ABSTRACT
Here we present an extensive genomic and genetic analysis of Escherichia coli strains of serotype O78 that represent the major cause of avian colisepticemia, an invasive infection caused by avian pathogenic Escherichia coli (APEC) strains. It is associated with high mortality and morbidity, resulting in significant economic consequences for the poultry industry. To understand the genetic basis of the virulence of avian septicemic E. coli, we sequenced the entire genome of a clinical isolate of serotype O78—O78:H19 ST88 isolate 789 (O78-9)—and compared it with three publicly available APEC O78 sequences and one complete genome of APEC serotype O1 strain. Although there was a large variability in genome content between the APEC strains, several genes were conserved, which are potentially critical for colisepticemia. Some of these genes are present in multiple copies per genome or code for gene products with overlapping function, signifying their importance. A systematic deletion of each of these virulence-related genes identified three systems that are conserved in all septicemic strains examined and are critical for serum survival, a prerequisite for septicemia. These are the plasmid-encoded protein, the defective ETT2 (E. coli type 3 secretion system 2) type 3 secretion system ETT2sepsis, and iron uptake systems. Strain O78-9 is the only APEC O78 strain that also carried the regulon coding for yersiniabactin, the iron binding system of the Yersinia high-pathogenicity island. Interestingly, this system is the only one that cannot be complemented by other iron uptake systems under iron limitation and in serum.
IMPORTANCE
Avian colisepticemia is a severe systemic disease of birds causing high morbidity and mortality and resulting in severe economic losses. The bacteria associated with avian colisepticemia are highly antibiotic resistant, making antibiotic treatment ineffective, and there is no effective vaccine due to the multitude of serotypes involved. To understand the disease and work out strategies to combat it, we performed an extensive genomic and genetic analysis of Escherichia coli strains of serotype O78, the major cause of the disease. We identified several potential virulence factors, conserved in all the colisepticemic strains examined, and determined their contribution to growth in serum, an absolute requirement for septicemia. These findings raise the possibility that specific vaccines or drugs can be developed against these critical virulence factors to help combat this economically important disease.
INTRODUCTION
Avian colisepticemia is a severe systemic disease of birds caused by septicemic Escherichia coli. These strains fall into the general group of extraintestinal pathogenic E. coli (ExPEC) bacteria that are involved in human septicemia as well. Many of the avian pathogenic E. coli (APEC) isolates are very similar to human septicemia isolates, raising questions about the zoonotic nature of the infective agent.
In birds, colisepticemia results in high morbidity and mortality (1–3). The mortality is either direct or by association with various disease conditions, such as various viral or mycoplasma diseases, as a secondary pathogen. Around the world, the disease is commonly associated with O serotypes O1, O2, and O78, with the latter two constituting about 80% of the cases. The bacteria associated with avian colisepticemia are highly antibiotic resistant, making antibiotic treatment ineffective. In addition, so far there is no effective vaccine. Therefore, the disease, which frequently occurs in chickens and turkeys, has a high economic toll to the poultry industry all over the world.
The bacteria involved in colisepticemia vary in their clonal origin (4), and there is a high degree of diversity in the virulence genes (5). Yet, there are several virulence factors that are conserved in all APEC strains, such as a large ColV plasmid (6). This plasmid carries many virulence-associated genes that together with the virulence genes carried on the chromosome promote pathogenesis. The virulence-related genes include the yersiniabactin gene cluster coding for the iron uptake system from the Yersinia high-pathogenicity island (HPI), genes involved in serum resistance such as the iss (increased serum survival) operon (7–10) and a degenerate type 3 secretion system (ETT2sepsis) previously shown to be important for virulence (11).
Here we performed genomic analyses of APEC strains isolated from poultry with avian colisepticemia which represent the major avian septicemic serotypes O78 and O2. The O78:H19 ST88 isolate 789 (O78-9) was fully sequenced. We then compared the genome sequence of APEC O78-9 with publicly available genomes: APEC O1, the draft genome sequence of APEC O2, and other APEC O78 strains. Comparative genome analysis demonstrated that individual APEC O78 isolates differ (i) in their overall virulence gene content and (ii) in the presence of mobile or mobilizable genetic elements harboring these virulence determinants. Nevertheless, we identified several genes or gene clusters that are conserved in the APEC septicemic isolates. It is assumed that these genes are critical for serum survival, which involves coping with the serum nutritional immunity—caused by the fact that various nutritional requirements are unavailable to pathogens—and the innate immunity of the host. In a previous paper (12), we showed that a major factor in the nutritional immunity is iron limitation. Furthermore, we showed that the expression of more than 80% of the serum-upregulated genes are controlled by the iron regulator Fur (12). Indeed, the sequenced APEC strains carry genes encoding several iron acquisition systems that appear to be overlapping. In order to determine which of these systems are important for septicemia, we constructed deletion mutants of isolate O78-9 and determined their ability to survive in serum. Here we show that there is a functional overlap of the iron acquisition systems, except for the yersiniabactin whose absence cannot be fully complemented by the other iron acquisition systems. Additional genes essential for survival in serum are the plasmid iss gene and ETT2sepsis, the degenerate type 3 secretion system ETT2 (E. coli type 3 secretion system 2).
RESULTS
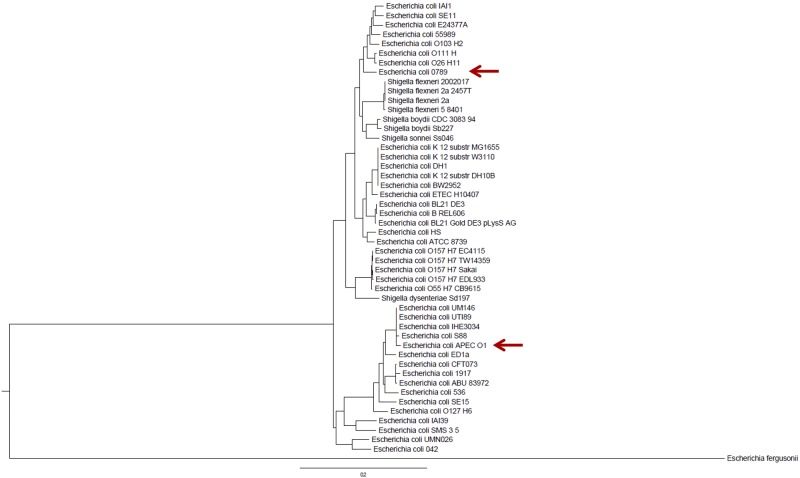
Positions of avian septicemic strains in the E. coli phylogenetic tree.
An E. coli phylogenetic tree was reconstructed using 1,548 conserved core gene clusters of 47 E. coli genomes. For this analysis, only bacteria with whole-genome sequences were used. As can be seen in Fig. 1, the avian septicemic strains belong to different clades. APEC O78:H19 ST88 isolate 789 (O78-9) clusters with a heterogenous group of E. coli isolates, including strains of serogroups O111 and O103, usually comprising many diarrheagenic E. coli isolates. In contrast, APEC O1 strains reside with the urinary tract infection (UTI) strains such as CFT073 (13) and strain 536 (14). These results clearly show that APEC constitute a group of bacteria that are phylogenetically distant although they cause the same disease and carry similar virulence factors.
FIG 1 .
Phylogenetic tree of E. coli. The tree is based on 1,548 conserved core gene clusters of 47 E. coli genomes. For this analysis, only bacteria with whole-genome sequences were used. The tree was rooted using Escherichia fergusonii. Scale units are in nucleotide replacements per site. The arrows mark strain O78:H19 ST88 isolate 789 (O78-9) and strain O1 (22). substr, substrain.
Genome of APEC O78-9 compared with the other published APEC genomes.
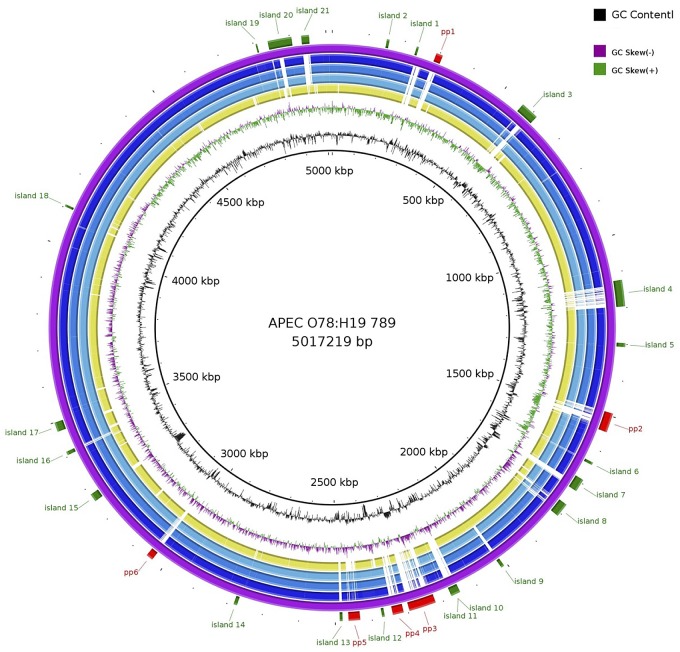
The genome of E. coli APEC O78:H19 ST88 isolate 789 includes the 5,017,219-bp chromosome (4,766 open reading frames [ORFs], 50.8% G+C content, and 90 tRNA loci) as well as the three plasmids p789-1 (144,859 bp), p789-2 (106,397 bp), and p789-3 (4,456 bp) (Fig. 2 and Table 1). Although the chromosomal sizes of all the sequenced O78 strains are comparable, O78-9 has more plasmid-encoded ORFs. The O78-9 genome contains 20 islands and seven bacteriophage-related genomic regions (Table 2).
FIG 2 .
Genome comparison between E. coli O78-9, different APEC strains, and E. coli K-12 MG1655. The circles from outside to inside: O78-9; O78:H9-ST23 strains c7122, IMT 2125, and NC21063; and E. coli K-12 strain MG1655. Conserved and variable regions were detected in the genomes of the different APEC strains. E. coli O78-9 was used as a reference. The chromosomal localization of regions specific for APEC O78-9 (islands and prophages) have been indicated. The map was created using the BLAST Ring Image Generator (BRIG) (67).
TABLE 1 .
Genome characteristics of different APEC O78 isolates
| Genome characteristic | Parameter value for the following APEC isolate: |
|||
|---|---|---|---|---|
| O78-9 (AC/1) (O78:H19 ST88) | NC20163 (O78:H9 ST23) | χ7122 (O78:H9 ST23) | IMT2125 (O78:H9 ST23) | |
| Length (bp) | 5,017,219 | 4,798,435 | 4,771,701 (4 contigs, 8 gaps) | 4,754,148 (15 contigs, 32 gaps) |
| G+C content (%) | 50.8 | 50.7 | 51.6 | 51.7 |
| No. of genes | 4,784 | 4,589 | 4,627 | 4,638 |
| No. of tRNAs | 90 | 88 | 86 | 78 |
| No. of rRNAs | 22 | 19 | 22 | 8 |
| No. of plasmids | 3 | 2 | 4 | 4 |
| Plasmids | pAPEC 789-1 (144,859 bp) = ColV | pAPEC-O78-1 (217,830 bp) | pχ7122-1 (103,275 bp) | pIMT2125-1 (118,067 bp; 2 gaps) |
| pAPEC 789-2 (106,397 bp) | pAPEC-O78-2 (113,260 bp) | pχ7122-2 (82,676 bp) | pIMT2125-2 (106,097 bp; 1 gap) | |
| pAPEC 789-3 (4,500 bp) | pχ7122-3 (56,676 bp) | pIMT2125-3 (4,107 bp) | ||
| pχ7122-4 (4,300 bp) | pIMT2125-4 (1,616 bp) | |||
TABLE 2 .
Genomic islands and bacteriophage-related genomic regions
| Position |
Size (bp) | Designation | Characteristic(s) or trait(s) encoded | |
|---|---|---|---|---|
| Start | End | |||
| 150034 | 157248 | 7,215 | Island 1 | Yad fimbriae |
| 236837 | 269652 | 32,815 | Island 2 | aspV-associated island, T6SS |
| 290228 | 308559 | 18,331 | Phage 1 | thrW-associated phage |
| 564684 | 615140 | 50,456 | Phage 2 | argU-associated island (bor-like) |
| 1123437 | 1196175 | 72,738 | Island 3 | PGA, AC/1 fimbriae, salmochelin, antigen 43 |
| 1296591 | 1307180 | 10,589 | Island 4 | |
| 1491934 | 1545302 | 53,368 | Phage 3 | |
| 1635793 | 1643068 | 7,275 | Island 5 | T6SS |
| 1689969 | 1725904 | 35,935 | Island 6 | Yde fimbriae, mercury resistance |
| 1771175 | 1809758 | 38,583 | Island 7 | |
| 2009004 | 2018168 | 9,164 | Island 8 | Major facilitator superfamily sugar transport system |
| 2148621 | 2174619 | 25,998 | Island 9 | leuZ-associated island |
| 2220926 | 2263405 | 42,479 | Phage 4 | serU-associated phage |
| 2264945 | 2295737 | 30,792 | Island 10 | asnT-associated HPI (yersiniabactin) |
| 2312011 | 2342101 | 30,090 | Phage 5 | Phage |
| 2364700 | 2372039 | 7,339 | Island 11 | O78 O antigen |
| 2432117 | 2464024 | 31,907 | Phage 6 | |
| 2480245 | 2487997 | 7,752 | Island 12 | Yeh fimbriae |
| 2772835 | 2782381 | 9,546 | Island 13 | argW-associated island |
| 3035749 | 3054798 | 19,049 | Phage 7 | ileY-associated phage |
| 3258021 | 3279955 | 21,934 | Island 14 | glyU-associated island, ETT2 |
| 3404120 | 3413945 | 9,825 | Island 15 | |
| 3473251 | 3499527 | 26,276 | Island 16 | |
| 4102977 | 4109122 | 6,145 | Island 17 | waa |
| 4805771 | 4809971 | 4,200 | Island 18 | |
| 4839584 | 4905463 | 65,879 | Island 19 | leuX-associated island (fec, glycine-betaine transport) |
| 4932397 | 4953623 | 21,226 | Island 20 | |
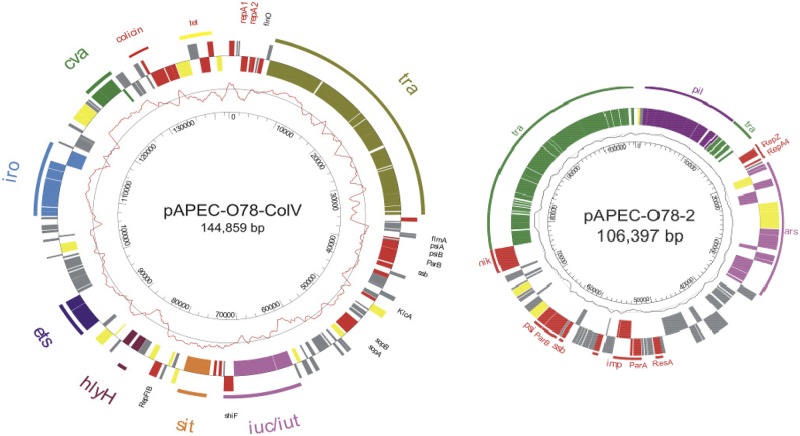
The ColV plasmids are characterized by an F-like transfer region similar to those of pAPEC-O1-ColBM, and pAPEC-O2-ColV, the E. coli K-12 F plasmid, and several F-like E. coli plasmids (Fig. 3). The ColV plasmid of strain O78-9 (p789-1) contains the following: a 72-kb region that comprises characteristic APEC virulence determinants, including the cva operon coding for colicin V production; the iss gene, involved in increased serum survival; ompT, encoding an outer membrane protease; and hlyF, a putative hemolysin previously identified on many other APEC virulence plasmids (6, 15–19). Like many other colicin plasmid-type APEC plasmids, p789-1 also contains the siderophore system determinants coding for aerobactin (iuc and iut), salmochelin (iroNBCDE), and the Salmonella iron transport (sit) ABC transport system. It also contains the etsAB genes (E. coli transport system), and the shiF gene previously found on a pathogenicity island (PAI) of Shigella flexneri. Furthermore, p789-1 carries a tetracycline resistance (tet) operon.
FIG 3 .
Map of the large virulence plasmids of APEC O78-9. The outer circle shows ORFs with functional classifications. The functional classifications are indicated by colors as follows: red, plasmid replication, stabilization, and maintenance; olive, plasmid transfer; yellow, mobile genetic elements; gray, unknown function. Virulence-associated operons cva (green), iro (blue), ets (dark blue), hlyF (brown), sit (orange), iuc or iut (pink) are indicated. The second and third circles show ORFs in the forward and reverse orientations, respectively. The fourth circle shows the G+C content plot. The innermost circle shows the scale. The map was created by using GenVision from DNASTAR.
The second plasmid, p789-2, carries the ars operon that confers resistance to heavy metals. It is similar to several other plasmids of members of the family Enterobacteriaceae (pND12_96 of porcine enterotoxigenic E. coli isolate ND12 [HQ114282] [GenBank accession numbers shown in brackets], pUMNK88_91 of porcine enterotoxigenic E. coli strain UMNK88 [CP002731], Shigella sonnei plasmid p9 [AB021078], Salmonella enterica plasmids pNF1358 [DQ017661], pR64 [AP005147], pCVM29188_101 [CP001121], and pSL476_91 [CP001118]). The function of the third plasmid, p789-3, is so far unknown. This plasmid carries only seven ORFs that are required for the maintenance and mobilization or that have not yet been functionally characterized. pO78-3 exhibits nucleotide sequence homology to plasmid pSN11/00Kan of Salmonella enterica subsp. Enterica serovar Newport strain SN11/00 (GQ470395) and plasmid pEC08-5 of extended-spectrum β-lactamase (ESBL)-producing Escherichia coli strain EC08 (JX238444) (GenBank accession numbers shown in parentheses).
We compared the completely closed chromosomal sequence of strain O78-9 with that of APEC O78:H9_ST23 strain NC20163 (20) (Fig. 2). Strain O78-9 possesses 363 singletons absent from the genome of APEC O78 NS20163, which has 219 genes which are absent from the APEC O78-9 chromosome. These singletons can often be allocated to prophage-like regions within the bacterial chromosome.
Two additional nonclosed APEC O78:H9-ST23 genome sequences (isolates 7122 and IMT2125) were recently published (21), and their sequences indicate that all APEC O78 strains belonging to the ST23 clonal complex are closely related. The ST23 strains and ST88 O78 isolate 789 differ mainly in the content of islands and bacteriophage-related genomic regions (Fig. 2 and Table 2). These genomic regions code for several virulence- or fitness-associated functions such as a type 6 secretion system, a type 3 secretion system (ETT2 [E. coli type 3 secretion system 2]), parts of a type 2 secretion system, several fimbriae (Yad, Yde, Yeh, and Fac), lipopolysaccharide (LPS) biosynthesis and the extracellular matrix component poly-N-acetylglucosamine (PGA) (Fig. 2 and Table 2). Interestingly, strain O78-9 differed markedly in the unique presence of chromosomal determinants coding for individual iron uptake systems—the salmochelin and yersiniabactin gene clusters (Tables 2 and 3). It should be noted that strain 78-9 carries two salmochelin gene clusters, one on the chromosome (island 3) and one on the plasmid (p789-1) (Fig. 2 and Table 3).
TABLE 3 .
Iron uptake determinants present in the genomes of different APEC strain:
| Iron uptake determinant(s) (genes) | Presence or absence of iron uptake determinant in the following APEC strain: |
|||||
|---|---|---|---|---|---|---|
| O78-9 (O78:H19 ST88) | O78:H9 ST23 [NC020163] | χ7122 (O78:H9 ST23) | IMT2125 (O78:H9 ST23) | O1:K1:H7 ST95 [NC008563] | O2 1772 | |
| Hemin uptake (chuAS) | − | − | − | − | + | + |
| Ferrous iron uptake (feoAB) | + | + | + | + | + | + |
| Ferrichrome uptake (fhuACDB) | + | + | + | + | + | + |
| Ferric citrate uptake (fecEDCBARI) | + | + | + | + | − | − |
| Enterobactin (ent-fes-fep) | + | + | + | + | + | + |
| Catecholate siderophore receptor (fiu) | + | + | + | + | + | + |
| Salmochelin (iroNEDCB) | p789-1 and island 3 | − | pχ7122-1 | − | pAPEC-O1-ColBM | + |
| Yersiniabactin (ybtSXQPA-irp2-irp1-ybtUTE) | + | − | − | − | + | + |
| Aerobactin (iucABCD) | p789-1 | − | pχ7122-1 | − | pAPEC-O1-ColBM | + |
| ABC iron transport system (eitABCD) | − | − | pχ7122-1 | − | pAPEC-O1-ColBM | + |
| ABC iron transport system (sitABCD) | p789-1 | pχ7122-1 | + | |||
Generally, multiple gene clusters coding for iron uptake systems can be found in APEC O78-9 and the O78:H9-ST23 isolates. All of these systems have been correlated with virulence. In strain O78-9, most of the siderophore systems are encoded by genes on the ColV plasmid. The chromosome contains the genes encoding enterobactin and salmochelin, and it also contains the genes coding for yersiniabactin—the iron acquisition system of the Yersinia high-pathogenicity island (Tables 2 and 3).
The APEC O78 strains also differ in their plasmid content (Table 1). Whereas strain O78-9 carried three plasmids, two plasmids were found in O78:H9-ST23 isolate NC20163, and four plasmids were detected in each of the APEC O78:H9 isolates χ7122 and IMT2125 (Table 1). All APEC strains possess large virulence-associated colicin plasmids (ColV) which were also found in other APEC serotypes, e.g., APEC O1 (22) and O2 (U. Dobrindt and E. Z. Ron, unpublished results).
These colicin plasmids of the APEC O78 strains harbor the determinants coding for the Salmonella iron transport (sit), aerobactin (iuc), and salmochelin (iro) siderophore systems, which can also be chromosomally encoded as parts of pathogenicity islands, e.g., in other extraintestinal pathogenic E. coli and Shigella strains.
iss genes.
The ColV plasmid contains the iss (increased serum survival) gene that has been identified to be required for serum resistance (7, 23). This gene is homologous to the bor gene of E. coli K-12 that codes for a small membrane-associated protein (24). The sequenced individual APEC isolates possess different numbers of iss alleles in their genomes. Whereas APEC O78-9 and APEC O78:H9-ST23 IMT2125 possess one chromosomal iss variant, the APEC O78:H9-ST23 isolates NC20163 and χ7122 carry two chromosomal iss alleles. Two chromosomal iss alleles can also be found in APEC O1:K1:H7 (9). Furthermore, iss gene variants can be found on large APEC virulence plasmids, e.g., on p789-1, pχ7122, pColBM, so that up to three iss copies can exist in an APEC genome. Unlike the three APEC O78:H9-ST23 isolates NC20163, χ7122, and IMT2125 that harbor three iss alleles each, strain O78-9 has only two alleles. One is located on p789-1, and one is on the chromosome (putative prophage 2). Whereas the different Iss protein variants of the APEC O78 strains exhibit only minor differences in their amino acid sequence (Fig. 4A), it is interesting to note that they differ in their upstream regions, including the putative promoters and transcription factor binding sites (Fig. 4B).
FIG 4 .
Comparison of Iss proteins and their upstream regions across different APEC strains. (A) Iss protein sequence. (B) iss regulatory sequence. Asterisks indicate positions which have a fully conserved residue. Colons indicate conservation between groups of strongly similar properties. chrom., chromosome.
In order to determine the role of the iss genes in serum survival of APEC strain O78-9, the iss gene located on p789-1 (ColV) was deleted. This deletion did not affect growth in minimal and rich media. However, the deletion had a significant effect on the ability of the bacteria to grow in serum as can be seen from the results shown in Fig. 5. These result indicated that removal of the plasmid-located iss gene rendered the strain sensitive to serum. Moreover, the results also showed that the chromosomal iss allele was not able to overcome the loss of the plasmid gene. Experiments to quantify the transcript levels of both iss genes by quantitative real-time PCR (qRT-PCR) indicated that deletion of the iss gene on p789-1 decreased the level of the iss transcript, but only by about 50% (Fig. 6). Consequently, the remaining transcripts detected in the p789-1Δiss mutant originate from the chromosomal iss gene. This suggests that this gene, although it was expressed under the conditions tested, could not compensate for the loss of the p789-1-located iss gene regarding serum survival. Moreover, the serum sensitivity remained even in bacteria overexpressing the chromosomal iss gene or the bor gene. Thus, the plasmid-encoded iss gene is essential for serum survival of O78-9. It is not yet understood why deletion of this gene could not be complemented by the chromosomal gene iss allele.
FIG 5 .
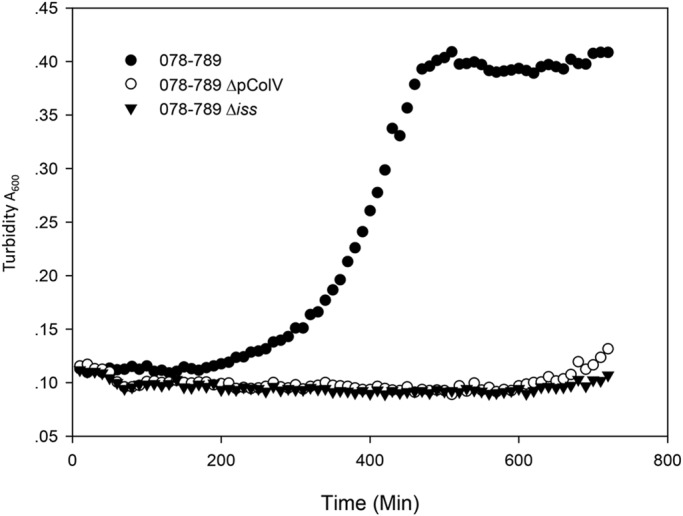
Growth of E. coli O78-9 and its iss mutants in 50% serum. For this experiment, we used E. coli strains O78-9, its mutant deleted for the iss gene (Δiss), and its derivative cured of the ColV plasmid (ΔpColV). Cultures were grown overnight in minimal MOPS medium and diluted into the same medium with or without 50% serum. Growth was monitored at an optical density or absorbance of 600 nm, and growth curves were generated on a Biotek Eon platform.
FIG 6 .
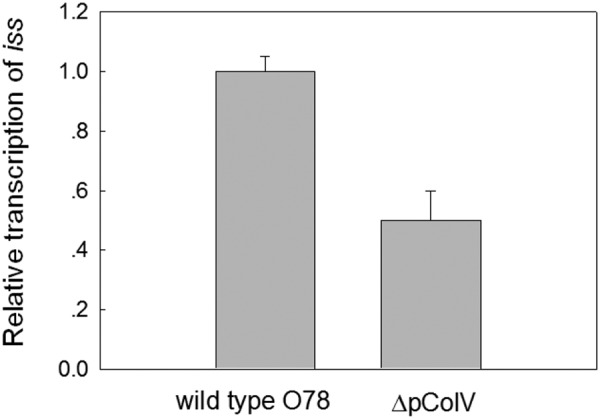
Levels of iss transcript. Cultures were grown in minimal MOPS medium. Transcript levels were determined by qRT-PCR as described in Materials and Methods.
Degenerate TTSS of E. coli O78-9.
The genome of strain O78-9 also codes for a degenerate form of the ETT2 system (E. coli type 3 secretion system 2), which is also present in other septicemic E. coli strains and referred to as ETT2sepsis (11). This type 3 secretion system has a large internal deletion and premature stop codons in several genes, which abolish the functions of the ETT2 needle complex, ATPase, and several other structural proteins. These genetic changes are conserved in all the APEC O78 strains (Fig. 7). It is therefore assumed that the biological role of ETT2sepsis may not involve classic secretion of effectors. Nevertheless, it was previously shown that removal of the putative genes for inner membrane ring formation results in reduced in vivo host mortality, indicating that ETT2sepsis, although degenerated, contributes to pathogenicity (11).
FIG 7 .
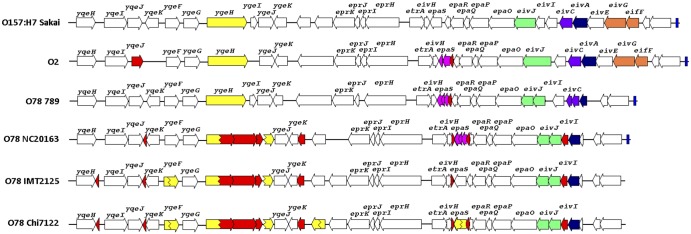
The operon coding for ETT2 (E. coli type 3 secretion system 2) in several E. coli. The genetic structure of the ETT2 determinants of APEC O2 and O78-9 isolates are compared with that of E. coli O157:H7 strain Sakai. Structural differences and ORFs with sequence alterations of the different ETT2 variants relative to the functional ETT2 gene cluster of E. coli O157:H7 strain Sakai have been highlighted. Different colors indicate ORFs affected by the genome plasticity. The ETT2 determinants of APEC O2 strain 1772, O78-9, and O78-H9-ST23 strains IMT2125 and χ7122 are compared.
Here we show that ETT2sepsis is essential for serum survival, as deletion mutants lacking the inner membrane ring of ETT2sepsis (ΔeprHIJK) are completely serum sensitive (Fig. 8). Although the precise role of ETT2sepsis in serum resistance is not fully elucidated, we have evidence that it affects bacterial surface properties, as deletion mutants differ from the wild type in outer surface properties such as pellicle formation (11).
FIG 8 .
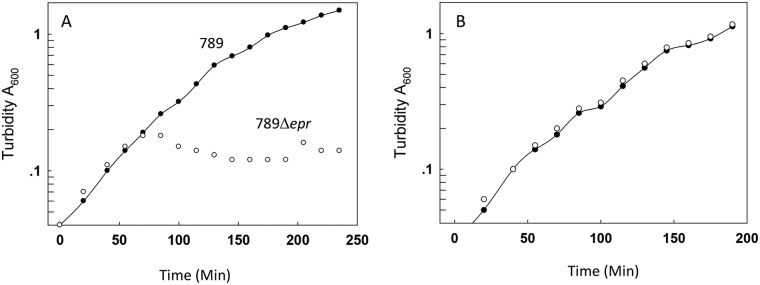
Effect of the ETT2 system on growth of E. coli O78-9 in serum. The experiment was conducted as described in the legend to Fig. 5. Cultures of E. coli O78-9 and its mutant deleted for the eprHIJK gene cluster of the ETT2 system were grown in minimal MOPS medium alone (B) or supplemented with 50% serum (A).
Iron binding systems.
Iron is a required component in the metabolism of bacteria, due to its roles in the catalytic centers of enzymes and in the electron transport chain. Although it is one of the most abundant chemical elements in nature, its biologically active form is scarcely available under physiological conditions (pH 7, aerobic environment) because of the rapid oxidation of Fe2+ to Fe3+ and the subsequent formation of insoluble hydroxide (25). The solubility of Fe3+ under these conditions is 10−9 M, which is far below the concentration of 10−7 M required for bacterial growth (26).
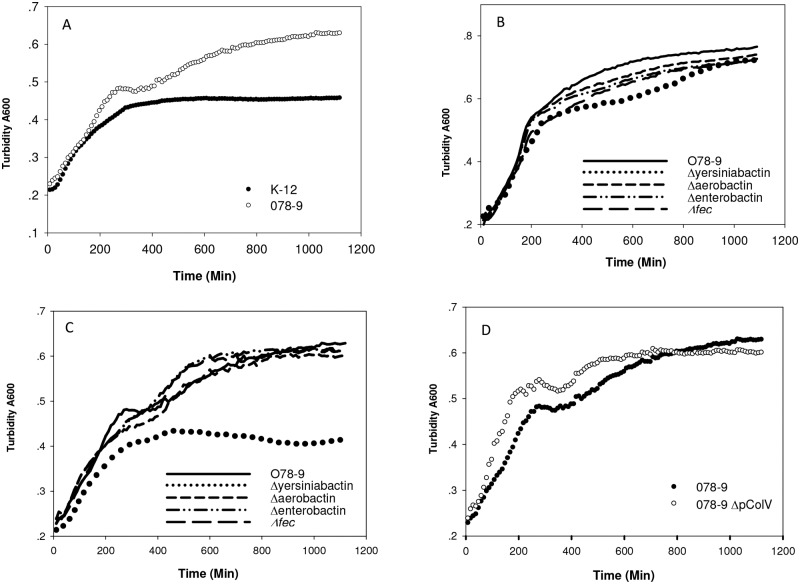
In eukaryotic cells, the iron ions are bound to special proteins such as transferrin and ferritin that release it, when needed, and thus maintain the free ferric ion (Fe3+) concentration at 10−24 M. As a result, septicemic bacteria must compete against these proteins, and they evolved a specific mechanism for the acquisition of iron in their host (27, 28). In Gram-negative bacteria, all iron sources are bound to highly specific proteins that serve as transporters across the outer membrane. E. coli, as well as other enteric bacteria, synthesizes the catecholic (dihydroxybenzoyl) serine trilactone—enterobactin, which is in fact the strongest iron chelator known, forming ferric enterobactin with an affinity of 10−52 M (29). However, under physiological conditions at pH 7.4, this value (picomolar) is 10−37.6, higher than that of transferrin, and is probably neutralized by albumin in serum (30) and by neutrophil gelatinase-associated lipocalin (NGAL) in epithelial cells. Hence, the enterobactin system is presumably insufficient for pathogenicity. The results shown in Fig. 9A indicate that E. coli K-12, which expresses only the enterobactin system, is inhibited by the presence of the iron chelator 2,2′-dipyridyl (Dipyridyl) (2,2′-bipyridine), while strain O78-9 grows very well under these conditions. This result could be explained by the fact that strain O78-9, as well as other avian septicemic strains, codes for other types of siderophore systems (31, 32). These systems include aerobactin (31), yersiniabactin (32), and IroN (33).
FIG 9 .
Growth of E. coli O78-9 and mutants in which genes coding for iron acquisition systems under iron limitation were deleted. Cultures were grown exponentially in LB medium as described in Materials and Methods. Iron depletion was with 0.4 mM 2,2′-dipyridyl. The bacteria tested were E. coli K-12 MG1655, O78-9 and its deletion mutants Δaerobactin, Δenterobactin, Δfec, and Δyersiniabactin cured of the ColV plasmid. (A) Growth of E. coli O78-9 and K-12 in the presence of Dipyridyl; (B) growth of O78-9 and its mutants in LB medium; (C) growth of O78-9 and its deletion strains in the presence of Dipyridyl; (D) growth of O78-9 and ΔColV in the presence of Dipyridyl.
Given the multiplicity of iron binding systems detected in APEC strain O78-9, it is reasonable to assume that not a single siderophore system is essential for iron acquisition under iron-limiting conditions. To test this possibility, we deleted each of the iron acquisition systems of strain O78-9 and determined their ability to grow in LB medium in the absence (Fig. 9B) or presence of the iron chelator 2,2′-dipyridyl (Fig. 9A, C, and D). The results shown in Fig. 9B indicate that all the deletion mutants grew at the same rate as the wild type in LB medium. Removal of each of the plasmid-encoded iron binding systems had no effect on growth even under iron limitation (Fig. 9C), suggesting the existence of a complete function overlap of these genes. In contrast, deletion of the chromosomal gene cluster coding for yersiniabactin considerably reduced the ability of the bacteria to multiply under iron-limiting conditions (Fig. 9C). In fact, under this condition, the yersiniabactin deletion mutant of strain O78-9 behaved comparably to E. coli K-12 (Fig. 9A). These results identified yersiniabactin as the critical gene for iron acquisition in O78-9. This conclusion was further supported by the finding that when yersiniabactin was present, even the deletion of the entire ColV plasmid does not render the bacteria sensitive to 0.4 mM 2,2′-didyridyl (Fig. 9D).
Iron limitation is a critical factor for bacterial survival in serum. The finding presented here, showing that yersiniabactin was essential under low-iron conditions suggested that deletion mutants of the yersiniabactin system will be inhibited in the presence of serum. Indeed, the results presented in Fig. 10 indicated that yersiniabactin is the only iron binding system whose deletion cannot be compensated by the presence of all the other iron binding systems during growth in serum.
FIG 10 .
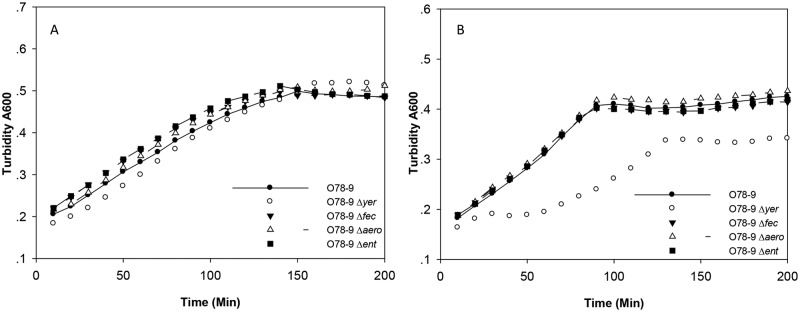
Serum survival of E. coli O78-9 and mutants in which the genes coding for iron acquisition systems were deleted. The culture conditions and bacterial strains and mutants were as described in the legend to Fig. 9. (A) Growth in LB medium alone; (B) growth of O78-9 in the presence of 50% serum.
DISCUSSION
Avian septicemic E. coli strains are highly virulent and cause considerable losses in the poultry industry, as they lead to high morbidity and mortality of chicken and turkeys. The common strains worldwide are E. coli serotype O1, O2 and O78 strains. The latter strains (of serotype O78) exhibit the highest level of virulence, some of them with a 50% lethal dose (LD50) of 102 (E. Z. Ron, unpublished data). Septicemic E. coli bacteria of serotype O78 are not restricted to birds. High mortality sepsis in newborn lambs and goats is caused by E. coli of serotype O78, which often also carries a plasmid encoding K-99 fimbriae (34–36). In humans, septicemic bacteria of serotype O78 were the cause of a newborn meningitis epidemic, which also had a high mortality rate (37, 38).
It should be noted that bacteria of serotype O78 differ significantly from those of serotypes O1 and O2 in their capsule. Serotype O1 and O2 strains carry genes encoding the K1 (sialic acid) capsule that is also present in Neisseria meningitidis and is known to be a strong virulence factor, as it protects the bacteria against the immune system (39–43). In contrast, serotype O78 strains do not have a polysaccharide capsule. Rather, they have a group 4 capsule (44), with a composition similar to that of the O antigen of lipopolysaccharide. This capsule also appears to be essential for pathogenicity, as shown by the fact that its absence—due to transposition (21, 45) or deletion (D. Biran, I. Rosenshine, and E. Z. Ron, unpublished data)—results in loss of virulence.
Here we show that the genomes of E. coli O78 strains exhibit a marked level of genome plasticity. This plasticity is evident in the presence and composition of plasmids, bacteriophages, and transposable element (Fig. 2 and 3 and Tables 1, 2, and 3). An interesting example is the iss gene. This gene is present in one or two copies on the bacterial chromosome, and additional copies can exist on colicin plasmids. Moreover, there are sequence differences in the ORFs coding for the proteins, and significant differences in the noncoding, upstream region (Fig. 4A and B). Yet, the amino acid compositions of the Iss protein are quite similar in all the copies of the gene in E. coli O78 as well as in E. coli O1 (9) and E. coli O2 (U. Dobrindt and E. Z. Ron, unpublished data). Even more surprising is the finding that removal of the plasmid-encoded iss gene renders the bacteria serum sensitive (Fig. 5). These results mean that the chromosomal copy of the gene does not complement the loss of the plasmid allele, although it is expressed. Moreover, even overexpression of the chromosomal gene does not complement the loss of the plasmid gene (data not shown).
An interesting question is whether the chromosomal iss gene has any activity. As of now we are not able to answer this question. Removal of the plasmid gene renders the bacteria completely serum sensitive, so that an additional deletion of the chromosomal iss gene has no phenotype. We tried to remove the chromosomal iss gene in bacteria carrying the ColV plasmid iss gene, but we were not able to obtain this deletion strain.
Another system present in avian strains of O78 serotype and required for serum survival is the degenerate type 3 secretion system—ETT2sepsis (11). This system is probably not involved in secretion. However, we could show that it is essential for serum survival (Fig. 8). The most plausible explanation for this finding is that the system is involved in the structure of the outer surface, which is important for serum survival. The involvement of ETT2sepsis in determining the structure of the bacterial outer surface is supported by experiments showing that ETT2sepsis is important for pellicle formation (11).
One of the factors required for septicemia is the ability to acquire iron, as the serum and tissues are poor in free iron, which is bound by various host proteins. It is therefore expected that septicemic bacteria will have efficient iron binding proteins, and indeed, all the examined bacteria had a large number of systems for iron acquisition. The gene coding for the iron binding proteins of the Yersinia high-pathogenicity island (46) is present only in a few avian septicemic strains (47, 48). Surprisingly, in strain APEC O78-9, this system is the most effective one for binding iron and is the only one that cannot be complemented by other siderophore systems (Fig. 9). Thus, the presence of this iron acquisition system is expected to provide positive selection for the septicemic bacteria expressing it. Although yersiniabactin appears to be the most important iron binding system, it should be noted that the additional systems of iron acquisition are expressed, as detected by transcriptomic and proteomic analysis, and are also upregulated in the absence of iron (12).
The comparative genomic study of avian septicemic E. coli indicates considerable genetic variability between these APEC isolates belonging to the same serotype. Several virulence- and fitness-associated traits have been attributed to the different virulence plasmids of, e.g., APEC O78:H9-ST23 strain χ7122. pχ7122-1, pχ7122-2, and pχ7122-3 encode traits that are required for host-pathogen interaction, including catabolism, systemic infection, biofilm formation, and tolerance to bile salts and acidic pH (49). Although the plasmids described for the APEC O78:H9-ST23 and the O78:H19-ST88 strains were not identical, they share the common feature of sequence similarity with plasmids of enterotoxigenic E. coli and other members of the Enterobacteriaceae such as Shigella and Salmonella. Genome sequence analysis of APEC O78:H9-ST23 strains χ7122 and IMT2125 revealed that based on their genome content, these avian isolates were more closely related to the genomes of human enterotoxigenic E. coli of sequence type 23 (ST23) than to APEC O1-ST95. Great variability has been described for their flexible gene pool, including genomic islands (18). Nevertheless, factors, including the group IV capsule, expressed by the O78:H9-ST23 isolates have been determined which contribute to their virulence, but which were not conserved in the APEC O1-ST95 isolate. Interestingly, only two genes involved in iron uptake mechanisms (entF and yddB) were shown to be negatively selected in a screen for respiratory tract colonization and systemic infection (18). Recently, a human multidrug-resistant E. coli ST131 isolate has been used to screen for bacterial factors required for serum resistance (50). Altogether, 315 essential genes for serum resistance have been identified in this isolate. The majority of the genes involved were shown to code for membrane proteins or to be involved in LPS and colonic acid biosynthesis. Genes that code for siderophore system determinants or that code for components of general transport mechanisms contributing to iron transport have not been selected in the serum resistome screening in E. coli ST131.
We show that serum resistance seems to be mediated by different strategies in individual E. coli pathogens. This is in accordance with the general finding that extraintestinal pathogenic E. coli, including APEC, use multiple strategies involving different sets of virulence-associated traits to cause disease. Serum resistance in APEC O78-9 which causes systemic infection in poultry may require different bacterial factors relative to serum resistance in a human urinary tract infection isolate of serotype O25b:H4-ST131. Our findings further underline the fact that APEC bacteria are a heterogenous group of extraintestinal pathogens and employ several strategies to cause infection in poultry. APEC bacteria often contain several genes coding for essential functions and even sometimes multiple alleles of these important genes. We could show that specific removal of one or more virulence-related functions involved in serum survival and systemic infection renders the bacteria sensitive to serum and reduces their virulence. These findings raise the possibility that specific vaccines or drugs can be developed against these critical septicemia factors to combat this economically important disease.
MATERIALS AND METHODS
Growth conditions.
Cultures were grown in morpholinepropanesulfonic acid (MOPS) minimal medium (51) or in Lennox broth (LB broth; Difco) (52). Exponentially growing cultures were diluted to an optical density at 600 nm (OD600) of 0.04 and examined with a Biotek Eon plate reader, and turbidity at 600 nm was measured every 20 min.
Construction of deletions and overexpressing mutants.
All site-specific gene knockouts were obtained by the method of Datsenko et al. (53). Briefly, competent wild-type E. coli O78:H19 ST88 isolate 789 (O78-9) bacteria were transformed with pKD46 plasmid. The transformants were grown in ampicillin-containing LB medium, induced by arabinose, made competent for electroporation, and stored at −70°C until used. Linear PCR product was made on the template of the kanamycin resistance cassette flanked by the FLP recognition target (FRT) sequences from the pKD4 plasmid according to the region to be deleted. The primers were designed to contain 36 nucleotides from the flanking region of the sequence to be deleted in strain O78-9 (Table 4). Kanamycin-resistant recombinants were screened by means of colony PCR. Unless stated otherwise, the pKD46 plasmid was cured by growth on LB medium at 42°C. Bacteria cured of the ColV plasmid were obtained by penicillin selection for bacteria unable to grow in the presence of tetracycline.
TABLE 4 .
Primers for deletion and qRT-PCR
| Primera | Sequence of primer (5′-3') |
|---|---|
| yersinP1 | TCGGTAAGACGTGCCATCAGGAGGAAGAATGATTTCGTGTAGGCTGGAGCTGCTTC |
| yersinP2 | GGTCGGTTTGCGCTTATTGGGCAGAATGGCGATAAC CATATGAATATCCTCCTTAG |
| fecP1 | GTGGTTTGGTTCTTACGGCCTGTGCAATCTACCTCAGTGTAGGCTGGAGCTGCTTC |
| fecP2 | ATGACTACGTGATAATTAACTTTTGATGCACTCCGCCATATGAATATCCTCCTTAG |
| entP1 | AGTTTTCCTGCAATCTCATTTAATTCTGTCGGGCTCGTGTAGGCTGGAGCTGCTT |
| entP2 | TCATTATTAAAGCCTTTATCATTTTGTGGAGGATGACATATGAATATCCTCCTTAG |
| aeroP1 | ATGTAGGCTGTCGCTTTTGATGGGCTTGCTGTCTTCGTGTAGGCTGGAGCTGCTTC |
| aeroP2 | TCCTGTCGACTTTACCACTCTCTCCCGGCTTATTATCATATGAATATCCTCCTTAG |
| issP1 | AGCGGAGTATAGATGCCAAAAGTGATAAAACCGAGCAATCCATATGAATATCCTCCTTA |
| issP2 | ATGCAGGATAATAAGATGAAAAAAATGTTATTTTCTGCCGGTGTAGGCTGGAGCTGCTTC |
| iss-F RT | AATTCCCGAAACGAAGAA |
| iss-R RT | ATGCTTATTACAAGGATG |
P1, primer 1; P2, primer 2. F, forward; R, reverse; RT, real time.
Overexpression strains were obtained by cloning the selected DNA fragments by PCR amplification using specific primers. The amplified fragment was digested with the appropriate restriction enzyme and ligated into a pBAD24 vector, digested with the same restriction enzymes, using Fast-link DNA ligase (Epicentre). The ligation was transformed into competent E. coli TG1 bacteria, and the resulting clones were screened by colony PCR. The final plasmid was extracted from the selected clone, and after sequence verification, it was transformed into the O78-9 strain.
RNA extraction and RT-PCR.
RNA from 500 µl of a bacterial culture was stabilized using the RNA Protect reagent (Qiagen) according to the manufacturer’s instructions. Total RNA was isolated using the RNeasy minikit (Qiagen) for total RNA isolation, and a RNase-free DNase set column (Qiagen) was used to eliminate DNA contamination. For real-time PCR (RT-PCR) experiments, 1 µg of total RNA was reverse transcribed using random hexamers (Amersham) by ImPromII reverse transcriptase (Promega). PCRs were performed using a 500 nM concentration of each gene-specific primer in a 10-µl volume with 1× SYBR green PCR master mix (Applied Biosystems). Reactions were run on an ABI 7700 instrument (Applied Biosystems) using the standard cycling parameters. Relative gene expression data analysis was carried out by the ΔΔCT method.
Genome sequence analysis.
Genomic sequencing was performed using a random shotgun approach. Total genomic DNA was sheared randomly (nebulizing), and the DNA fragments in the range of 1.5 kb to 4 kb were cloned into pCR2.1 TOPO cloning vectors (Invitrogen) to establish a shotgun library. The inserts of the recombinant plasmids were sequenced from both ends using the 96-capillary high-throughput ABI 3730xl DNA analyzer. Approximately 55,000 sequences were processed first with PHRED and Pregap4, assembled into contigs by using the PHRAP assembly tool and edited with Gap4, which is a part of the STADEN package software. PCR-based techniques and primer walking on recombinant plasmids were applied in order to close remaining sequence gaps. Annotation was done using the ERGO tool (54). Open reading frames (ORFs) were predicted with YACOP (55). For annotation, all proteins were screened against Swiss-Prot data and publicly available protein sequences from other completed genomes. All predictions were then verified and manually modified using the ERGO software package (Integrated Genomics, Inc., Chicago, IL) (56) and GenDB (57). Horizontally acquired genomic regions were determined using ISLANDVIEWER (58). Complete genome comparisons were done with ACT (59) and MAUVE (60, 61) based on replicon-specific nucleotide BLAST (62) and with reciprocal protein-based BLAST comparisons to selected E. coli genomes using EDGAR (63).
Computation of E. coli core genome.
The E. coli set of orthologs was defined by identifying pairwise reciprocal tBLASTx best hits (64). We demand at least 95% identity in nucleotide sequence for the region of homology identified by tBLASTx, the high-scoring segment pairs (hsp). These hsp may be only a fraction of the total length of the proteins, and we thus also request that the hsp, excluding gaps, is not less than 50% of the length of each protein. Finally, we also request that the lengths of the two proteins would not differ by more than 20%.
Computation of the E. coli phylogenetic species tree.
We performed multiple-sequence alignments (MSAs) for the 1,548 E. coli core genes using the PRANK program (65). The concatenated MSA over all core genes was used as input to the PhyML program (66) in order to reconstruct the species tree.
Nucleotide sequence accession number.
The E. coli strain O78-9 genome sequence reported in this paper has been deposited in the GenBank database under accession numbers CP010315 (E. coli 78-9 chromosome), CP010316 (p789-1), CP010317 (p789-2), and CP010318 (p789-3).
ACKNOWLEDGMENTS
This project was supported by a DIP grant from the German-Israeli Project Cooperation (RO 2612/1-1), by the EC Network of Excellence EuroPathoGenomics (CEE LSHB-CT-2005-512061), by the ERA-NET Pathogenomics project COLIRISK, by the EMIDA ERA-NET project CombatColiBacillosis (grant 0316039), and by the Israeli Ministry of Health. We thank the Alfried Krupp Kolleg for a fellowship for S.H.
Footnotes
Citation Huja S, Oren Y, Trost E, Brzuszkiewicz E, Biran D, Blom J, Goesmann A, Gottschalk G, Hacker J, Ron EZ, Dobrindt U. 2015. Genomic avenue to avian colisepticemia. mBio 6(1):e01681-14. doi:10.1128/mBio.01681-14.
REFERENCES
- 1. Kabir SML. 2010. Avian colibacillosis and salmonellosis: a closer look at epidemiology, pathogenesis, diagnosis, control and public health concerns. Int. J. Environ. Res. Public Health 7:89–114. doi: 10.3390/ijerph7010089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dho-Moulin M, Fairbrother JM. 1999. Avian pathogenic Escherichia coli (APEC). Vet. Res. 30:299–316. [PubMed] [Google Scholar]
- 3. Mellata M, Dho-Moulin M, Dozois CM, Curtiss R, Brown PK, Arné P, Brée A, Desautels C, Fairbrother JM. 2003. Role of virulence factors in resistance of avian pathogenic Escherichia coli to serum and in pathogenicity. Infect. Immun. 71:536–540. doi: 10.1128/IAI.71.1.536-540.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Adiri RS, Gophna U, Ron EZ. 2003. Multilocus sequence typing (MLST) of Escherichia coli O78 strains. FEMS Microbiol. Lett. 222:199–203. doi: 10.1016/S0378-1097(03)00295-7. [DOI] [PubMed] [Google Scholar]
- 5. Mokady D, Gophna U, Ron EZ. 2005. Extensive gene diversity in septicemic Escherichia coli strains. J. Clin. Microbiol. 43:66–73. doi: 10.1128/JCM.43.1.66-73.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Johnson TJ, Siek KE, Johnson SJ, Nolan LK. 2006. DNA sequence of a ColV plasmid and prevalence of selected plasmid-encoded virulence genes among avian Escherichia coli strains. J. Bacteriol. 188:745–758. doi: 10.1128/JB.188.2.745-758.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Binns MM, Mayden J, Levine RP. 1982. Further characterization of complement resistance conferred on Escherichia coli by the plasmid genes traT of R100 and iss of ColV,I-K94. Infect. Immun. 35:654–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nolan LK, Horne SM, Giddings CW, Foley SL, Johnson TJ, Lynne AM, Skyberg J. 2003. Resistance to serum complement, iss, and virulence of avian Escherichia coli. Vet. Res. Commun. 27:101–110. doi: 10.1023/A:1022854902700. [DOI] [PubMed] [Google Scholar]
- 9. Johnson TJ, Wannemuehler YM, Nolan LK. 2008. Evolution of the iss gene in Escherichia coli. Appl. Environ. Microbiol. 74:2360–2369. doi: 10.1128/AEM.02634-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Foley SL, Horne SM, Giddings CW, Robinson M, Nolan LK. 2000. Iss from a virulent avian Escherichia coli. Avian Dis. 44:185–191. doi: 10.2307/1592523. [DOI] [PubMed] [Google Scholar]
- 11. Ideses D, Gophna U, Paitan Y, Chaudhuri RR, Pallen MJ, Ron EZ. 2005. A degenerate type III secretion system from septicemic Escherichia coli contributes to pathogenesis. J. Bacteriol. 187:8164–8171. doi: 10.1128/JB.187.23.8164-8171.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huja S, Oren Y, Biran D, Meyer S, Dobrindt U, Bernhardt J, Becher D, Hecker M, Sorek R, Ron EZ. 2014. Fur is the master regulator of the extraintestinal pathogenic Escherichia coli response to serum. mBio 5(4):e01460-14. doi: 10.1128/mBio.01460-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Welch RA, Burland V, Plunkett G III, Redford P, Roesch P, Rasko D, Buckles EL, Liou SR, Boutin A, Hackett J, Stroud D, Mayhew GF, Rose DJ, Zhou S, Schwartz DC, Perna NT, Mobley HL, Donnenberg MS, Blattner FR. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 99:17020–17024. doi: 10.1073/pnas.252529799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hochhut B, Wilde C, Balling G, Middendorf B, Dobrindt U, Brzuszkiewicz E, Gottschalk G, Carniel E, Hacker J. 2006. Role of pathogenicity island-associated integrases in the genome plasticity of uropathogenic Escherichia coli strain 536. Mol. Microbiol. 61:584–595. doi: 10.1111/j.1365-2958.2006.05255.x. [DOI] [PubMed] [Google Scholar]
- 15. Johnson TJ, Wannemeuhler YM, Scaccianoce JA, Johnson SJ, Nolan LK. 2006. Complete DNA sequence, comparative genomics, and prevalence of an IncHI2 plasmid occurring among extraintestinal pathogenic Escherichia coli isolates. Antimicrob. Agents Chemother. 50:3929–3933. doi: 10.1128/AAC.00569-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Peigne C, Bidet P, Mahjoub-Messai F, Plainvert C, Barbe V, Médigue C, Frapy E, Nassif X, Denamur E, Bingen E, Bonacorsi S. 2009. The plasmid of Escherichia coli strain S88 (O45:K1:H7) that causes neonatal meningitis is closely related to avian pathogenic E. coli plasmids and is associated with high-level bacteremia in a neonatal rat meningitis model. Infect. Immun. 77:2272–2284. doi: 10.1128/IAI.01333-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mellata M, Maddux JT, Nam T, Thomson N, Hauser H, Stevens MP, Mukhopadhyay S, Sarker S, Crabbé A, Nickerson CA, Santander J, Curtiss R. 2012. New insights into the bacterial fitness-associated mechanisms revealed by the characterization of large plasmids of an avian pathogenic E. coli. PLoS One 7:e29481. doi: 10.1371/journal.pone.0029481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mellata M, Touchman JW, Curtiss R. 2009. Full sequence and comparative analysis of the plasmid pAPEC-1 of avian pathogenic E. coli chi7122 (O78:K80:H9). PloS One 4:e4232. doi: 10.1371/journal.pone.0004232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mellata M, Maddux JT, Nam T, Thomson N, Hauser H, Stevens MP, Mukhopadhyay S, Sarker S, Crabbé A, Nickerson CA, Santander J, Curtiss R III. 2012. New insights into the bacterial fitness-associated mechanisms revealed by the characterization of large plasmids of an avian pathogenic E. coli. PLoS One 7:e29481. doi: 10.1371/journal.pone.0029481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mangiamele P, Nicholson B, Wannemuehler Y, Seemann T, Logue CM, Li G, Tivendale KA, Nolan LK. 2013. Complete genome sequence of the avian pathogenic Escherichia coli strain APEC O78. Genome Announc. 1(2):e0002613. doi: 10.1128/genomeA.00026-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dziva F, Hauser H, Connor TR, van Diemen PM, Prescott G, Langridge GC, Eckert S, Chaudhuri RR, Ewers C, Mellata M, Mukhopadhyay S, Curtiss R III, Dougan G, Wieler LH, Thomson NR, Pickard DJ, Stevens MP. 2013. Sequencing and functional annotation of avian pathogenic Escherichia coli serogroup O78 strains reveal the evolution of E. coli lineages pathogenic for poultry via distinct mechanisms. Infect. Immun. 81:838–849. doi: 10.1128/IAI.00585-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Johnson TJ, Kariyawasam S, Wannemuehler Y, Mangiamele P, Johnson SJ, Doetkott C, Skyberg JA, Lynne AM, Johnson JR, Nolan LK. 2007. The genome sequence of avian pathogenic Escherichia coli strain O1:K1:H7 shares strong similarities with human extraintestinal pathogenic E. coli genomes. J. Bacteriol. 189:3228–3236. doi: 10.1128/JB.01726-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Binns MM, Davies DL, Hardy KG. 1979. Cloned fragments of the plasmid ColV,I-K94 specifying virulence and serum resistance. Nature 279:778–781. doi: 10.1038/279778a0. [DOI] [PubMed] [Google Scholar]
- 24. Barondess JJ, Beckwith J. 1995. bor gene of phage lambda, involved in serum resistance, encodes a widely conserved outer membrane lipoprotein. J. Bacteriol. 177:1247–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Faraldo-Gómez JD, Sansom MS. 2003. Acquisition of siderophores in gram-negative bacteria. Nat. Rev. Mol. Cell Biol. 4:105–116. doi: 10.1038/nrm1015. [DOI] [PubMed] [Google Scholar]
- 26. Braun V, Braun M. 2002. Iron transport and signaling in Escherichia coli. FEBS Lett. 529:78–85. doi: 10.1016/S0014-5793(02)03185-X. [DOI] [PubMed] [Google Scholar]
- 27. Ratledge C. 2002. Regulation of lipid accumulation in oleaginous micro-organisms. Biochem. Soc. Trans. 30:1047–1050. [DOI] [PubMed] [Google Scholar]
- 28. Ratledge C, Wynn JP. 2002. The biochemistry and molecular biology of lipid accumulation in oleaginous microorganisms. Adv. Appl. Microbiol. 51:1–51. doi: 10.1016/S0065-2164(02)51000-5. [DOI] [PubMed] [Google Scholar]
- 29. Gehring AM, Bradley KA, Walsh CT. 1997. Enterobactin biosynthesis in Escherichia coli: isochorismate lyase (EntB) is a bifunctional enzyme that is phosphopantetheinylated by EntD and then acylated by EntE using ATP and 2,3-dihydroxybenzoate. Biochemistry 36:8495–8503. doi: 10.1021/bi970453p. [DOI] [PubMed] [Google Scholar]
- 30. Hantke K, Nicholson G, Rabsch W, Winkelmann G. 2003. Salmochelins, siderophores of Salmonella enterica and uropathogenic Escherichia coli strains, are recognized by the outer membrane receptor IroN. Proc. Natl. Acad. Sci. U. S. A. 100:3677–3682. doi: 10.1073/pnas.0737682100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lafont JP, Dho M, D’Hauteville HM, Bree A, Sansonetti PJ. 1987. Presence and expression of aerobactin genes in virulent avian strains of Escherichia coli. Infect. Immun. 55:193–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schubert S, Picard B, Gouriou S, Heesemann J, Denamur E. 2002. Yersinia high-pathogenicity island contributes to virulence in Escherichia coli causing extraintestinal infections. Infect. Immun. 70:5335–5337. doi: 10.1128/IAI.70.9.5335-5337.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Russo TA, McFadden CD, Carlino-MacDonald UB, Beanan JM, Barnard TJ, Johnson JR. 2002. IroN functions as a siderophore receptor and is a urovirulence factor in an extraintestinal pathogenic isolate of Escherichia coli. Infect. Immun. 70:7156–7160. doi: 10.1128/IAI.70.12.7156-7160.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kjelstrup CK, Arnesen LP, Granquist EG, L’Abée-Lund TM. 2013. Characterization of Escherichia coli O78 from an outbreak of septicemia in lambs in Norway. Vet. Microbiol. 166:276–280. doi: 10.1016/j.vetmic.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 35. Babai R, Blum-Oehler G, Stern BE, Hacker J, Ron EZ. 1997. Virulence patterns from septicemic Escherichia coli O78 strains. FEMS Microbiol. Lett. 149:99–105. [DOI] [PubMed] [Google Scholar]
- 36. Chérifi A, Contrepois M, Picard B, Goullet P, Orskov I, Orskov F. 1994. Clonal relationships among Escherichia coli serogroup O78 isolates from human and animal infections. J. Clin. Microbiol. 32:1197–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Czirok E, Milch H, Madar J, Semjen G. 1977. Characterization of Escherichia coli serogroups causing meningitis, sepsis and enteritis. I. Serological properties and animal pathogenicity of O18, O78 and O83 isolates. Acta Microbiol. Acad. Sci. Hung. 24:115–126. [PubMed] [Google Scholar]
- 38. Milch H, Czirók E, Madár J, Semjén G. 1977. Characterization of Escherichia coli serogroups causing meningitis, sepsis and enteritis. II. Classification of Escherichia coli O78 strains by phage sensitivity, colicin type and antibiotic resistance. Acta Microbiol. Acad. Sci. Hung. 24:127–137. [PubMed] [Google Scholar]
- 39. Jann K, Jann B. 1987. Polysaccharide antigens of Escherichia coli. Rev. Infect. Dis. 9(Suppl 5):S517–S526. doi: 10.1093/clinids/9.Supplement_5.S517. [DOI] [PubMed] [Google Scholar]
- 40. Goller CC, Seed PC. 2010. Revisiting the Escherichia coli polysaccharide capsule as a virulence factor during urinary tract infection: contribution to intracellular biofilm development. Virulence 1:333–337. doi: 10.4161/viru.1.4.12388. [DOI] [PubMed] [Google Scholar]
- 41. Bliss JM, Silver RP. 1996. Coating the surface: a model for expression of capsular polysialic acid in Escherichia coli K1. Mol. Microbiol. 21:221–231. doi: 10.1046/j.1365-2958.1996.6461357.x. [DOI] [PubMed] [Google Scholar]
- 42. Pluschke G, Mayden J, Achtman M, Levine RP. 1983. Role of the capsule and the O antigen in resistance of O18:K1 Escherichia coli to complement-mediated killing. Infect. Immun. 42:907–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Silver RP, Aaronson W, Vann WF. 1988. The K1 capsular polysaccharide of Escherichia coli. Rev. Infect. Dis. 10(Suppl 2):S282–S286. doi: 10.1093/cid/10.Supplement_2.S282. [DOI] [PubMed] [Google Scholar]
- 44. Peleg A, Shifrin Y, Ilan O, Nadler-Yona C, Nov S, Koby S, Baruch K, Altuvia S, Elgrably-Weiss M, Abe CM, Knutton S, Saper MA, Rosenshine I. 2005. Identification of an Escherichia coli operon required for formation of the O-antigen capsule. J. Bacteriol. 187:5259–5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dziva F, Hauser H, Connor TR, van Diemen PR, Prescott G, Chaudhuri RR, Ewers C, Mellata M, Mukhopadhyay S, Curtiss R III, Dougan G, Wieler LH, Thomson NR, Pickard DJ, Stevens MP. 2013. Sequencing and functional annotation of avian pathogenic Escherichia coli serogroup O78 strains reveals the evolution of E. coli lineages pathogenic for poultry via distinct mechanisms. Infect. Immun. 81:838–849. doi: 10.1128/IAI.00585-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Carniel E. 2001. The Yersinia high-pathogenicity island: an iron-uptake island. Microbes Infect. 3:561–569. doi: 10.1016/S1286-4579(01)01412-5. [DOI] [PubMed] [Google Scholar]
- 47. Bach S, de Almeida A, Carniel E. 2000. The Yersinia high-pathogenicity island is present in different members of the family Enterobacteriaceae. FEMS Microbiol. Lett. 183:289–294. doi: 10.1111/j.1574-6968.2000.tb08973.x. [DOI] [PubMed] [Google Scholar]
- 48. Gophna U, Oelschlaeger TA, Hacker J, Ron EZ. 2001. Yersinia HPI in septicemic Escherichia coli strains isolated from diverse hosts. FEMS Microbiol. Lett. 196:57–60. doi: 10.1111/j.1574-6968.2001.tb10540.x. [DOI] [PubMed] [Google Scholar]
- 49. Mellata M, Ameiss K, Mo H, Curtiss R III. 2010. Characterization of the contribution to virulence of three large plasmids of avian pathogenic Escherichia coli chi7122 (O78:K80:H9). Infect. Immun. 78:1528–1541. doi: 10.1128/IAI.00981-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Phan MD, Peters KM, Sarkar S, Lukowski SW, Allsopp LP, Gomes Moriel D, Achard ME, Totsika M, Marshall VM, Upton M, Beatson SA, Schembri MA. 2013. The serum resistome of a globally disseminated multidrug resistant uropathogenic Escherichia coli clone. PLoS Genet. 9:e1003834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Neidhardt FC, Bloch PL, Smith DF. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sezonov G, Joseleau-Petit D, D’Ari R. 2007. Escherichia coli physiology in Luria-Bertani broth. J. Bacteriol. 189:8746–8749. doi: 10.1128/JB.01368-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Staden R, Beal KF, Bonfield JK. 2000. The Staden package, 1998. Methods Mol. Biol. 132:115–130. [DOI] [PubMed] [Google Scholar]
- 55. Tech M, Merkl R. 2003. YACOP: enhanced gene prediction obtained by a combination of existing methods. In Silico Biol. 3:441–451. [PubMed] [Google Scholar]
- 56. Overbeek R, Larsen N, Walunas T, D’Souza M, Pusch G, Selkov E Jr, Liolios K, Joukov V, Kaznadzey D, Anderson I, Bhattacharyya A, Burd H, Gardner W, Hanke P, Kapatral V, Mikhailova N, Vasieva O, Osterman A, Vonstein V, Fonstein M, Ivanova N, Kyrpides N. 2003. The ERGO genome analysis and discovery system. Nucleic Acids Res. 31:164–171. doi: 10.1093/nar/gkg148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Meyer F, Goesmann A, McHardy AC, Bartels D, Bekel T, Clausen J, Kalinowski J, Linke B, Rupp O, Giegerich R, Pühler A. 2003. GenDB—an open source genome annotation system for prokaryote genomes. Nucleic Acids Res. 31:2187–2195. doi: 10.1093/nar/gkg312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Langille MG, Brinkman FS. 2009. IslandViewer: an integrated interface for computational identification and visualization of genomic islands. Bioinformatics 25:664–665. doi: 10.1093/bioinformatics/btp030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Carver T, Berriman M, Tivey A, Patel C, Böhme U, Barrell BG, Parkhill J, Rajandream MA. 2008. Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics 24:2672–2676. doi: 10.1093/bioinformatics/btn529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Darling AC, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Darling AE, Tritt A, Eisen JA, Facciotti MT. 2011. Mauve assembly metrics. Bioinformatics 27:2756–2757. doi: 10.1093/bioinformatics/btr451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Blom J, Albaum SP, Doppmeier D, Pühler A, Vorhölter FJ, Zakrzewski M, Goesmann A. 2009. EDGAR: a software framework for the comparative analysis of prokaryotic genomes. BMC Bioinformatics 10:154. doi: 10.1186/1471-2105-10-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 65. Löytynoja A, Goldman N. 2005. An algorithm for progressive multiple alignment of sequences with insertions. Proc. Natl. Acad. Sci. U. S. A. 102:10557–10562. doi: 10.1073/pnas.0409137102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 67. Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. 2011. BLAST Ring Image Generator (Brig): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]