcinoma with known HPV-16 tumor status from the Johns Hopkins Medical Institutions and Greater Baltimore Medical Center (from 1999 through 2010), 93 patients were identified with a complete set of pretreatment and posttreatment plasma or saliva samples, of which 81 patients had HPV-16–positive tumors and 12 patients had HPV-16–negative tumors. Real-time quantitative polymerase chain reaction was used to detect HPV-16 E6 and E7 DNA in saliva and plasma samples.
MAIN OUTCOMES AND MEASURES
Main outcomes included sensitivity, specificity, negative predictive value of combined saliva and plasma pretreatment HPV-16 DNA status for detecting tumor HPV-16 status, as well as the association of posttreatment HPV DNA status with clinical outcomes, including recurrence-free survival and overall survival.
RESULTS
The median follow-up time was 49 months (range, 0.9–181.0 months). The sensitivity, specificity, negative predictive value, and positive predictive value of combined saliva and plasma pretreatment HPV-16 DNA status for detecting tumor HPV-16 status were 76%, 100%, 42%, and 100%, respectively. The sensitivities of pretreatment saliva or plasma alone were 52.8%and 67.3%, respectively. In a multivariable analysis, positive posttreatment saliva HPV status was associated with higher risk of recurrence (hazard ratio [HR], 10.7; 95% CI, 2.36–48.50) (P = .002). Overall survival was reduced among those with posttreatment HPV-positive status in saliva (HR, 25.9; 95% CI, 3.23–208.00) (P = .002) and those with HPV-positive status in either saliva or plasma but not among patients with HPV-positive status in plasma alone. The combined saliva and plasma posttreatment HPV-16 DNA status was 90.7%specific and 69.5%sensitive in predicting recurrence within 3 years.
CONCLUSIONS AND RELEVANCE
Using a combination of pretreatment plasma and saliva can increase the sensitivity of pretreatment HPV-16 status as a tool for screening patients with HPV-16–positive OPSCC. In addition, analysis of HPV-16 DNA in saliva and plasma after primary treatment may allow for early detection of recurrence in patients with HPV-16–positive OPSCC.
While the overall incidence of head and neck cancer is decreasing in the United States, recognized cases of oropharyngeal squamous cell carcinoma (OPSCC) are on the rise. This is predominantly owing to an epidemic of oropharyngeal cancer related to high-risk human papillomavirus (HPV). Prior studies cite a rising proportion of OPSCC cases related to HPV, with literature supporting 50% or greater being HPV-16 related.1–3 Recently, oral HPV infection has been shown to have a prevalence of 7% in the general population with a bimodal distribution.4 Oral HPV infection is more prevalent in the male compared with female population, with a prevalence ratio of 2.3 and a peak incidence of up to 10% in men aged 55 to 64 years. Within the general population, approximately 1% are infected with the high-risk subtype HPV-16.4 In addition, both retrospective and prospective studies have demonstrated an improved overall survival in HPV-16–positive OPSCC vs HPV-16–negative OPSCC counterparts; an outcome believed to hold true for both surgical and nonsurgical treatment modalities.2,5,6
The detection of primary OPSCC and recurrence following completion of therapy is often delayed because of the challenging anatomy of the areas of the oropharynx that can harbor tumor. Thus, development of a surveillance tool for OPSCC may allow for earlier detection of recurrent lesions and further improve outcomes in this subset of patients. Studies have shown that high-risk HPV-16 integration results in production of the viral oncoproteins E6 and E7, which promote tumor progression by inactivating the p53 and retinoblastoma tumor suppressor gene products.7–9 Furthermore, previous studies have shown the feasibility of quantitative polymerase chain reaction (PCR) in detecting E6 and E7 from oral salivary rinses as well as serum and suggested its use in disease surveillance for HPV-16–related OPSCC.10–12
Due to the high prevalence of oral HPV infection in the population, we investigated the role of HPV-16 DNA detection as a biomarker for OPSCC disease status. The aim of our study was to evaluate the HPV-16 status in salivary and plasma samples of patients with OPSCC using quantitative PCR for HPV-16 E6 and E7 DNA and correlate the results with disease outcome.
Methods
Study Patients
The study protocol was approved by the institutional review board of the Johns Hopkins Hospital. The Johns Hopkins Head and Neck database was queried for patients with head and neck squamous cell carcinoma (HNSCC) of unknown primary or of the oropharynx. The initial cohort included 158 patients from both Johns Hopkins Hospital and Greater Baltimore Medical Center, Baltimore, Maryland, from1999 through 2010. Subsequently, 93 patients were identified who had a complete set of pretreatment and posttreatment saliva or plasma samples, had documented HPV-16 tumor status or tumor samples available for analysis of HPV-16 status, and had provided written informed consent for the study. The remaining 65 patients were excluded owing to missing pretreatment or posttreatment saliva and/or plasma samples. All patients had undergone primary treatment with curative intent.
Tissue, Saliva, and Plasma
Tumor samples, salivary rinses, and plasma samples of 93 patients were obtained. Pretreatment oral salivary rinses were performed as previously described (Chuang et al12) with 20 mL of normal saline gargled twice, for 20 and 10 seconds, respectively. Pretreatment blood samples were collected in citrate-containing blood tubes for plasma. Surveillance salivary rinses and plasma samples were obtained in the office at a follow-up clinic at 3-month intervals, starting 3 months after completion of therapy. All posttreatment samples of patients who developed recurrence were collected prior to the date of recurrence detection.
DNA Extraction
Microdissected tissues and saliva were extracted using techniques described previously.13 Plasma samples were centrifuged at 1000 rpm for 10 minutes, and the top layer was collected. DNA from microdissected fresh tissues, plasma, and saliva were extracted with phenol-chloroform, precipitated in 100% ethanol, centrifuged at 5100 rpm for 45 minutes, washed in 70% ethanol twice, dissolved in LoTE buffer (10mM TRIS hydrochloride, 1mM EDTA buffer, pH 8), and stored at −20°C. The samples were diluted to 50 ng in each reaction and analyzed by quantitative PCR (Taqman HT 7900; Applied Biosystems).
Quantitative PCR
Quantitative PCR was performed for HPV E6, E7, and β-actin using the Taqman HT 7900 system. Specific primers and probes previously designed to amplify the E6 and E7 regions of HPV-16 were used.
E6 forward primer, 5′-TCAGGACCCACAGGAGCG-3′; E6 reverse primer, 5′-CCTCACGTCGCAGTAACTGTTG-3′; E6 Taqman probe, 5′-(FAM)-CCCAGAAAGTTACCACAGTTATGCACAGAGCT-(TAMRA)-3′.
E7 forward primer, 5′-CCGGACAGAGCCCATTACAA-3′; E7 reverse primer, 5′-CGAATGTCTACGTGTGTGCTTTG-3′; E7 Taqman probe, 5′-(FAM)-CGCACAACCGAAGCGTAGAGTCACACT-(TAMRA)-3′.
β-actin forward primer, 5′-TCACCCACACTGTGCCCATCTACGA-3′; β-actin reverse primer, 5′-CAGCGGAACCGCTCATTGCCAATGG-3′; and β-actin TagMan probe, 5′-(FAM)-ATGCCCTCCCCCATGCCATCCTGCGT-(TAMRA)-3′.
All samples were run in triplicates. Standard curves for the HPV-16 viral copy number were developed using the CaSki (American Type Culture Collection) cell line. This cell line is known to have 600 copies per genome of HPV-16 DNA. Standard curves were developed for HPV-16 E6 and E7 using DNA extracted from CaSki cells, serially diluted into 50 ng, 5 ng, 0.5 ng, 0.05 ng, and 0.005 ng. β-actin was used as a housekeeping gene (2 copies per genome), and a standard curve was developed using above concentrations of DNA extracted from CaSki cells.
Data Analysis
Descriptive statistics were used to summarize the characteristics of the study population. The differences in demographics of the patient population based on tumor HPV-16 status were analyzed using an exact χ2 test. Tumor samples with 0.1 copy or more per genome and salivary and plasma samples with 0.001 copy or more genome were considered as HPV-16 positive. When using the combined saliva and plasma method to determine HPV-16 status, we considered it positive if either the saliva was positive alone, the plasma was positive alone, or both the saliva and the plasma were positive simultaneously. Sensitivity, specificity, positive predictive value, and negative predictive value were estimated on pretreatment samples in detection of tumor HPV-16 status.
The associations of HPV status in saliva and plasma and recurrence-free survival (RFS) or overall survival (OS) in patients with HPV-positive tumors were assessed with univariate and multivariable Cox proportional hazard models. Recurrence-free survival was defined as the time from the date of completion of treatment to the date of disease recurrence and was censored at the earliest of date of last follow-up or death if the patient did not have recurrent disease. Overall survival was defined as the time from the date of completion of treatment to the date of death from any cause. For patients who did not die, OS was censored at their last follow-up date. Hazard ratios (HRs) and 95% CIs were computed with Cox proportional hazard models. Overall survival and RFS were summarized using Kaplan-Meier curves. Time-dependent sensitivity and specificity for cumulative disease recurrence were estimated using a semiparametric approach that allows the censoring process to depend on the marker.14 All statistical tests were 2-sided and considered statistically significant at P < .05. The analysis was performed using statistical software R (version 2.15.1) and SAS (version 9.3, SAS Institute Inc).
Results
Study Population
We examined 93 patients with OPSCC or unknown primary SCC who had pretreatment and posttreatment samples of saliva and/or plasma using quantitative PCR. Demographics of the cohort and the risk factors of the patients grouped by HPV-16 status are summarized in Table 1. Of 93 patients, 81 were determined to have an HPV-16–positive tumor based on in situ hybridization or HPV-16 detection by quantitative PCR. The tumor HPV status of the 6 patients with unknown primary SCC was determined using the metastatic neck specimen. The 2 groups were similar with respect to age, sex, and site of primary tumor. However, there was higher percentage of nonsmokers in the HPV-16–positive patients. Patients with HPV-16–positive disease also presented with higher N classification (N2 or N3).With a median follow-up time of 49 months (range, 0.9–181.0 months), 19 of 93 patients (20%) developed recurrence after completing their treatment, with 14 patients with HPV-16–positive tumors and 5 patients with HPV-16–negative tumors. None of the patients with unknown primary SCC developed recurrence.
Table 1.
Patient Demographics and Tumor Characteristics by HPV-16 Status
| Characteristic | Total Patients, No. (%) (N = 93) |
Tumor HPV Status, No. (%) |
P Value | |
|---|---|---|---|---|
| Positive (n = 81) |
Negative (n = 12) |
|||
| Sex | ||||
| Male | 81 (87.1) | 72 (88.9) | 9 (75.0) | .18 |
| Female | 12 (12.9) | 9 (11.1) | 3 (25.0) | |
| Age, y | ||||
| >60 | 28 (12.9) | 23 (28.4) | 5 (41.7) | .50 |
| ≤60 | 65 (87.1) | 58 (71.6) | 7 (58.3) | |
| Race | ||||
| White | 86 (92.5) | 77 (95.1) | 9 (75.0) | .008 |
| African American | 4 (4.3) | 1 (1.2) | 3 (25.0) | |
| Other | 3 (3.2) | 3 (3.7) | 0 | |
| Tumor site | ||||
| Oropharynx | 87 (93.5) | 75 (92.6) | 12 (100) | >.99 |
| Unknown primary | 6 (6.5) | 6 (7.4) | 0 | |
| Drinking status | ||||
| Never | 59 (63.4) | 54 (66.7) | 5 (41.7) | .17 |
| Former | 7 (7.5) | 5 (6.2) | 2 (16.7) | |
| Current | 26 (27.9) | 21 (25.9) | 5 (41.7) | |
| Unknown | 1 (1.1) | 1 (1.2) | 0 | |
| Smoking status | ||||
| Never | 38 (40.9) | 36 (44.4) | 2 (16.7) | .02 |
| Former | 35 (37.6) | 31 (38.3) | 4 (33.3) | |
| Current | 19 (20.4) | 13 (16.1) | 6 (50.0) | |
| Unknown | 1 (1.1) | 1 (1.2) | 0 | |
| Primary therapy | ||||
| Radiation only | 2 (2.1) | 1 (1.2) | 1 (8.3) | .27 |
| Surgery only | 58 (62.4) | 50 (61.7) | 8 (66.7) | |
| Chemoradiation | 33 (35.5) | 30 (37.0 | 3 (25.0) | |
| T classification | ||||
| T0/T1/T2 | 76 (81.7) | 67 (82.7) | 9 (75.0) | .46 |
| T3/T4 | 17 (18.3) | 14 (17.3) | 3 (25.0) | |
| N classification | ||||
| N0/N1 | 14 (15.1) | 9 (11.1) | 5 (41.7) | .02 |
| N2/N3 | 79 (84.9) | 72 (88.9) | 7 (58.3) | |
Abbreviations: HPV, human papillomavirus; N, nodal; T, tumor.
Sensitivities, Specificities, and Predictive Values for Detection of Tumor HPV-16 Status
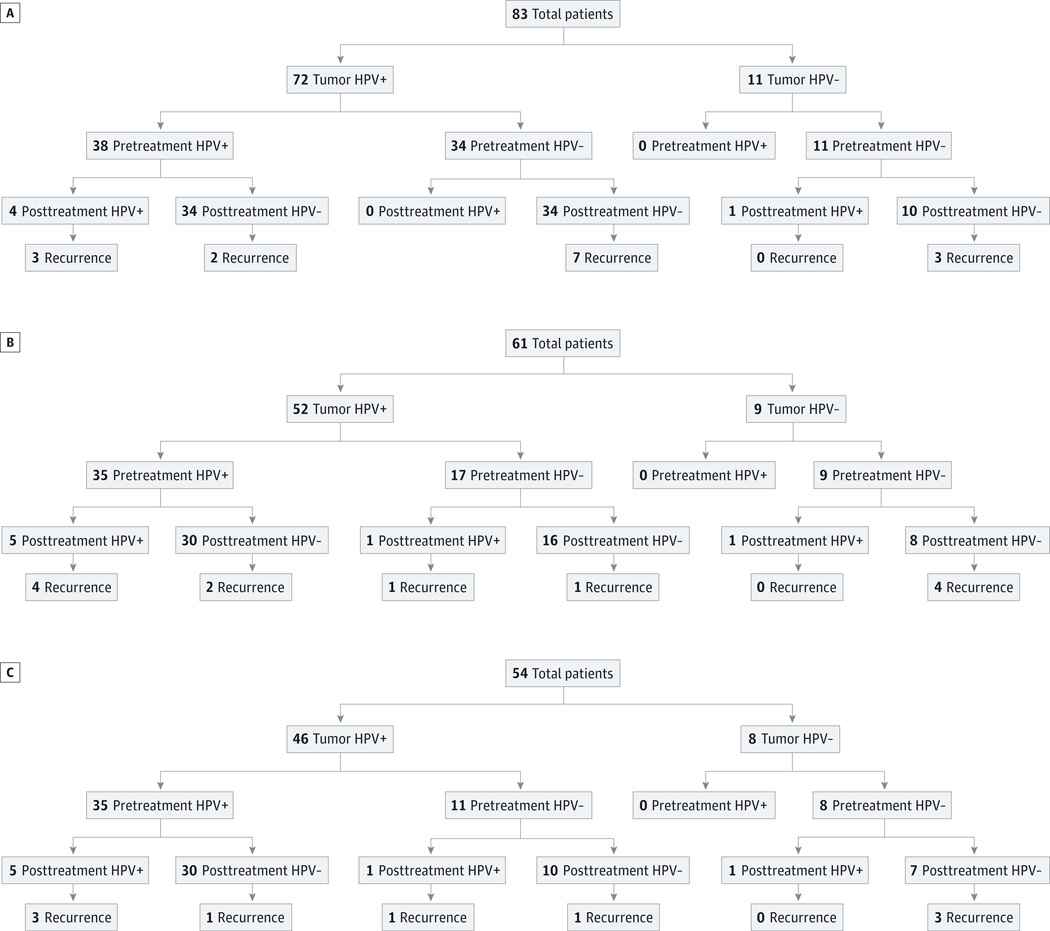
Patients with HPV-16–positive and HPV-16–negative tumors were categorized into 3 groups for analysis: those with pretreatment and posttreatment saliva samples, those with pretreatment and posttreatment plasma samples, and those with parallel pretreatment and posttreatment saliva and plasma samples. Subsequently, tumor HPV-16 status and treatment outcomes were correlated with HPV-16 status determined by quantitative PCR in either saliva samples, plasma samples, or both. Figure 1 demonstrates the breakdown of our patient population in 3 groups by pretreatment screening sample HPV-16 status, posttreatment surveillance sample HPV-16 status, and disease recurrence. Overall, there were 83 patients who had a complete set of pretreatment and posttreatment saliva samples (Figure 1A). Seventy-two patients (87%) had HPV-16–positive tumors and 11 patients (13%) had HPV-16–negative tumors. No HPV-16 DNA was detected by quantitative PCR in pretreatment saliva samples from patients with HPV-16–negative tumors (specificity, 100%). Of the 11 patients with HPV-16–negative tumors, 1 patient had HPV-16 DNA detected in posttreatment saliva sample, and this patient did not go on to develop recurrence with a follow-up duration of 7 years. Of the 72 patients with HPV-16–positive tumor, 38 patients had HPV-16 DNA detected in pretreatment saliva samples, with a sensitivity of 53%. Of 38 patients with HPV-16–positive pretreatment saliva, 4 had HPV-16 DNA detected in posttreatment saliva samples, and 3 of these 4 patients developed recurrence with a follow-up time range of 17.0 to 53.0 months. The remaining 34 patients with HPV-16–positive pretreatment saliva had HPV-16–negative posttreatment saliva samples, and 2 of these patients developed recurrence with a follow-up time range of 3.4 to 111.0 months. Finally, 7 patients with HPV-16–positive tumors who did not have detectable HPV-16 DNA in pretreatment saliva samples or posttreatment saliva samples developed recurrence with a follow-up time range of 15.5 to 97.0 months.
Figure 1. Layout of Patient Groups by Human Papillomavirus Type 16 Status.
A, Saliva pretreatment and posttreatment samples; B, plasma pretreatment and posttreatment samples; and C, combined saliva and plasma pretreatment and posttreatment samples. HPV+ indicates human papillomavirus positive; and HPV−, human papillomavirus negative.
Figure 1B illustrates the layout of the patient population with a complete set of pretreatment and posttreatment plasma samples by tumor HPV-16 status and their disease status. There were a total of 61 patients in this group: 52 patients (85%) with HPV-16–positive tumors and 9 patients (15%) with HVP-16– negative tumors. Once again, no patient with HPV-16 negative tumor had HPV-16 DNA detected in screening plasma samples collected pretreatment (specificity, 100%).Of the 9 patients with HPV-16–negative tumors, 1 again had HPV-16 DNA detected in posttreatment plasma sample and did not have recurrence with a follow-up duration of 7 years. Of 52 patients with HPV-16–positive tumors, 35 had detectable HPV-16 DNA in pretreatment plasma, making the sensitivity of screening HPV-16 status in plasma 67%. Of the 35 patients with HPV-16–positive pretreatment plasma, 5 had HPV-16 detected in posttreatment plasma samples, and 4 of these went on to develop recurrence with a follow-up time range of 16.0 to 60.5 months. Thirty patients did not have HPV-16 detected in posttreatment plasma sample, but 2 of these patients developed recurrence during a 3.4- to 121.0-month follow-up period. Interestingly, 1 patient with HPV-16–positive tumor had no HPV-16 DNA detected in pretreatment plasma but had detectable HPV-16 DNA in posttreatment plasma and was found to have a recurrence during a 7-year follow-up period.
There were a total of 54 patients with both sets of pretreatment and posttreatment saliva and plasma samples, of which 46 (85%) had HPV-16–positive tumors and 8 (15%) had HPV-16–negative tumors (Figure 1C). No patient with HPV-16– negative tumors had HPV-16 DNA detected in either pretreatment plasma or pretreatment saliva samples (specificity, 100%). Of 46 patients with HPV-16–positive tumors, 35 patients had HPV-16 DNA detected in either pretreatment plasma or pretreatment saliva or both (sensitivity, 76%). There were 6 patients with HPV-16–positive tumor who developed recurrence posttreatment during the follow-up duration of 31.0 to 87.0 months. Four of these patients had detectable HPV-16 DNA in either posttreatment saliva sample or plasma sample or both, while 2 patients did not have any detectable HPV-16 DNA in either posttreatment saliva or plasma.
The sensitivity, specificity, positive predictive value, and negative predictive value of HPV-16 status under different detection sources are summarized in Table 2. When using a pretreatment detection source to screen patients for HPV-16–positive tumor, the sensitivity increased from 52.8% with saliva HPV-16 status only to 67.3% with plasma HPV-16 status only and finally to 76.1% when using combined saliva and plasma HPV-16 status. The specificity and positive predictive value of pretreatment HPV-16 status were 100% regardless of the detection source used. The negative predictive value of pretreatment HPV-16 status increased from 24.4% with saliva HPV-16 status to 34.6% with plasma HPV-16 status to 42.1% with combined plasma and saliva HPV-16 status.
Table 2.
Change in Sensitivity, Specificity, and Predictive Values of Pretreatment and Posttreatment HPV-16 Status Under Different Detection Source
| Detection Source | Sensitivity, % | Specificity, % | PPV, % | NPV, % |
|---|---|---|---|---|
| Pretreatmenta | ||||
| Saliva only | 52.8 | 100 | 100 | 24.4 |
| Plasma only | 67.3 | 100 | 100 | 34.6 |
| Combined | 76.1 | 100 | 100 | 42.1 |
| Posttreatmentb | ||||
| Saliva only | 18.8 | 96.6 | NA | NA |
| Plasma only | 55.1 | 95.6 | NA | NA |
| Combined | 69.5 | 90.7 | NA | NA |
Abbreviations: HPV, human papillomavirus; NA, not applicable; NPV, negative predictive value; PPV, positive predictive value.
Clinical outcome: tumor HPV status.
Clinical outcome: tumor recurrence within 3 years among patients with HPV-16-positive tumors.
Posttreatment Saliva and Plasma HPV Status and Clinical Outcome
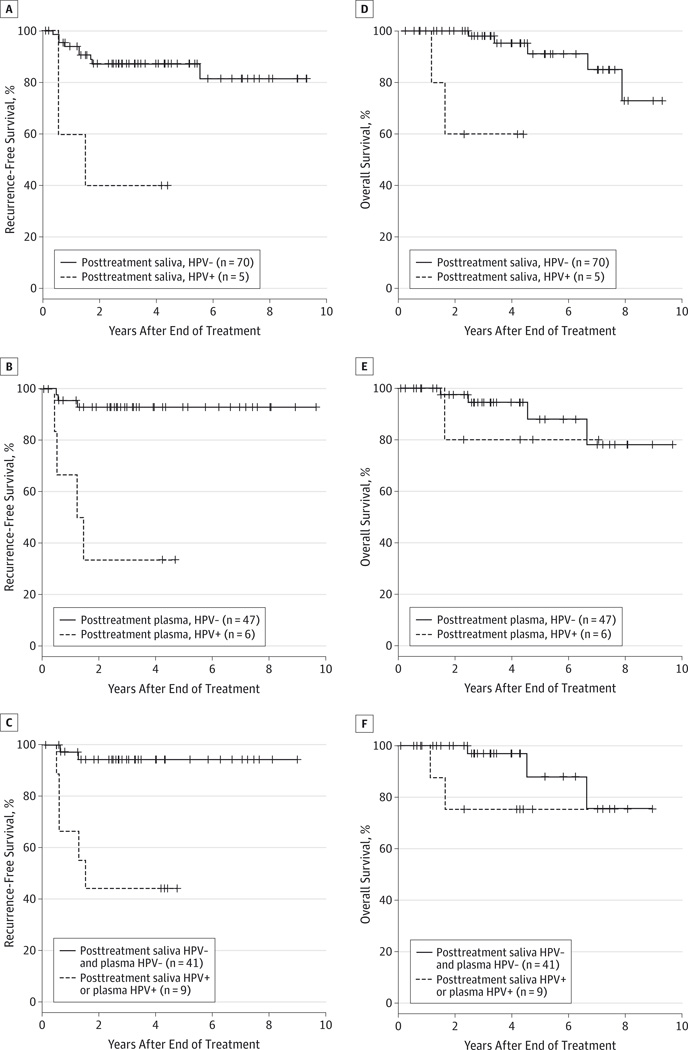
In univariate analysis, both HPV DNA positivity in posttreatment saliva and HPV DNA positivity in posttreatment plasma in patients with HPV-16–positive tumors were significantly associated with worse RFS (HR,7.14; 95% CI, 1.88–27.10 [P = .004]; and HR, 12.7; 95% CI, 2.84–57.10 [P < .001], respectively), compared with those with HPV DNA–negative status in saliva and plasma. When using a combination of posttreatment saliva and plasma samples to determine the HPV DNA status, patients with HPV DNA–positive status in either saliva or plasma had significantly higher risk of disease recurrence compared with patients with HPV DNA negative status in both saliva and plasma (HR, 14.0; 95% CI, 2.72–72.60) (P = .002).
In multivariable analysis, patients with posttreatment saliva HPV-positive status were associated with higher risk of recurrence after adjusting for alcohol use, smoking status, T classification, and N classification (HR, 10.7; 95% CI, 2.36–48.50) (P = .002). Patients with posttreatment plasma HPV-positive status demonstrated a significant association with increased risk of disease recurrence after adjusting for alcohol use, smoking status, and N classification (HR, 41.5; 95% CI, 4.2–407.0) (P = .001). When combining posttreatment plasma and saliva samples, patients with HPV-positive status in either plasma or saliva had worse RFS after adjusting for alcohol use, smoking status, and N classification (HR, 24.4; 95% CI, 3.58–166.00) (P = .001). The HRs for RFS are summarized in Table 3, and Kaplan-Meier curves for RFS stratified by HPV status in saliva and plasma are shown in Figure 2.
Table 3.
Univariate and Multivariable Analysis of Association Between HPV Status and Clinical Outcomes
| Variable | Recurrence-Free Survivala | Overall Survivalb | ||
|---|---|---|---|---|
| HR (95% CI) | P Value | HR (95% CI) | P Value | |
| Univariate Analysis | ||||
| Age | 0.99 (0.40–1.06) | .85 | 0.99 (0.91–1.09) | .86 |
| Sex | ||||
| Male vs female | 0.71 (0.16–3.17) | .65 | 0.78 (0.10–6.38) | .82 |
| Alcohol | ||||
| Former vs none | 4.10 (1.08–15.60) | .04 | ||
| Current vs none | 0.81 (0.22–3.06) | .76 | ||
| Smoking | ||||
| Former vs none | 0.67 (0.20–2.30) | .53 | 0.20 (0.02–1.61) | .13 |
| Current vs none | 1.64 (0.42–6.44) | .48 | NA | NA |
| Primary therapy | ||||
| Chemoradiation vs surgery | 0.77 (0.23–2.50) | .66 | 0.81 (0.16–4.19) | .80 |
| T classification | ||||
| T3–4 vs T0–2 | 7.62 (2.64–22.00) | <.001 | 4.05 (0.96–17.10) | .06 |
| N classification | ||||
| N2–3 vs N0–1 | 0.380 (0.11–1.36) | .14 | 0.71 (0.09–9.95) | .76 |
| Posttreatment saliva HPV | ||||
| Positive vs negative | 7.14 (1.88–27.10) | .004 | 16.8 (2.33–121.70) | .005 |
| Posttreatment plasma HPV | ||||
| Positive vs negative | 12.7 (2.84–57.10) | <.001 | 2.10 (0.23–18.90) | .51 |
| Posttreatment plasma/saliva HPV | ||||
| Either positive vs both negative | 14.0 (2.72–72.60) | .002 | 3.68 (0.61–22.10) | .16 |
| Multivariable Analysis c | ||||
| Posttreatment saliva HPV | ||||
| Positive vs negative | 10.7 (2.36–48.50) | .002 | 25.9 (3.23–208.00) | .002 |
| Posttreatment plasma HPV | ||||
| Positive vs negative | 41.5 (4.20–407.00) | .001 | 3.33 (0.03–37.00) | .33 |
| Posttreatment plasma/saliva HPV | ||||
| Either positive vs both negative | 24.4 (3.58–166.00) | .001 | 21.2 (1.85–242.00) | .01 |
Abbreviations: HR, hazard ratio; HPV, human papillomavirus.
The final model for recurrence-free survival was adjusted for alcohol use, smoking status, T classification, and N classification.
The final model for overall survival was adjusted for smoking status, T classification, and N classification.
Multivariable analysis was performed for each detection source separately.
Figure 2. Kaplan-Meier Curves for Recurrence-Free Survival and Overall Survival.
A, Recurrence-free survival is illustrated among patients with human papillomavirus-positive (HPV+) tumors stratified by posttreatment saliva HPV DNA status. B, Recurrence-free survival is illustrated among patients with HPV+ tumors stratified by posttreatment plasma HPV DNA status. C, Recurrence-free survival is illustrated among patients with HPV+ tumors stratified by both posttreatment saliva and plasma HPV DNA status. D, Overall survival is illustrated among patients with HPV+ tumors stratified by posttreatment saliva HPV DNA status. E, Overall survival is illustrated among patients with HPV+ tumors stratified by posttreatment plasma HPV DNA status. F, Overall survival is illustrated among patients with HPV+ tumors stratified by both posttreatment saliva and plasma HPV DNA status. HPV− indicates human papillomavirus negative.
Eight patients (10%) with HPV-positive tumor were documented and confirmed to be dead, of which 5 patients died of disease while 3 patients died from unrelated causes. Of the 5 patients who died of disease, 2 developed local recurrence, 1 developed locoregional recurrence, 1 developed regional and distant metastasis, and the final patient developed distant metastasis. In univariate analysis, OS for patients with HPV-positive tumor was reduced among those with posttreatment saliva HPV-positive status (HR, 16.8; 95% CI, 2.33–121.70) (P = .005). The risk of death was unchanged in patients with posttreatment plasma HPV positive status (HR, 2.10; 95% CI, 0.23–18.90) (P = .51). HPV DNA–positive status in either saliva or plasma did not have any significant association with OS (HR, 3.68; 95% CI, 0.61–22.10) (P = .16) compared with patients with HPV DNA–negative status in both saliva and plasma.
In multivariable analysis, posttreatment saliva HPV DNA positivity (HR, 25.9; 95% CI, 3.23–208) (P = .002) retained a significant association with increased risk of death in patients with HPV-positive HNSCC (Table 3). Human papillomavirus DNA status using posttreatment plasma was not statistically significant association with OS. However, when combining saliva HPV DNA status with plasma HPV DNA status, patients with HPV-positive status in either source had significantly worse OS with an HR of 21.2 (95% CI, 1.85–242.00) (P = .01). Kaplan-Meier curves for OS stratified by HPV status are shown in Figure 2.
Sensitivities and Specificities for Prediction of Disease Recurrence Within 3 Years
Time-dependent sensitivity and specificity for cumulative disease recurrence within 3 years were estimated for patients with HPV-16-positive tumors. The sensitivity and specificity of posttreatment salivary HPV-16 status in HPV-16–positive tumors in predicting recurrence within 3 years were 18.8% and 96.6%, respectively. The sensitivity and specificity of surveillance plasma HPV-16 status were 55.1% and 95.6%, respectively. When combining of posttreatment saliva and plasma samples, the presence of HPV-16 DNA was 90.7% specific and 69.5% sensitive in predicting locoregional or metastatic recurrence within 3 years in this patient population. Thus, the sensitivity of posttreatment HPV-16 status increased from 18.8% with saliva HPV-16 status to 55.1% with plasma HPV-16 status to 69.5% with combined plasma and saliva HPV-16 status in this patient population of patients with HPV-16–positive tumors (Table 2).
Discussion
In addition to smoking and alcohol use, high-risk HPV subtypes, especially HPV-16 subtype, have emerged as an important etiologic and prognostic factor in patients with OPSCC.1,2 The detection of primary OPSCC and recurrence following completion of therapy is often delayed owing to the challenging, difficult-to-examine areas of the tonsils and base of tongue. Therefore, the development of a saliva-based or plasma-based screening and surveillance test for HPV-16–positive OPSCC can aid in early detection of primary cancer and recurrence prior to development of clinical symptoms. With the application of real-time quantitative PCR technique, as little as 0.001 copy per genome HPV-16 DNA can be detected in patient’s salivary and plasma samples. With a high prevalence of up to 10% of men aged 55 to 64 years with a high risk of HPV infection, developing a strategy for early detection of OPSCC can be challenging.4 The aim of our study was to determine the feasibility of using pretreatment and posttreatment HPV-16 status in plasma and saliva as a biomarker for HPV-related OPSCC disease status.
In a large cross-sectional study by Gillison et al,4 the prevalence of oral HPV infection has been shown to be 7% in the population and 1% for the high-risk HPV-16 subtype. Thurs far, only a few studies have shown any correlation between pretreatment HPV-16 status and HNSCC cancer using real-time quantitative PCR. In one study by Capone et al,10 6 of 15 HPV-16 positive tumors (40%) had HPV-16 DNA detected in serum. Zhao et al11 found that HPV-16 DNA was detected in 50% (21 of 42) of the salivary rinse samples from patients with HPV-16–positive tumors. In terms of using HPV-16 status as a surveillance tool, only 1 study has shown any correlation between posttherapy HPV-16 status insaliva and recurrence using quantitative PCR. In Chuang et al,12 2 of 20 patients with pretreatment HPV-16–positive status had HPV-16 DNA detected in surveillance saliva, and both went on to develop recurrence. The sensitivity, specificity, positive predictive value, and negative predictive value in this study were 50%, 100%, 100%, and 88.89%, respectively.12 With a cohort of 93 patients (81 patients with HPV-16–positive tumors and 12 patients with HPV-16–negative tumors), our study is the largest study to date to determine the feasibility of early detection of HPV-16 positive primary cancer and recurrence using quantitative PCR technique.
In the present study, patients with HPV-16–positive tumors were nonsmokers and tended to present with advanced stage disease with lymph node metastasis. When using pretreatment HPV-16 status to screen for HPV-16–positive OPSCC, we found that HPV-16 DNA detection in both plasma and saliva rinses demonstrated 100% specificity with a 100% positive predictive value. The sensitivity for salivary-based screening test was 52.8%, which was similar to the rates reported previously.11,12 The sensitivity HPV-16 status for predicting the presence of HPV-16–positive OPSCC improved to 67.3% when using a plasma-based screening test compared with using a saliva-based test. A sensitivity of 76.1% was achieved by using a combined approach using both plasma-based and saliva-based screening tests.
Positron emission tomography–computed tomography (PET-CT) is routinely used as a surveillance tool in patients with HNSCC treated with chemoradiation. A recent study examined the reliability of 3-month posttreatment PET-CT in patients with HPV-associated OPSCC.15 This study found the sensitivity of a 3-month posttreatment PET-CT to be 33% for the primary site and 63% for the regional site of recurrence. The negative predictive values were reported to be high at 91%to 98%. Our study investigated the role of saliva or plasma-based PCR testing in posttherapy surveillance of disease recurrence within 3-year period. While specificity stayed high (>90%) with each method of detection, the sensitivity of posttreatment HPV-16 status was better with a plasma-based surveillance test (55.1%) compared with that of a saliva-based test (18.8%) and highest with a combined plasma and saliva based surveillance test (69.5%). Thus, while the sensitivities of surveillance saliva and plasma HPV-16 status alone are lower compared with the sensitivity of a 3-month posttreatment PET-CT, the sensitivity achieved by combining the surveillance saliva and plasma HPV-16 status is better when compared with the sensitivity of a 3-month posttreatment PET-CT.
Interestingly, 1 patient with HPV-16 positive tumor tested false negative on pretreatment screening test based on both saliva and plasma. However, HPV-16 DNA was detected in the posttreatment plasma sample, and the patient went on to develop recurrence. This patient initially presented with T1N1M0 OPSCC and was treated with surgical resection without any adjuvant therapy. The patient developed local recurrence 15 months after treatment. The surveillance posttreatment plasma sample was positive for HPV-16 DNA 7 months prior to detection of recurrence. One possible explanation is that the initial tumor burden was low enough that HPV-16 E6 and E7 DNA was undetectable. Another hypothesis is that with a negative predictive value of 24.4% for pretreatment saliva HPV-16 status and 34.6% for pretreatment plasma HPV-16 status, the pretreatment negative HPV-16 status in this patient was simply a false negative. Regardless, a plasma-based surveillance test may potentially still be used to monitor for recurrence in patients with HPV-16–positive tumor even if pretreatment screening test was not able to detect HPV-16 DNA.
Another interesting outlier is a patient with HPV-negative OPSCC who had negative HPV-16 DNA status in both pretreatment saliva and plasma but subsequently became HPV-16 positive in posttreatment saliva and plasma samples but did not develop recurrence. Our hypothesis for this false positive posttreatment HPV-16 status is that based on a 1% prevalence rate of oral HPV-16 infection in the general population, as reported in Gillison et al,4 one would expect 1 patient to test positive for HPV-16 DNA in a study cohort of 93 patients.
The median lead time from time of HPV-16 DNA detection in saliva or plasma to time of clinical detection of tumor recurrencewas4.4months in our cohort. Our hypothesis is that the presence of detectable HPV-16 DNA in surveillance sample could represent microscopic residual disease or micrometastasis in plasma. It could also represent the patient’s overall immune incompetency and the patient’s inability to clear HPV-infected cells. We also investigated any potential relationship between the source of HPV DNA (saliva vs plasma) and site of recurrence (local, regional, or distant).We hypothesized that HPV DNA status in saliva would correlate with local recurrence in the oropharynx, while HPV DNA status in plasma would predict regional or distant metastasis. However, owing to a small number patients with recurrence as well as multiple sites of recurrence in patients, we did not find any statistically significant association between the source of HPV DNA and the site of recurrence. To our knowledge, our study is the first to evaluate the prognostic value of HPV-16 status in salivary and plasma samples of patients with HPV-positive OPSCC using quantitative PCR for HPV-16 E6 and E7 DNA. In both univariate and multivariable analysis associating HPV status in saliva and plasma and RFS, both HPV DNA positivity in posttreatment saliva and HPV DNA positivity in posttreatment plasma were significantly associated with worse RFS. When combining saliva HPV DNA status with plasma HPV DNA status, patients with HPV-positive status in either source had higher risk of disease recurrence compared with patients with HPV DNA–negative status in both saliva and plasma. For patients with HPV-positive tumor, OS was reduced among those with posttreatment saliva HPV-positive status in both univariate and multivariable analysis. However, the risk of death was unchanged in patients with posttreatment plasma HPV positive status. In addition, when combining saliva HPV DNA status with plasma HPV DNA status, patients with HPV-positive status in either source had significantly worse OS.
Our current study was based on a limited retrospective cohort. A larger scale, prospective study designed to study the feasibility of plasma and saliva-based screening and surveillance test using quantitative HPV-16 PCR would be needed to confirm the findings of our study. Quantitative HPV-16 PCR can also be combined with methods detecting other biomarkers of genetic and epigenetic alterations specific to HPV-16– positive OPSCC to achieve even better sensitivity and specificity. If validated, detection of HPV-16 DNA using a plasma and saliva-based PCR testing may emerge as an important surveillance biomarker in identifying a select group of patients with previously treated HPV-related OPSCC who may require treatment intensification.
Conclusions
Using a combination of pretreatment plasma and salivary samples can increase the sensitivity of pretreatment HPV-16 status as a screening tool for HPV-related OPSCC compared with using either sample alone. Patients with HPV-positive OPSCC who have HPV-16 DNA detected in surveillance salivary samples are at significantly higher risk of recurrence and death. In addition, when combining saliva HPV DNA status with plasma HPV DNA status, patients with HPV-positive status in either source had significantly worse RFS and OS. Thus, quantitative PCR detection of HPV DNA in posttreatment surveillance saliva and plasma samples can function as a valuable prognostic biomarker of RFS and OS in patients with HPV-positive OPSCC.
Acknowledgments
Funding/Support: This work was supported by grant P50 CA19032 from the National Cancer Institute/National Institute of Dental and Craniofacial Research Head & Neck Cancer Specialized Program of Research Excellence.
Role of the Sponsor: The sponsor had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Footnotes
Author Contributions: Drs Ahn and Califano had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Ahn, Chan, Koch, Califano.
Acquisition, analysis, or interpretation of data: All authors.
Drafting of the manuscript: Ahn, Chan, Wang, Khan, Califano.
Critical revision of the manuscript for important intellectual content: Ahn, Zhang, Bishop, Westra, Koch, Califano.
Statistical analysis: Zhang, Wang, Califano.
Obtained funding: Koch, Califano.
Administrative, technical, or material support: Ahn, Chan, Khan, Westra, Koch, Califano.
Study supervision: Koch, Califano.
Conflict of Interest Disclosures: None reported.
Previous Presentation: This research was presented at the American Head & Neck Society 2013 Annual Meeting; April 12, 2013; Orlando, Florida.
Contributor Information
Sun M. Ahn, Department of Otolaryngology–Head and Neck Surgery, Johns Hopkins Medical Institutions, Baltimore, Maryland.
Jason Y. K. Chan, Department of Otolaryngology–Head and Neck Surgery, Johns Hopkins Medical Institutions, Baltimore, Maryland.
Zhe Zhang, Division of Biostatics and Bioinformatics, Johns Hopkins School of Medicine, Baltimore, Maryland.
Hao Wang, Division of Biostatics and Bioinformatics, Johns Hopkins School of Medicine, Baltimore, Maryland.
Zubair Khan, Department of Otolaryngology–Head and Neck Surgery, Johns Hopkins Medical Institutions, Baltimore, Maryland.
Justin A. Bishop, Department of Pathology, Johns Hopkins Medical Institutions, Baltimore, Maryland.
William Westra, Department of Pathology, Johns Hopkins Medical Institutions, Baltimore, Maryland.
Wayne M. Koch, Department of Otolaryngology–Head and Neck Surgery, Johns Hopkins Medical Institutions, Baltimore, Maryland.
Joseph A. Califano, Department of Otolaryngology–Head and Neck Surgery, Johns Hopkins Medical Institutions, Baltimore, Maryland; Milton J. Dance Head and Neck Center, Greater Baltimore Medical Center, Baltimore, Maryland.
REFERENCES
- 1.Licitra L, Perrone F, Bossi P, et al. High-risk human papillomavirus affects prognosis in patients with surgically treated oropharyngeal squamous cell carcinoma. J Clin Oncol. 2006;24(36):5630–5636. doi: 10.1200/JCO.2005.04.6136. [DOI] [PubMed] [Google Scholar]
- 2.Fakhry C, Westra WH, Li S, et al. Improved survival of patients with human papillomavirus-positive head and neck squamous cell carcinoma in a prospective clinical trial. J Natl Cancer Inst. 2008;100(4):261–269. doi: 10.1093/jnci/djn011. [DOI] [PubMed] [Google Scholar]
- 3.Chaturvedi AK, Engels EA, Pfeiffer RM, et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol. 2011;29(32):4294–4301. doi: 10.1200/JCO.2011.36.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gillison ML, Broutian T, Pickard RK, et al. Prevalence of oral HPV infection in the United States, 2009–2010. JAMA. 2012;307(7):693–703. doi: 10.1001/jama.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gillison ML. HPV and prognosis for patients with oropharynx cancer. Eur J Cancer. 2009;45(suppl 1):383–385. doi: 10.1016/S0959-8049(09)70058-9. [DOI] [PubMed] [Google Scholar]
- 6.Gillison M. HPV and its effect on head and neck cancer prognosis. Clin Adv Hematol Oncol. 2010;8(10):680–682. [PubMed] [Google Scholar]
- 7.Werness BA, Levine AJ, Howley PM. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science. 1990;248(4951):76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- 8.Kessis TD, Slebos RJ, Nelson WG, et al. Human papillomavirus 16 E6 expression disrupts the p53-mediated cellular response to DNA damage. Proc Natl Acad Sci U S A. 1993;90(9):3988–3992. doi: 10.1073/pnas.90.9.3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sisk EA, Soltys SG, Zhu S, Fisher SG, Carey TE, Bradford CR. Human papillomavirus and p53 mutational status as prognostic factors in head and neck carcinoma. Head Neck. 2002;24(9):841–849. doi: 10.1002/hed.10146. [DOI] [PubMed] [Google Scholar]
- 10.Capone RB, Pai SI, Koch WM, et al. Detection and quantitation of human papillomavirus (HPV) DNA in the sera of patients with HPV-associated head and neck squamous cell carcinoma. Clin Cancer Res. 2000;6(11):4171–4175. [PubMed] [Google Scholar]
- 11.Zhao M, Rosenbaum E, Carvalho AL, et al. Feasibility of quantitative PCR-based saliva rinse screening of HPV for head and neck cancer. Int J Cancer. 2005;117(4):605–610. doi: 10.1002/ijc.21216. [DOI] [PubMed] [Google Scholar]
- 12.Chuang AY, Chuang TC, Chang S, et al. Presence of HPV DNA in convalescent salivary rinses is an adverse prognostic marker in head and neck squamous cell carcinoma. Oral Oncol. 2008;44(10):915–919. doi: 10.1016/j.oraloncology.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johns MM, III, Westra WH, Califano JA, Eisele D, Koch WM, Sidransky D. Allelotype of salivary gland tumors. Cancer Res. 1996;56(5):1151–1154. [PubMed] [Google Scholar]
- 14.Heagerty PJ, Lumley T, Pepe MS. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics. 2000;56(2):337–344. doi: 10.1111/j.0006-341x.2000.00337.x. [DOI] [PubMed] [Google Scholar]
- 15.Vainshtein JM, Spector ME, Stenmark MH, et al. Reliability of post-chemoradiotherapy F-18-FDG PET/CT for prediction of locoregional failure in human papillomavirus-associated oropharyngeal cancer. Oral Oncol. 2014;50(3):234–239. doi: 10.1016/j.oraloncology.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]