ABSTRACT
We recently described the architecture of the Epstein-Barr virus (EBV) fusion-triggering complex consisting of the EBV B cell receptor human leukocyte antigen (HLA) class II and the EBV-encoded proteins gp42 and gH/gL. The architecture of this structure positioned the main body of gp42, comprising the C-type lectin domain (CTLD), away from the membrane and distant from where the membrane-bound form of gp42 might be tethered. gp42 is a type II membrane glycoprotein, with functional gp42 formed by cleavage near the gp42 amino-terminal transmembrane domain. This cleavage results in an approximately 50-amino-acid unstructured region that is responsible for binding gH/gL with nanomolar affinity. Our previous studies had shown that membrane-bound gp42 is not functional in B cell fusion. To investigate whether we could restore gp42 function by extending it from the membrane, we introduced one, two, and four structured immunoglobulin-like domains from muscle protein titin into a membrane-bound form of gp42 and tested function in binding to gHgL and HLA class II and function in fusion. We hypothesized that cleavage of gp42 generates a soluble functional form that relieves steric hindrance imposed on gHgL by membrane-bound gp42. All of the linker mutants had a dominant-negative effect on gp42 function, indicating that gp42 fusion function could not be restored simply by the addition of one to four titin domains.
IMPORTANCE
Epstein-Barr virus (EBV) is associated with numerous diseases from benign mononucleosis to Burkitt’s and Hodgkin’s lymphoma, nasopharyngeal and gastric carcinoma, and lymphoproliferative disorders in patients with immune dysfunction resulting from immune suppression. Among the glycoproteins important for fusion, gp42, along with gH/gL, determines EBV tropism between epithelial and B cells. The function of gp42 is dependent on N-terminal cleavage, since membrane-bound gp42 cannot mediate fusion. We further investigated whether insertion of a linker into membrane-bound gp42 would relieve steric hindrance imposed on membrane-bound gp42 and restore fusion function. However, adding one, two, or four structured immunoglobulin-like domains to membrane gp42 did not restore fusion activity, indicating that the architecture and membrane orientation of the B cell fusion-triggering complex of EBV may be easily perturbed and that gp42 cleavage is essential for B cell fusion.
INTRODUCTION
Epstein-Barr virus (EBV) (also called human herpesvirus 4 [HHV-4]) is an enveloped gammaherpesvirus and one of only eight human herpesviruses (1). The two major cell types that EBV infects are epithelial cells and B cells. EBV enters these different cell types by the concerted effort of three or four essential viral glycoproteins, depending on the cell type (1, 2). Virions produced from B cells contain gB and gHgL which mediate fusion of virions with epithelial cells (1, 2). In contrast, virions produced from epithelial cells also contain gp42 in addition to gB and gH/gL (1, 2). It is gp42 that activates entry into B cells by binding to human leukocyte antigen (HLA) class II (1–4), whereas gH/gL binding to integrins allows entry into epithelial cells (1, 2, 5–7). gp42 binding to gHgL also inhibits entry into epithelial cells, and thus serves as a viral tropism switch (5).
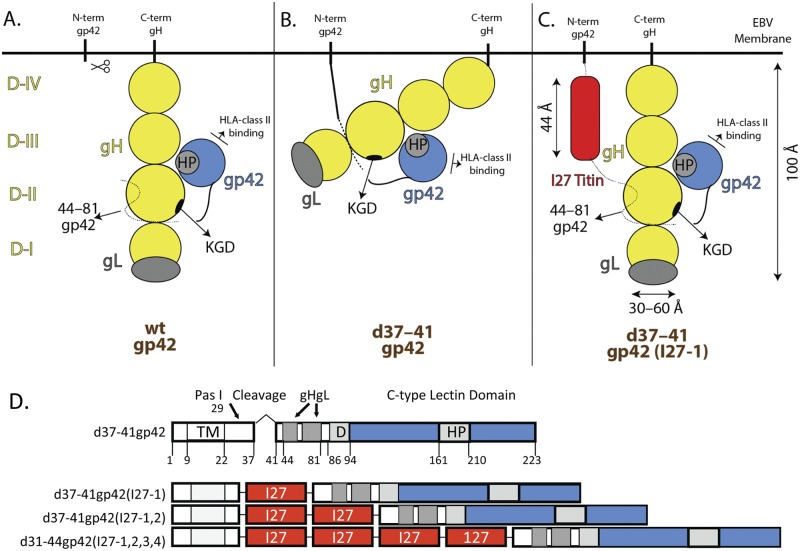
When gp42 is synthesized, it is a 223-amino-acid type II membrane protein (1, 8). A soluble form of gp42 is generated by proteolytic cleavage in the endoplasmic reticulum (ER) with cleavage occurring around amino acids 40, 41, and 42 (9). Deletion of the predicted cleavage site (residues 37 to 41) results in a membrane-bound form of gp42 that significantly abrogates B cell fusion (10), verifying the results of earlier studies indicating that soluble gp42 (sgp42) functions in B cell fusion (11, 12). Soluble gp42 mutants, containing an EBV gB signal sequence followed by gp42 N-terminal deletions up to residue 46, are functional for fusion (10). Deletions up to residue 52, however, are not well tolerated, as this affects the region within gp42 (amino acids 44 to 81) that is important for gH/gL binding (10, 13). In addition to being essential for B cell fusion, gp42 inhibits epithelial cell fusion (5, 11) presumably by binding an overlapping region in gH/gL that also binds the receptor for epithelial cell fusion (11, 14, 15). Mutational studies have shown that amino acids within residues 44 to 81 of the N terminus of soluble gp42 interact with domain II (DII) of gHgL, and cocrystallization studies between gp42 and HLA-DR1 have shown that the gp42 C-type lectin domain (CTLD) (solid blue circle in Fig. 1A) interacts with HLA class II (13, 16). Crystallization studies have shown that the EBV gHgL structure is comprised of four sequential semiautonomous domains (DI, DII, DIII, and DIV) and gL forms a stable heterodimer with gH and is integral to DI folding and structure (17). The prominent KGD loop in DII has been implicated in binding residues 62 to 66 of gp42 (Fig. 1A), as well as the gHgL epithelial receptor integrin αvβ6, αvβ8, or αvβ5 (7, 15). Membrane-bound gp42 (Fig. 1B), which contains a deletion of the predicted cleavage site from residues 37 to 41 (d37-41gp42), efficiently binds gHgL but is unable to mediate fusion (10, 18).
FIG 1 .
Schematic of gp42 and gH/gL interactions. The four domains of gHgL are shown in yellow: domain I (DI) (residues 18 to 65), DII (residues 66 to 344) with the KGD motif indicated as a black circle, DIII (residues 345 to 529), DIV (residues 530 to 679), transmembrane domain (residues 680 to 698), and the cytoplasmic tail (residues 699 to 706). Wild-type (wt) gL (gray) (residues 24 to 137) is within DI of gH. The N terminus of gp42 (N-term gp42) is shown as a dotted line (residues 44 to 81) where it interacts with gHgL and as a solid line (residues 82 to 94) where the putative dimerization domain exits. The C terminus (C-term) of gp42 contains a C-type lectin domain (CTLD) and is shown as a solid blue circle. The hydrophobic pocket (HP) of gp42 is indicated as a gray circle within the CTLD. (A) Functional gp42-gHgL complex with soluble gp42. sgp42 is able to interact with DII both at the N and C termini as well as interact with HLA (human leukocyte antigen) class (II). (B) Inhibited gp42-gHgL complex with membrane-bound gp42. The putative bent conformation of gHgL due to restriction by membrane-bound gp42 may prevent or reduce binding to other viral proteins, such as gB or viral receptors, such as HLA class II. (C) Addition of one copy of the 27th immunoglobulin-like domain (I27) (44 Å) of the muscle protein titin which may restore the normal configuration of gH/gL and binding to other viral proteins and receptors. (D) Schematic of d37-41gp42 containing zero, one, two, or four structured titin I27 linker clones. One, two, or four copies of the 27th immunoglobulin-like domain for the muscle protein titin were cloned into a unique PasI site (residue 29) of gp42, just upstream of the deleted cleavage site (residue 37 to 41) and just downstream of the transmembrane domain (TM) (residue 9 to 22). Each I27 linker clone is 92 amino acids long and roughly 44 Å (red). The gHgL binding domain (residues 47 to 81), putative dimerization domain (D) (residues 86 to 94), C-type lectin domain (residues 95 to 223) (blue), and hydrophobic pocket (HP) (residues 161 to 210) are also shown.
We recently studied the assembly and architecture of the EBV B cell entry complex by electron microscopy (19). In this structure, gp42 binds to gHgL and HLA class II. It is the binding of gp42 to the B cell receptor HLA class II that is thought to trigger B cell fusion by the concerted action of gHgL and gB (1, 2). This structure, which likely represents an intermediate state of EBV entry, suggests that the B cell fusion-triggering complex may bring the two membrane bilayers of the virion and cell into proximity, orienting critical regions of the N- and C-terminal ends of gHgL to promote the activation of gB and efficient membrane fusion (19). Furthermore, we found that in this structure, gHgL also interacts with a functionally important hydrophobic pocket on gp42 (19). In our current studies, we sought to determine whether gp42 cleavage is essential for function, allowing flexibility so that the protein components of the B cell fusion-triggering complex can interact efficiently with each other to enable the successful triggering of membrane fusion.
RESULTS
Construction of d37-41gp42 titin linker insertion mutants.
To investigate whether membrane-bound gp42 sterically inhibits gHgL, shown bent and constrained in Fig. 1B, we designed a number of gp42 mutants (Fig. 1D). In the constrained conformation, interaction of gHgL with other viral glycoproteins, such as gB, or viral receptors may be prevented or reduced, resulting in a reduction in viral membrane fusion with the host cell membrane. To determine whether potential steric inhibition of membrane-bound gp42 could be relieved by increasing the distance or spacing of gp42 from the membrane, we inserted structural linkers immediately downstream of the gp42 transmembrane domain (red boxes in Fig. 1C and D). We chose to insert one, two, or four copies of the 27th immunoglobulin-like (I27) domain of the muscle protein titin. The titin Ig-like structure has been solved by both nuclear magnetic resonance (NMR) and crystallization, indicating the following: (i) The titin sequence consists mostly of repetitive motifs of tandem immunoglobulin-like (Ig-like) modules that are conformationally rigid segments interspersed with pliant hinges. (ii) The Ig-like domains consist of a beta sandwich formed by two four-stranded sheets which results in a domain with the N and C termini on opposite sides of the domain separated by 44 Å. (iii) The Ig-like domains fold independently in solution (20–23). In addition, the naturally occurring I27 domains have previously been successfully used to study the spacing requirements for proteasome binding and the initiation of protein degradation (22). Thus, addition of one to four titin I27 domains would space membrane-bound gp42 anywhere from approximately 44 to 176 Å from the membrane, and a particular spacing may be predicted to alleviate steric hindrance and allow for functional interaction with gHgL or other viral glycoproteins and cellular receptor(s) to promote fusion.
d37-41gp42 titin linker insertion mutants are expressed.
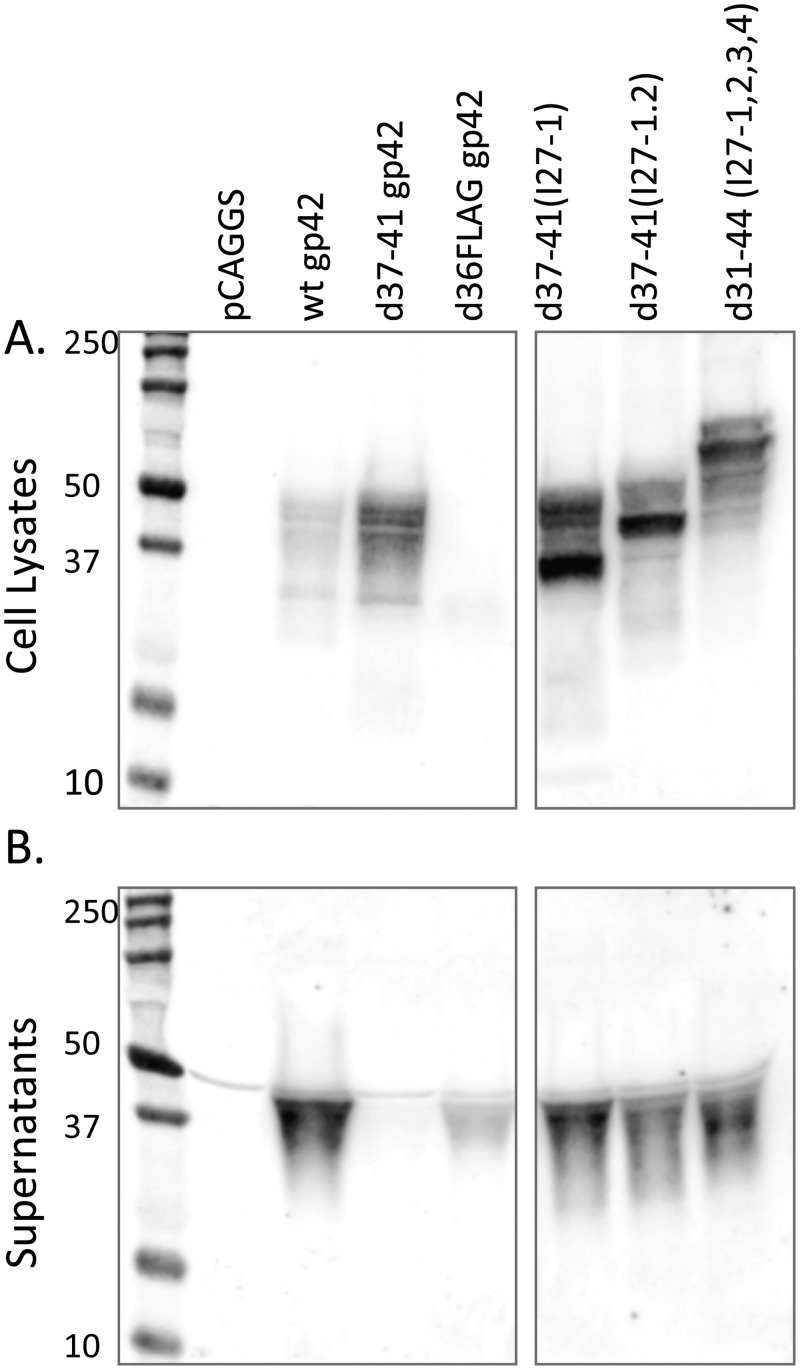
Plasmids encoding each of the linker mutants were transfected into Chinese hamster ovary cells (CHO-K1) cells, and protein expression was monitored in cell lysates (Fig. 2A) and medium supernatants (Fig. 2B) 48 h posttransfection by SDS-PAGE, followed by Western blotting using a polyclonal antibody specific for gp42. As expected, wild-type (wt) gp42 produced both soluble gp42 (sgp42) and cell-associated gp42 with most of the gp42 detected in the medium supernatants. Also as expected, a previously published soluble gp42 (d36FLAGgp42) (24) produced only soluble gp42, whereas d37-41gp42 predominantly produced cell-associated gp42. The linker mutants produced cell-associated gp42 of approximately the predicted size; however, surprisingly, soluble gp42 was also produced for each of the mutants. Deletion of a larger region (residues 22 to 46) around the cleavage site and insertion of the linkers did not alter this result but decreased expression, so these clones were not analyzed further (data not shown), and we decided to focus on the first group of mutants (Fig. 1D) that had higher levels of expression.
FIG 2 .
All of the gp42 linker mutants are expressed on the cell surface and are cleaved and secreted. CHO-KI cells were transiently transfected in Opti-MEM medium (Gibco) using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Cells were transfected overnight with 4 µg each vector alone (pCAGGS), wild-type gp42, d37-41gp42, d36FLAGgp42, d37-41(I27-1), d37-41(I27-1,2), and d31-44 (I27-1,2,3,4), followed by a medium change to Ham’s F-12 medium containing 10% fetal bovine serum and 1% penicillin-streptomycin. Forty-eight hours posttransfection, the supernatants were collected, and the cells were lysed in Triton X-100 lysis buffer containing protease inhibitors. (A and B) Cell lysates (A) and culture supernatants (B) were analyzed by 12% SDS-PAGE and Western blotting with polyclonal anti-gp42 antibody (PB114; R. Longnecker) and secondary IRDye 800CW-conjugated goat (polyclonal) anti-rabbit IgG (H+L) (catalog no. 926-32211; Li-Cor Biosciences) as previously described. Blots were washed and analyzed with Li-Cor Biosciences Odyssey infrared imaging studio software. The positions of molecular size markers (in kilodaltons) are indicated to the left of the blots. Although the blots in panels A and B were from the same gel, intervening lanes that were not applicable to the experiment shown were removed.
d37-41gp42 titin linker insertion mutants are expressed on the surface regardless of the presence of gHgL or gB and are not functional in cell-cell fusion.
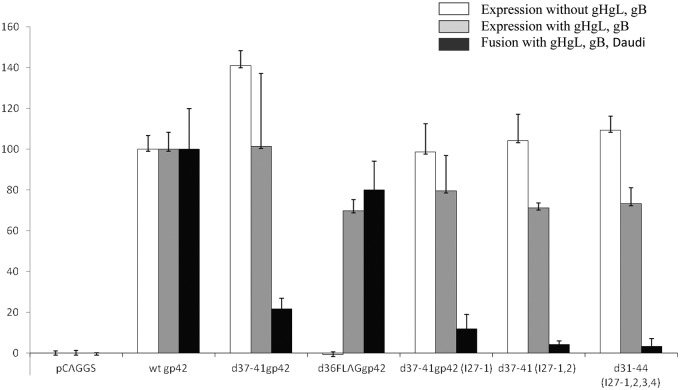
To study whether the gp42 linker mutants were expressed on the cell surface, their expression was analyzed in the presence and absence of gHgL and gB by cell-based enzyme-linked immunosorbent assay (cELISA). We cotransfected CHO-K1 cells with plasmids encoding vector, wt gp42, d37-41gp42, d37-41gp42 (I27-1), d37-41gp42 (I27-1,2) and d31-44gp42 (I27-1,2,3,4) with or without gHgL and gB. We found that the presence or absence of gHgL and gB did not make a significant difference in the surface expression of any of the membrane-bound mutants which were expressed at levels similar to that of wt gp42 (set at 100%) (Fig. 3). For a control, the soluble d36FLAGgp42 was expressed on the surface only in the presence of gHgL. In addition, we confirmed that the linker mutants were expressed on the surface of the cell by biotinylating the cell surface and performing immunoprecipitation with antibody to gp42 (F-2-1) (data not shown).
FIG 3 .
The gp42 linker mutants are expressed independently of gHgL expression and are not functional in B cell fusion. CHO-KI cells were transiently transfected with 2 µg of the gp42 linker mutants either in the presence or absence of gHgL and gB (0.5 µg each) and T7 luciferase (0.8 µg). Eighteen hours posttransfection, 40,000 cells were transferred to each well of a black 96-well plate and overlaid with 40,000 Daudi B cells for fusion assay. Eighteen hours after overlay, cells were washed with phosphate-buffered saline (PBS) and lysed for 10 min with 50 µl of passive lysis buffer (Promega) per well. Luciferase activity was measured with a PerkinElmer Victor plate reader immediately after addition of 50 µl/well of luciferase reagent (Promega). Eighteen hours posttransfection, 80,000 cells were transferred to each well of a clear 96-well plate, and eighteen hours later, surface expression was determined by cELISA using anti-gp42 (3H3), fixation, secondary biotinylated anti-mouse IgG antibody (Sigma), tertiary streptavidin-horseradish peroxidase (GE Healthcare) and TMB (3,3=,5,5=-tetramethylbenzidine) after TMB one-component substrate (BioFX). Absorbance readings were taken at 380 nm using a PerkinElmer Victor plate reader. Binding was normalized to wild-type gp42 binding levels, which were set at 100%. Data shown are representative of the results from three independent experiments.
However, when we tested the ability of the mutants to mediate fusion of Daudi B cells expressing T7 polymerase in the presence of gHgL, gB, and a T7 polymerase-driven luciferase reporter, we found that d37-41gp42, d37-41gp42 (I27-1), d37-41gp42 (I27-1,2) and d31-44gp42 (I27-1,2,3,4) were all significantly reduced in fusion (Fig. 3). As expected from previous studies, d37-41gp42 fusion function was greatly reduced compared to wt gp42 (10). d37-41gp42 (I27-1), d37-41gp42 (I27-1,2), and d31-44gp42 (I27-1,2,3,4) mediated fusion at levels less than 20% of wt gp42. Addition of one to four I27 domains to the N terminus of sgp42 (d36FLAGgp42) did not significantly alter fusion compared to wild-type sgp42 (data not shown), indicating that the addition of linkers alone does not make gp42 nonfunctional.
d37-41gp42 titin linker insertion mutants bind to exogenously expressed gHgL and HLA class II.
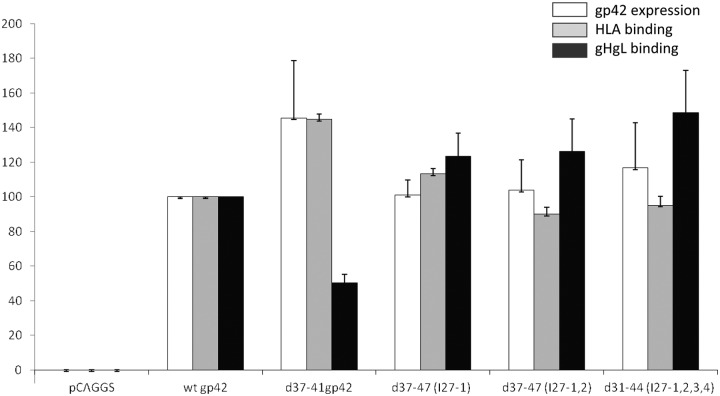
To determine whether the reduction in fusion was due to altered or decreased binding to gHgL or HLA class II, CHO-K1 cells were transfected with a previously published soluble form of gHgL (sgHgL) (25) and sgH/gL culture supernatants were collected to obtain sgH/gL. The sgH/gL along with purified HLA-DQ2 (αII) (or sDQ2-αII) (19) was used to monitor binding to each of the d37-41gp42 titin linker insertion mutants expressed in CHO-K1 cells. Following transfection with the vector control, wt gp42, d37-41gp42, d37-41gp42 (I27-1), d37-41gp42 (I27-1,2), and d31-44gp42 (I27-1,2,3,4), the transfected cells were overlaid 4 h later with sgHgL or sDQ2-αII for 1 h at 4°C. Bound protein was then determined by cELISA using appropriate antibodies recognizing HLA class DQ2-αII or sgH/gL. All the linker mutants bound HLA class II and gHgL at levels similar to those of wt gp42 (Fig. 4). However, d37-41gp42 bound HLA class II with slightly higher efficiency (gray bars) and bound gHgL approximately 50% of wt gp42 (black bars). This suggests that adding one to four titin linkers does alleviate some steric hindrance and allows improved binding to gHgL compared to when no spacer is inserted in the case of d37-41gp42. We confirmed that the lack of gHgL binding was not due to decreased expression of gHgL by cELISA and immunoprecipitation (data not shown). There was a minimal decrease in the expression for the linker mutants when gH/gL and gB were present and no decrease for d37-41gp42 compared to wt gp42.
FIG 4 .
The gp42 linker mutants bind exogenous gHgL and HLA class II comparable to wild-type gp42. CHO-K1 cells were transiently transfected with each of the d37-41 gp42 linker mutants. Twenty-four hours later, the cells were overlaid with sDQ2-αII (HLA class II) purified protein or sgHgL supernatants (isolated 48 h posttransfection). Protein was overlaid for 1 h at 4°C, and unbound protein was washed away. Binding of HLA class II and sgHgL was determined by cELISA using anti-HLA class II DQ antibody (1a3) (catalog no. ab24265; Abcam) and anti-FLAG-M2 (catalog no. F1804; Sigma), secondary biotinylated anti-mouse IgG antibody, tertiary streptavidin-HRP and TMB substrate and compared to expression using anti-gp42 antibody (3H3). Absorbance readings were taken at 380 nm using a PerkinElmer Victor plate reader. Binding was normalized to wild-type gp42 binding levels, which were set at 100%. Data shown are the average values of three independent experiments.
d37-41gp42 titin linker insertion mutants act in a dominant-negative fashion and compete with wild-type gp42 during B cell fusion.
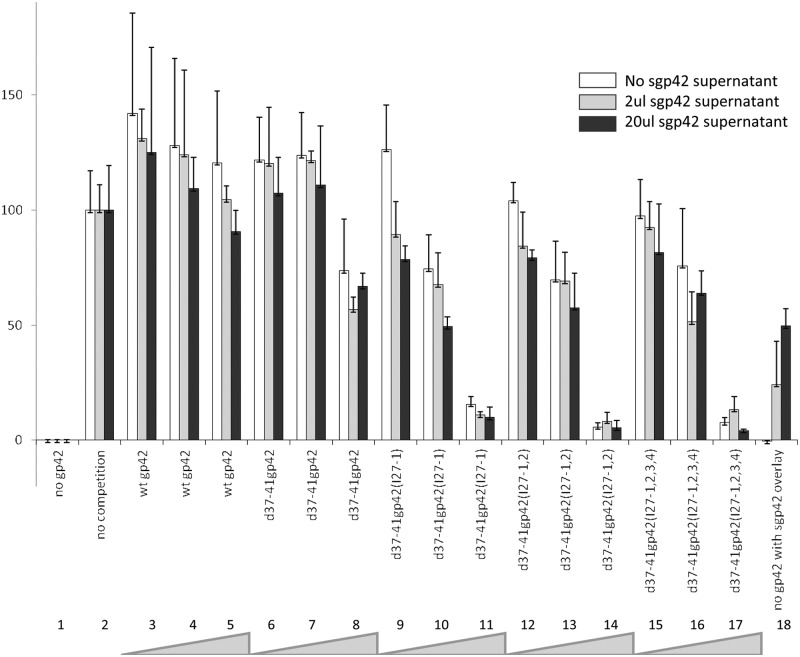
We postulated that since the linker mutants lack function in fusion but still bind HLA class II and gHgL, they may compete with wild-type gp42 during fusion. To determine whether the mutants compete with wt gp42 for fusion in a dose-dependent manner, Daudi B cells expressing HLA class II and T7 polymerase were cocultured with CHO-KI cells as effector cells containing vector alone (Fig. 5, lanes 1 and 18) or wt gp42 (0.5 µg) (lanes 2 to 17) and gH (0.5 µg), gL (0.5 µg), gB (0.5 µg), and T7 polymerase (0.8 µg) (lanes 1 to 18). Figure 5, lane 2, represents wild-type fusion without any competing gp42 added (white bar). Lanes 3 to 5 (white bars) show slight competition with increasing wild-type gp42 (0.01, 0.1, and 1.0 µg). Lanes 6 to 8 (white bars) show that with the highest concentration of d37-41gp42 (1.0 µg [lane 8]), there is approximately 25% reduction in fusion. In contrast, lanes 9 to 11, 12 to 14, and 15 to 17 (white bars) show that with the highest concentration (1.0 µg) of d37-41gp42 (I27-1), d37-41gp42 (I27-1,2), and d31-44gp42 (I27-1,2,3,4), respectively, there is approximately 90% reduction in fusion, suggesting that the gp42 linker mutants are better at blocking fusion than the d37-41gp42 mutant and that this effect is dose dependent.
FIG 5 .
The gp42 linker mutants act in a dominant-negative fashion and compete with wild-type gp42 during B cell fusion. CHO-KI cells were transiently transfected with 0.5 µg of wild-type gp42 and increasing amounts of each of the gp42 linker mutants (0.01 µg, 0.10 µg, and 1.0 µg) indicated by the thickness of the gray wedge at the bottom of the figure in the presence of gH (0.5 µg), gL (0.5 µg), gB (0.5 µg), and T7 polymerase (0.8 µg). Twenty-four hours posttransfection, cells were detached and overlaid with Daudi B cells expressing HLA class II and T7 promoter. Zero, 2 or 20 µl of sgp42 supernatant was added at the time of overlay (white, gray, and black bars). Twenty hours after the overlay, fusion activity was assessed by the addition of passive lysis buffer, followed by the addition of luciferase substrate. Luciferase activity was measured with a PerkinElmer Victor plate reader. Data shown are results representative of the results of three independent experiments. Error bars represent standard deviations for the normalized values.
To determine whether the dominant-negative effect was alleviated by adding sgp42, we transfected CHO-KI cells with d36FLAGgp42 and isolated soluble gp42 from supernatants 48 h posttransfection. Either 2 or 20 µl of supernatant (gray bars and black bars, respectively, in Fig. 5) was added to duplicate and triplicate plates of the fusion assay described above (with the exception of lane 1). We found that increasing the amount of sgp42 increased the level of fusion in a dose-dependent manner when no gp42 was transfected in the plate (lane 18, compare white, gray, and black bars). However, fusion did not increase with addition of sgp42 when any of the membrane-bound gp42s were transfected first (lanes 6 to 17 [compare white, gray, and black bars]). This confirms that d37-41gp42 and the linker mutants work in a dominant-negative manner, and once gHgL is bound by membrane-bound gp42, sgp42 cannot compete for binding.
DISCUSSION
We recently reported that deletion of the gp42 N-terminal cleavage site blocks gp42 function in fusion (10), indicating that when bound to a membrane by the transmembrane domain, gp42 does not function in fusion, despite binding to gHgL. This is in contrast with gD, the functional homolog of gp42 found in herpes simplex virus (HSV) that functions both as a soluble form and a membrane-bound form, although at reduced levels when soluble (26). Presumably gp42 is cleaved in the ER and is membrane bound by virtue of being tethered to gHgL, which contains a transmembrane domain.
To determine whether membrane-bound gp42 could be made functional by increasing separation from the membrane, the naturally occurring structured immunoglobulin-like domain I27 from the multidomain muscle protein titin was inserted between the gp42 transmembrane domain and the gHgL binding region of gp42. While I27 is rigid in nature, the N and C termini that connect each I27 domain have some flexibility (20–23). We predicted that the addition of one, two, and four I27 domains would approximately match, double, or quadruple the wild-type gp42 spacing from the membrane and thus potentially relieve any steric hindrance imposed on gHgL by being membrane bound. Our results show that while all the gp42 linker mutants are expressed on the surface, cleavage occurs even in the absence of the canonical cleavage site. Because the cleavage products were identical in size, it is likely that the cleavage occurs after the linker additions. We tried to alleviate this problem by making another series of insertion mutants with a larger deletion of the region that is cleaved in gp42, but these mutants also had some cleavage and were poorly expressed. Of the mutants we studied in detail, each of the soluble gp42 variants produced were functional in fusion when added as an overlay to gHgL/gB-expressing cells (data not shown). However, cotransfection of the linker mutants together with wild-type gH/gL and gB resulted in significantly reduced fusion compared to wild-type gp42 or sgp42. These results suggest that the membrane-bound linker mutants bind gHgL prior to cleavage and that this binding is not readily replaced by functional, soluble gp42, even though it is produced by the mutants. This finding is reinforced by the observation that the titin linker mutants effectively act in a dominant-negative manner, even more efficiently than the d37-41gp42 cleavage mutant, perhaps due to their wild-type-like levels of binding gHgL as well as HLA. Overall, our results suggest that the structured titin linkers that match, double, or quadruple the presumed wild-type gp42 spacing from the membrane do not alleviate the steric hindrance imposed on gHgL by membrane-bound gp42 to sufficiently promote fusion similar to that induced by wild-type gp42 or sgp42. These results indicate that the architecture of the B cell fusion-triggering complex (19) has additional, specific requirements for membrane fusion to proceed. Tethering gp42 to the membrane through its N terminus may generate a steric block that could interfere with membrane fusion by blocking the recruitment of gB or potentially altering other gHgL interactions that promote gB activation.
MATERIALS AND METHODS
Cells and antibodies.
Chinese hamster ovary cells (CHO-K1) were grown in 75-cm2 cell culture flasks (Corning) in Ham’s F-12 medium (BioWhittaker) supplemented with 10% fetal bovine serum (HyClone) and 1% penicillin-streptomycin (BioWhittaker). Trypsin-Versene (BioWhittaker) was used to detach adherent cells. Polyclonal anti-gp42 antibody serum (PB114) was used as previously described (27). Monoclonal antibody 3H3 (anti-gp42) was obtained as previously described (11). The HLA class II DQ monoclonal antibody (1a3) (catalog no. ab24265; Abcam) and anti-FLAG-M2 (catalog no. F1804; Sigma-Aldrich Chemical Company) were used to detect HLA class II and gp42, respectively. Monoclonal anti-FLAG M2 antibody (F1804) and polyclonal anti-FLAG antibody (F7425) were obtained from Sigma-Aldrich Chemical Company.
Plasmids.
The 27th immunoglobulin-like (I27) domain of muscle protein titin (schematically represented in Fig. 1D) was PCR amplified with sequence-specific primers containing PasI-modified ends and cloned so that one, two, and four copies of I27 (22) were obtained. The I27 domains were unidirectionally cloned into a unique PasI site (residue 29) of d37-41gp42 in the pCAGGS plasmid. The linkers were cloned just downstream of the transmembrane domain which ends at residue 22 and upstream of the gHgL binding domain which begins at residue 44. Other functional regions of gp42 were not disturbed. The ligated products were transformed into competent Escherichia coli DH5α and selected on plates containing ampicillin. DNA was isolated from overnight cultures using the Qiagen miniprep kit, digested to confirm the presence of the insert and correct orientation, and sequenced with primers internal and adjacent to the site of insertion and in both directions by the Northwestern Genomic Core Facility to confirm the resulting sequences. The resulting linker insertion sequences are shown in Table 1. Large-scale DNA preparations were isolated using Qiagen Endo-Free plasmid maxiprep kit and used in subsequent experiments. EBV gL in pCAGGS was previously described (28).
TABLE 1 .
d37‐41gp42 titin linker insertion mutantsa
| Mutant gp42 | Length of insert (no of amino acids) | Sequence of linker insert |
|---|---|---|
| d37‐41(I27‐1) | 92 | ALIEVEKPLYGVLVFVGETAHFEIELSEPDVHGQWKLKGQPLTASPDAEIIEDGKKHILILHNAQLGMTGEVSFQAANAKSAANLKVKELPR |
| d37-41 (I27-1,2) | 184 | ALIEVEKPLYGVLVFVGETAHFEIELSEPDVHGQWKLKGQPLTASPDAEIIEDGKKHILILHNAQLGMTGEVSFQAANAKSAANLKVKELPRRSLIEVEKPLYGVLVFVGETAHFEIELSEPDVHGQWKLKGQPLTASPDAEIIEDGKKHILILHNAQLGMTGEVSFQAANAKSAANLKVKELPR |
| d31-44 (I27-1,2,3,4) | 368 | ALIEVEKPLYGVLVFVGETAHFEIELSEPDVHGQWKLKGQPLTASPDAEIIEDGKKHILILHNAQLGMTGEVSFQAANAKSAANLKVKELPRALIEVEKPLYGVLVFVGETAHFEIELSEPDVHGQWKLKGQPLTASPDAEIIEDGKKHILILHNAQLGMTGEVSFQAANAKSAANLKVKELPRALIEVEKPLYGVLVFVGETAHFEIELSEPDVHGQWKLKGQPLTASPDAEIIEDGKKHILILHNAQLGMTGEVSFQAANAKSAANLKVKELPRALIEVEKPLYGVLVFVGETAHFEIELSEPDVHGQWKLKGQPLTASPDAEIIEDGKKHILILHNAQLGMTGEVSFQAANAKSAANLKVKELPR |
a Titin immunoglobulin-like 127 domains were PCR amplified from clones kindly provided by Andreas Matouschek (21). Following PCR, bands representing one, two, three, and four titin domainss were digested with PasI and ligated into a unique PasI in d37-41gp42.
Transfection and Western blotting.
CHO-K1 cells were transfected in Opti-MEM (Gibco) medium using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Briefly, 24 h after plating the cells in a six-well plate, various combinations of expression vectors were transfected with Lipofectamine in Opti-MEM overnight. The medium was changed 16 h posttransfection to complete Ham’s F-12 medium, and cells and culture supernatants were collected at 48 h posttransfection or used for cell binding or cell fusion assays. For protein analysis, cells were detached with Versene, washed with phosphate-buffered saline (PBS), and lysed using a 1% Triton X-100 buffer containing protease inhibitors (1 ml of lysis buffer/10 million cells). Culture supernatants were collected prior to cell detachment and spun down to pellet detached cells. Supernatants and lysates were run on 12% Bio-Rad Criterion gels in sodium dodecyl sulfate (SDS) sample buffer at 90 V for l.5 h. Proteins were transferred to Whatman Optitran 0.45-µm nitrocellulose membrane in transfer buffer at 100 V for 90 min. Blots were blocked in Tris-buffered saline with 5% milk for 1 h at room temperature or overnight at 4°C and then incubated for 2 h at room temperature with polyclonal anti-gp42 antibody serum (PB114) 1:2,000 (Fig. 2) in blocking solution, as previously described (27, 29). The blots were washed, and IRDye 800CW-conjugated goat (polyclonal) anti-rabbit IgG (H+L) (catalog no. 926-32211; Li-Cor Biosciences) diluted 1:10,000 in blocking solution was applied for 1 h at room temperature with an aluminum foil cover. The blots were washed and analyzed with Li-Cor Biosciences Odyssey infrared imaging studio software. For gp42 competition experiments, Daudi B cells expressing HLA class II and T7 polymerase were cocultured with CHO-KI cells as effector cells containing vector alone or wt gp42 (0.5 µg) and gH (0.5 µg), gL (0.5 µg), gB (0.5 µg), and T7 polymerase (0.8 µg). Increasing amounts of wild-type gp42 or the d37-41gp42 titin linker insertion mutants (0.01, 0.1, and 1.0 µg) were then added into each transfection mixture, and inhibition was monitored by using a cell-cell fusion assay.
Cell-cell fusion assay.
CHO-K1 cells were transiently transfected as described above. The medium was changed 16 h posttransfection, the cells were detached with Versene, and 37,500 cells per well in triplicate were transferred to duplicate 96-well plates. One plate was used for cELISA (described above), and the other plate was overlaid with equal numbers of CHO-K1 target cells transfected with T7 polymerase luciferase reporter plasmid and relevant test plasmids and Daudi B cells expressing T7 polymerase. The total volume was adjusted to 150 µl with complete Ham’s F-12 medium. Eighteen to 20 h after the cells were laid over the plate, the cells were washed with PBS and lysed for 10 min with 50-µl passive lysis buffer (Promega) per well. Luciferase activity was measured with a PerkinElmer Victor plate reader immediately after the addition of 50 µl/well of luciferase reagent (Promega).
HLA class II and gH/gL binding.
cELISA was used to determine soluble gH (sgH) binding or soluble HLA class DQ2-αII (sDQ2-αII). Briefly, CHO-K1 cells were cotransfected in a six-well dish with each of the FLAG-tagged mutants and wild-type gp42, gB, or wild-type gH. The medium was changed 16 h posttransfection, and 1 h later, the cells were detached with Versene, counted using a Beckman Coulter Z1 particle counter, 37,500 cells were transferred to each well of a 96-well plate, and the total volume was adjusted to 150 µl with complete Ham’s F-12 medium. Twenty-four hours later, the cells were washed once with PBS. To determine whether the significantly reduced fusion was due to altered or decreased binding to either gHgL or HLA class II, we transfected CHO-KI cells with a previously purified sDQ2-αII (HLA class II) that was kindly provided by Elizabeth Mellins at Stanford University and used to monitor HLA class II binding (19), and a previously described published soluble gHgL (sgHgL) collected from culture supernatant was used to monitor gH/gL binding (25). CHO-KI cells were transfected with the vector control, wt gp42, d37-41gp42, d37-41gp42 (I27-1), d37-41gp42 (I27-1,2), and d31-44gp42 (I27-1,2,3,4), and 24 h later, they were overlaid with sgHgL or HLA class II for 1 h at 4°C. Bound protein was then determined by cELISA either using anti-HLA class II DQ antibody (1a3) (catalog no. ab24265; Abcam) and anti-FLAG-M2 (catalog no. F1804; Sigma) which recognized the epitope-tagged gH/gL. The cells were washed, fixed, and incubated with biotinylated goat anti-mouse IgG (Sigma) for 30 min at room temperature, followed by incubation with streptavidin-horseradish peroxidase (HRP) (GE Healthcare) for 30 min at room temperature and with 3,3′,5,5′-tetramethylbenzidine (TMB) one-component HRP substrate (BioFX). Absorbance readings were taken at 380 nm using a Wallac-Victor plate reader (PerkinElmer).
ACKNOWLEDGMENTS
We thank the members of Andreas Matouschek’s lab, particularly Susan Fishbain, for providing us with clones containing various copies of titin I27. We thank Elizabeth Mellins’s lab at Stanford University for providing us with purified HLA-DQ2 protein. We thank the members of R. Longnecker’s and T. S. Jardetzky’s laboratories for their help and support. We thank Lindsey Hutt-Fletcher for providing key reagents and Nanette Susmarski for excellent technical support. Finally, we thank Karthik Sathiyamoorthy and Britta Mohl for carefully reading the manuscript and helping to design Fig. 1.
This research was supported by grant AI076183 (R.L. and T.S.J.) from the National Institute of Allergy and Infectious Diseases, grants CA117794 (R.L. and T.S.J.) and CA133063 (R.L. and C.L.R.) from the National Cancer Institute, and by fellowships 12POST9380013 and 14POST18600021 (J.C.) from the American Heart Association. R.L. is the Dan and Bertha Research Professor at Northwestern University.
Footnotes
Citation Rowe CL, Chen J, Jardetzky TS, Longnecker R. 2015. Membrane anchoring of Epstein-Barr virus gp42 inhibits fusion with B cells even with increased flexibility allowed by engineered spacers. mBio 6(1):e02285-14. doi:10.1128/mBio.02285-14.
REFERENCES
- 1.Longnecker R, Kieff E, Cohen JI. 2013. Epstein-Barr virus, p 1898–1959. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Connolly SA, Jackson JO, Jardetzky TS, Longnecker R. 2011. Fusing structure and function: a structural view of the herpesvirus entry machinery. Nat Rev Microbiol 9:369–381. doi: 10.1038/nrmicro2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haan KM, Kwok WW, Longnecker R, Speck P. 2000. Epstein-Barr virus entry utilizing HLA-DP or HLA-DQ as a coreceptor. J Virol 74:2451–2454. doi: 10.1128/JVI.74.5.2451-2454.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Q, Spriggs MK, Kovats S, Turk SM, Comeau MR, Nepom B, Hutt-Fletcher LM. 1997. Epstein-Barr virus uses HLA class II as a cofactor for infection of B lymphocytes. J Virol 71:4657–4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borza CM, Hutt-Fletcher LM. 2002. Alternate replication in B cells and epithelial cells switches tropism of Epstein-Barr virus. Nat Med 8:594–599. doi: 10.1038/nm0602-594. [DOI] [PubMed] [Google Scholar]
- 6.Chesnokova LS, Hutt-Fletcher LM. 2011. Fusion of Epstein-Barr virus with epithelial cells can be triggered by αvβ5 in addition to αvβ6 and αvβ8, and integrin binding triggers a conformational change in glycoproteins gHgL. J Virol 85:13214–13223. doi: 10.1128/JVI.05580-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chesnokova LS, Nishimura SL, Hutt-Fletcher LM. 2009. Fusion of epithelial cells by Epstein-Barr virus proteins is triggered by binding of viral glycoproteins gHgL to integrins αvβ6 or αvβ8. Proc Natl Acad Sci U S A 106:20464–20469. doi: 10.1073/pnas.0907508106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Q, Turk SM, Hutt-Fletcher LM. 1995. The Epstein-Barr virus (EBV) gene product associates with the gH and gL homologs of EBV and carries an epitope critical to infection of B cells but not of epithelial cells. J Virol 69:3987–3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ressing ME, van Leeuwen D, Verreck FA, Keating S, Gomez R, Franken KL, Ottenhoff TH, Spriggs M, Schumacher TN, Hutt-Fletcher LM, Rowe M, Wiertz EJ. 2005. Epstein-Barr virus gp42 is posttranslationally modified to produce soluble gp42 that mediates HLA class II immune evasion. J Virol 79:841–852. doi: 10.1128/JVI.79.2.841-852.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sorem J, Jardetzky TS, Longnecker R. 2009. Cleavage and secretion of Epstein-Barr virus glycoprotein 42 promote membrane fusion with B lymphocytes. J Virol 83:6664–6672. doi: 10.1128/JVI.00195-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirschner AN, Omerovic J, Popov B, Longnecker R, Jardetzky TS. 2006. Soluble Epstein-Barr virus glycoproteins gH, gL, and gp42 form a 1:1:1 stable complex that acts like soluble gp42 in B-cell fusion but not in epithelial cell fusion. J Virol 80:9444–9454. doi: 10.1128/JVI.00572-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X, Kenyon WJ, Li Q, Müllberg J, Hutt-Fletcher LM. 1998. Epstein-Barr virus uses different complexes of glycoproteins gH and gL to infect B lymphocytes and epithelial cells. J Virol 72:5552–5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu F, Marquardt G, Kirschner AN, Longnecker R, Jardetzky TS. 2010. Mapping the N-terminal residues of Epstein-Barr virus gp42 that bind gH/gL by using fluorescence polarization and cell-based fusion assays. J Virol 84:10375–10385. doi: 10.1128/JVI.00381-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen J, Jardetzky TS, Longnecker R. 2013. The large groove found in the gH/gL structure is an important functional domain for Epstein-Barr virus fusion. J Virol 87:3620–3627. doi: 10.1128/JVI.03245-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen J, Rowe CL, Jardetzky TS, Longnecker R. 2012. The KGD motif of Epstein-Barr virus gH/gL is bifunctional, orchestrating infection of B cells and epithelial cells. mBio 3(1):e00290-11. doi: 10.1128/mBio.00290-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mullen MM, Haan KM, Longnecker R, Jardetzky TS. 2002. Structure of the Epstein-Barr virus gp42 protein bound to the MHC class II receptor HLA-DR1. Mol Cell 9:375–385. doi: 10.1016/S1097-2765(02)00465-3. [DOI] [PubMed] [Google Scholar]
- 17.Matsuura H, Kirschner AN, Longnecker R, Jardetzky TS. 2010. Crystal structure of the Epstein-Barr virus (EBV) glycoprotein H/glycoprotein L (gH/gL) complex. Proc Nat Acad Sci U S A 107:22641–22646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kirschner AN, Lowrey AS, Longnecker R, Jardetzky TS. 2007. Binding-site interactions between Epstein-Barr virus fusion proteins gp42 and gH/gL reveal a peptide that inhibits both epithelial and B-cell membrane fusion. J Virol 81:9216–9229. doi: 10.1128/JVI.00575-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sathiyamoorthy K, Jiang J, Hu YX, Rowe CL, Mohl BS, Chen J, Jiang W, Mellins ED, Longnecker R, Zhou ZH, Jardetzky TS. 2014. Assembly and architecture of the EBV B cell entry triggering complex. PLoS Pathog 10:e1004309. doi: 10.1371/journal.ppat.1004309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Politou AS, Gautel M, Pfuhl M, Labeit S, Pastore A. 1994. Immunoglobulin-type domains of titin: same fold, different stability? Biochemistry 33:4730–4737. doi: 10.1021/bi00181a604. [DOI] [PubMed] [Google Scholar]
- 21.Improta S, Politou AS, Pastore A. 1996. Immunoglobulin-like modules from titin I-band: extensible components of muscle elasticity. Structure 4:323–337. doi: 10.1016/S0969-2126(96)00036-6. [DOI] [PubMed] [Google Scholar]
- 22.Inobe T, Fishbain S, Prakash S, Matouschek A. 2011. Defining the geometry of the two-component proteasome degron. Nat Chem Biol 7:161–167. doi: 10.1038/nchembio.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.von Castelmur E, Marino M, Svergun DI, Kreplak L, Ucurum-Fotiadis Z, Konarev PV, Urzhumtsev A, Labeit D, Labeit S, Mayans O. 2008. A regular pattern of Ig super-motifs defines segmental flexibility as the elastic mechanism of the titin chain. Proc Natl Acad Sci U S A 105:1186–1191. doi: 10.1073/pnas.0707163105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rowe CL, Matsuura H, Jardetzky TS, Longnecker R. 2011. Investigation of the function of the putative self-association site of Epstein-Barr virus (EBV) glycoprotein 42 (gp42). Virology 415:122–131. doi: 10.1016/j.virol.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rowe CL, Connolly SA, Chen J, Jardetzky TS, Longnecker R. 2013. A soluble form of Epstein-Barr virus gH/gL inhibits EBV-induced membrane fusion and does not function in fusion. Virology 436:118–126. doi: 10.1016/j.virol.2012.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Atanasiu D, Saw WT, Cohen GH, Eisenberg RJ. 2010. Cascade of events governing cell-cell fusion induced by herpes simplex virus glycoproteins gD, gH/gL, and gB. J Virol 84:12292–12299. doi: 10.1128/JVI.01700-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McShane MP, Mullen MM, Haan KM, Jardetzky TS, Longnecker R. 2003. Mutational analysis of the HLA class II interaction with Epstein-Barr virus glycoprotein 42. J Virol 77:7655–7662. doi: 10.1128/JVI.77.13.7655-7662.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haan KM, Lee SK, Longnecker R. 2001. Different functional domains in the cytoplasmic tail of glycoprotein B are involved in Epstein-Barr virus-induced membrane fusion. Virology 290:106–114. doi: 10.1006/viro.2001.1141. [DOI] [PubMed] [Google Scholar]
- 29.Fan Q, Longnecker R. 2010. The Ig-like v-type domain of paired Ig-like type 2 receptor alpha is critical for herpes simplex virus type 1-mediated membrane fusion. J Virol 84:8664–8672. doi: 10.1128/JVI.01039-10. [DOI] [PMC free article] [PubMed] [Google Scholar]