Abstract
It is believed that transferring the C4 engine into C3 crops will greatly increase the yields of major C3 crops. Many efforts have been made since the 1960s, but relatively little success has been achieved because C4plant traits, referred to collectively as C4 syndrome, are very complex, and little is known about the genetic mechanisms involved. Unfortunately, there exists no ideal genetic model system to study C4 syndrome. It was previously reported that the Haloxylon species have different photosynthetic pathways in different photosynthetic organs, cotyledons and assimilating shoots. Here, we took advantage of the developmental switch from the C3 to the C4 pathway to study the genetic mechanisms behind this natural transition. We compared the transcriptomes of cotyledons and assimilating shoots using mRNA-Seq to gain insight into the molecular and cellular events associated with C4 syndrome. A total of 2959 differentially expressed genes [FDR≤0.001 and abs (|log2(Fold change)|≥1)] were identified, revealing that the transcriptomes of cotyledons and assimilating shoots are considerably different. We further identified a set of putative regulators of C4 syndrome. This study expands our understanding of the development of C4 syndrome and provides a new model system for future studies on the C3-to- C4 switch mechanism.
Introduction
Photosynthetic CO2 fixation is a fundamental life process involving the conversion of solar energy into chemical energy that can be later released to fuel the activity of an organism. The ancestral photosynthetic CO2-fixation process is C3 photosynthesis. The first organic product of CO2 fixation is a three-carbon compound. C3 photosynthesis and its key enzyme Rubisco (ribulose-1,5-bisphosphate carboxylase/oxygenase) evolved early in the history of life, when there was no oxygen in the atmosphere and atmospheric CO2 levels were significantly high. As atmospheric CO2 dropped and O2 accumulated, Rubisco inefficiency began to limit C3 photosynthesis because the active site of Rubisco does not completely discriminate between CO2 and O2, leading to the catalysis of two competitive reactions: photosynthetic CO2 assimilation and photorespiratory CO2 loss, especially under hot, dry and/or saline conditions that enhance photorespiration. Approximately 30 million years ago, an abrupt drop in atmospheric [CO2] further reduced the efficiency of C3 photosynthesis and triggered the evolution of C4 photosynthesis, in which CO2 is initially fixed into a four-carbon compound and concentrated around Rubisco [1–6]. C4 photosynthesis has evolved >60 times and occurs in approximately 7500 species of flowering plants [7–9]. The transition from C3 to C4 plants was the green revolution of nature. Although C4 plants comprise only 3% of land plant species, they account for some 25% of global terrestrial carbon fixation [3,4,7,10,11].
C4 photosynthesis is a complex trait that combines biochemical, physiological and anatomical characteristics, the so-called C4 syndrome [12,13]. Other than four single-celled C4 lineages, the vast majority of C4 plants possess Kranz anatomy and C4 syndrome [8,9], indicative of convergent evolution [7]. Traditional biochemical and modern molecular biological studies showed that all of the proteins required for the core C4 cycle are present in C3 plants [14]. Based on this evidence, it is hypothesized that no significant genetic changes are required for the transition from C3 to C4 photosynthesis [15,16]. The first attempt at introducing C4 photosynthesis into C3 plants was made in Atriplex through conventional interspecific hybridization of photosynthetic types to reduce photorespiration and increase photosynthetic capacity [17–22]. The results indicated that F1 hybrids of Atriplex rosea (C4, NAD-ME type) × Atriplex triangularis (C3) were more similar to their C3 parents in physiology and failed to form well-developed Kranz anatomy. Similar hybridization studies were conducted in the genera Flaveria, Panicum, Moricandia, and Brassica, but none proved fruitful at converting C3 species into functional C4 types [23]. Due to infertility, few hybrids have been developed beyond the F1 generation. In the few advanced generations studied in Atriplex and Flaveria hybrids, correlations among photosynthetic traits were low, indicating that C4 photosynthesis is a combination of independent biochemical, physiological and anatomical characteristics [23]. Recently, a progress report was released on oat-maize addition lines showing that the addition of individual maize chromosomes to the C3 species oat caused increases in vein density but did not confer functional C4 photosynthesis [24]. Therefore, introducing a fully developed C4 photosynthesis pathway into C3 plants through interspecific hybridization or even genetic engineering is far more practical than previously thought [25].
In nature, there exist examples of switches from a C3 pathway to a two-celled C4 pathway triggered by external or internal signals. The former example includes the freshwater amphibious leafless sedge Eleocharis vivipara, which can switch from a C3 pathway to a C4 pathway with Kranz anatomy after induction by environmental changes and exogenous application of abscisic acid (ABA) [26–28]. The latter examples were reported in Haloxylon and Salsola species that have different photosynthetic pathways in different photosynthetic organs [29,30]. Haloxylon aphyllum and H. persicum of Chenopodiaceae have C3 photosynthesis in cotyledons and C4 photosynthesis in assimilating shoots (the main photosynthetic organs) with a typical Salsoloid-type Kranz anatomy [29]; the same phenomenon has been observed in Salsola gemmascens of the genus Salsola (Chenopodiaceae), in which cotyledons exhibit C3-type photosynthesis, while leaves perform NAD-malic enzyme (NAD-ME) C4-type photosynthesis with a Salsoloid-type Kranz anatomy [30]. However, this manner of C3-to-C4 switches has not received much attention, and there were no follow-up studies after their discovery. Investigations into the developmental genetic mechanisms controlling the different photosynthetic types in cotyledons and leaves or other photosynthetic organs will illuminate the genetic regulatory network of C4 syndrome.
With the development of new technologies, more studies have begun to analyze C4 syndrome at the systems biology level. To understand C4 formation, mesophyll cells and bundle sheath cells in the leaf blade of maize were used as a model system for C4 differentiation. The cells were separated and analyzed for transcriptional changes by microarray analysis and next-generation sequencing [31–33]; 21% of genes were differentially expressed between mesophyll cells and bundle sheath cells [32]. Recently, John et al. sequenced RNA isolated from the mesophyll cells and bundle sheath cells of Setaria viridis and found the significant convergence of cell-specific gene expression in S. viridis and maize [34]. Such studies deepen our understanding of C4 syndrome. In 2011, two research groups used mRNA-Seq analysis of closely related C3 and C4 species for which gene expression is altered, and these groups identified genes associated with the C4 pathway [35,36]. Up to 603 and 3582 transcripts differed in abundance between C3 and C4 leaves in the genera Cleome and Flaveria, respectively. While these two experiments were designed to identify C4-related transcriptomic gene expression changes, it is difficult to tell if the observed variation in transcript abundance was associated with differences between the species or C4 photosynthesis. However, the comparative transcriptomics of C3 cotyledons and C4 assimilating shoots in this study will identify more C4-specific genes than before because there is no genetic variation between these different species; more importantly, the natural developmental C3-to-C4 transition may give clues concerning its master switch.
Materials and Methods
Plant Growth and Harvesting
Seeds of Haloxylon ammodendron were provided by the Turpan Eremophyte Botanic Garden, Chinese Academy of Sciences in Turpan, Xinjiang, China (http://english.egi.cas.cn/rs/sr/tdbg/). The seeds were incubated and germinated on moist filter paper in Petri dishes. After germination, seedlings were planted in sand in a greenhouse maintained at 30/20°C day/night, 70% relative humidity and a photoperiod of 12 h light/12 h dark under a light intensity of 1000 µE m–2 s–1. Cotyledons were fully expanded after 2 days growing in sand and sampled for all analyses. Assimilating shoots were collected from plants at 10 days of age. For mRNA-Seq, samples were taken from 10–15 individual plants during the middle of the light period, immediately frozen in liquid nitrogen, and stored at—80°C until use.
Light Microscopy
Samples of fully expanded cotyledons and assimilating shoots were fixed in Formalin–acetic acid–alcohol (FAA) for 24 hours, dehydrated through an alcohol series, cleared with xylene, and embedded in paraffin wax. Cross-sections were obtained using a microtome. For light microscopy, semi-thin sections were stained with safranin O solution and studied under DIC microscope (BX51, Olympus, Japan) equipped with an LM Digital Camera (DP70, Olympus).
Stable Carbon Isotope Analysis
Stable carbon isotope ratios (13C/12C) were quantified in cotyledons and dried assimilating shoots from plants grown in the greenhouse. Then, 1–2 cm segments of the middle regions of fully expanded cotyledons or assimilating shoots were collected. All samples were oven-dried at 65°C for 48 h to a constant weight.
The measurements of stable carbon isotope ratios were carried out at the Chinese Academy of Forestry’s Stable Isotope Laboratory (Beijing, China) using a Flash EA1112 HT elemental analyzer (Thermo Scientific) coupled with a Delta V advantage isotope ratio mass spectrometer (Thermo Scientific). Stable carbon isotope ratios were expressed as δ13C (‰), calculated as follows:
where Rsample and Rstandard are the 13C/12C ratios for an individual sample and the reference standard (Pee Dee Belemnite), respectively.
RNA Preparation and Sequencing
Total RNA was prepared with TRIzol according to the manufacturer’s instructions (Invitrogen Life Technologies, Shanghai, China). Following extraction, total RNA was purified using the RNeasy Mini Kit from Qiagen (Shanghai, China), including on-column DNase digestion (Qiagen, Shanghai, China). Purified RNA was checked for integrity and quality using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). The cDNA library was constructed for sequencing as described in the Illumina TruSeq RNA sample preparation v2 guide (Catalog # RS-930–1021). Sequencing was performed on an Illumina HiSeq 2000.
Mapping and Quantification of the Sequence Reads
We filtered and examined the quality of the raw sequence reads as described by Xu et al. [37]. The first 10 bases in each read were trimmed off before the mapping process.
Mapping and Quantification of the Sequence Reads from Cotyledons and Leaves of Arabidopsis. Clean reads were mapped onto the latest Arabidopsis thaliana genome assembly (http://www.phytozome.net/arabidopsis.php) using the Bowtie2 (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml) [38]. The best hit of each read with a maximum of three nucleotide mismatches was used. For each Arabidopsis Genome Initiative (AGI) code, the number of matching reads was counted, and the raw digital gene expression counts were normalized using the RPKM (Reads Per Kilobase per Million mapped reads) method [39,40].
Mapping and Quantification of the Sequence Reads from the Cotyledons and Assimilating Shoots of H. ammodendron. Clean reads were mapped onto the coding sequences of the latest A. thaliana genome assembly (http://www.phytozome.net/arabidopsis.php) using BLAT [41]. Alignments were performed in the protein space, and the best hit for each read was retained. For each Arabidopsis Genome Initiative (AGI) code, the number of matching reads was counted, and the raw digital gene expression counts were normalized using the RPKM method [39,40].
The differential expression between samples was statistically accessed by the R/Bioconductor package edgeR [42]. Genes with FDR≤0.001 and |log2(Fold change)|≥1 were considered significant.
Overrepresentation Analysis
To identify functional categories with significant differences between cotyledons and assimilating shoots, we performed an overrepresentation analysis using the GO Term Enrichment of AmiGO (http://amigo.geneontology.org/amigo) [43]. We used all detected transcripts in cotyledons and assimilating shoots as the background set and TAIR as the filter.
Results
Photosynthetic Features of Assimilating Shoots and Cotyledons
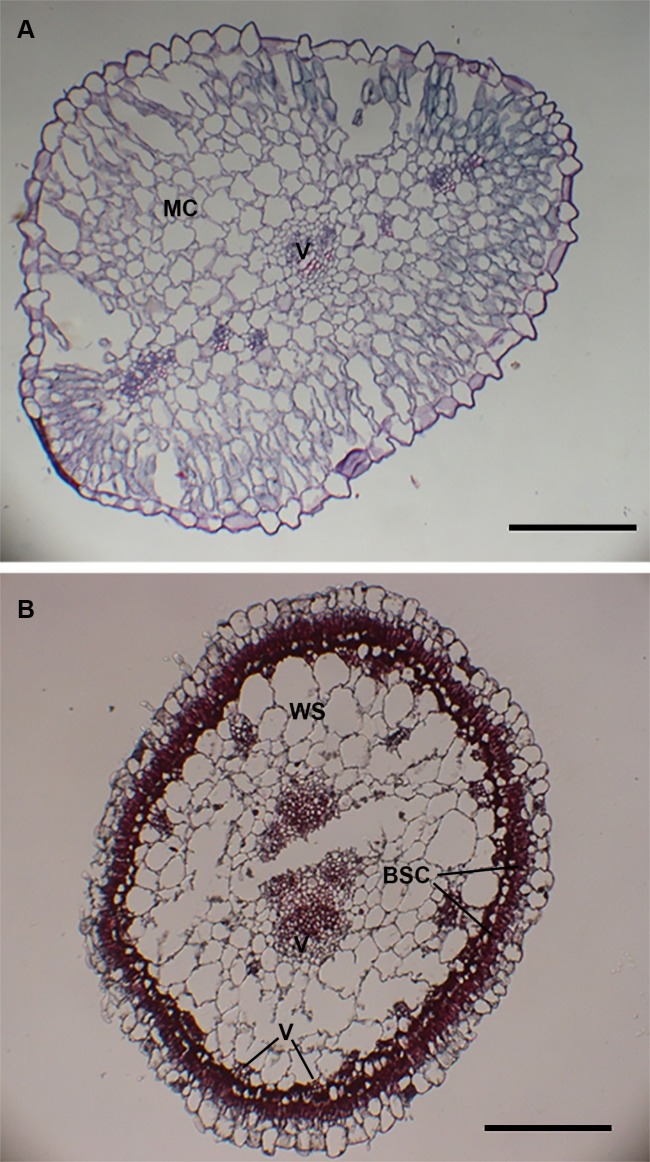
Haloxylon species have an unusual photosynthetic apparatus. The true leaves are reduced, and the young annual cylindrical shoots (assimilating shoots) are the main photosynthetic tissues. Fifteen years ago, Pyankov et al. [29] discovered that two Haloxylon species, H. aphyllum and H. persicum, have a C4 type of photosynthesis in assimilating shoots with Kranz anatomy, whereas leaf-like cotyledons lack Kranz-anatomy and incorporate CO2 via C3 photosynthesis, as observed through analyses of stable carbon isotope ratios, anatomy, primary photosynthetic products, and activities of carbon metabolism enzymes. We examined the anatomy assimilating shoots and cotyledons in H. ammodendron and confirmed that H. ammodendron, similarly to the other two Haloxylon species, uses different types of photosynthesis in assimilating shoots and cotyledons, as shown in Fig. 1. The cotyledons have no Kranz-type anatomy and several layers of mesophyll cells around only a few vascular bundles (Fig. 1A). The assimilating shoots have Salsoloid-type Kranz anatomy with two continuous layers of chlorenchyma (a layer of palisade mesophyll cells and an inner layer of bundle sheath cells) on the periphery and water-storage parenchyma in the center (Fig. 1B). The main vascular bundle occupies the central position, and only the small, peripheral vascular bundles are in contact with bundle sheath cells (Fig. 1B).
Figure 1. Transverse sections of a cotyledon (A) and an assimilating shoot (B) of Haloxylon ammodendron.
MC, mesophyll cell; BSC, bundle sheath cell; WS, water storage tissue; V, vascular tissue. Scale bars represent 100 μm.
Stable carbon isotope ratios are used to distinguish the photosynthetic CO2-fixing pathways of plants [44–46]. To further confirm the photosynthetic types of cotyledons and assimilating shoots, we analyzed the stable carbon isotope ratios by measuring δ13C values. The δ13C value for H. ammodendron cotyledons was-15.58±0.72 ‰ (mean ± SE, n = 3), similar to that of H. aphyllum and H. persicum reported by Pyankov et al. (-17.5 ‰) [29]. Unexpectedly, the assimilating shoots exhibited a more negative δ13C value of-21.89±1.05 ‰ (mean ± SE, n = 3).
Major Transcriptional Changes
To identify differences in transcript abundance related to C4 syndrome, the transcriptomes of H. ammodendron assimilating shoots and cotyledons were compared. cDNA libraries of assimilating shoots and cotyledons were constructed and sequenced using the Illumina HiSeq 2000 platform, resulting in 30,287,044 and 36,971,687 reads, respectively, with a mean read length of 101 nucleotides. After trimming adapters and filtering out low-quality reads, 29,558,368 reads from assimilating shoots and 36,070,605 reads from cotyledons were retained for further analysis. Clean reads were mapped onto the coding sequences of the latest A. thaliana genome assembly (http://www.phytozome.net/arabidopsis.php) using BLAT [41]. 27037741 reads (~75.0%) from cotyledons and 23188916 reads (~78.5%) from assimilating shoots could be mapped onto the Arabidopsis transcriptome.
mRNA-Seq analysis comparing the transcriptomes of H. ammodendron assimilating shoots and cotyledons yielded 2959 differentially expressed genes [FDR≤0.001 and abs (|log2(Fold change)|≥1)], with 1852 and 1107 more abundant transcripts in assimilating shoots and cotyledons, respectively (see Table A in S1 File). To test whether these differentially expressed transcripts are enriched in functional categories, we performed overrepresentation analysis using the GO Term Enrichment of AmiGO. The significantly up-regulated transcripts in assimilating shoots were enriched in several fundamental biological process categories, including methylation, cytokinesis, DNA replication, cell wall organization or biogenesis, biosynthetic process, anatomical structure morphogenesis, signaling pathway and developmental process (see Table B in S1 File). Down-regulated GO categories are less abundant than up-regulated ones, mainly in response to endogenous stimulus (see Table C in S1 File). Because we used TAIR as a filter and Arabidopsis is a typical C3 plant, no functional C4 class was detected.
C4 Cycle Genes Were Up-regulated in Assimilating Shoots
Transcript analysis of known C4 genes showed that all genes necessary for the core C4 cycle of NADP-ME type plants were significantly up-regulated in assimilating shoots compared with cotyledons (Table 1). Among these genes, NADP-ME was most significantly different, with a 12.9- (At5g11670), 12.7- (At2g19900) or 9.3- (At1g79750) fold higher transcript abundance in assimilating shoots. The second biggest difference came from PEPC, with a 12.1- (At2g42600) or 10.5- (At1g53310) fold up-regulation. Additionally, transcripts encoding AspAT (At4g31990) and the chloroplastidic MDH (At5g58330) were up-regulated 8.5- and 2.1-fold, respectively. Transcripts encoding PPDK (At4g15530) were increased 2.7-fold.
Table 1. Transcript abundance of C4 cycle genes and C4-related transporters.
| Gene ID | Protein | Ha-AS(RPKM) | Ha-C(RPKM) | Fold Change | log2(Ha-AS/Ha-C) |
|---|---|---|---|---|---|
| At4g15530 | PPDK | 5768.71 | 2133.50 | 2.70 | 1.44 |
| At1g53310 | PEPC | 1081.51 | 102.90 | 10.51 | 3.39 |
| At2g42600 | PEPC | 1201.30 | 99.31 | 12.10 | 3.60 |
| At1g68750 | PEPC | 2.84 | 6.96 | 0.41 | -1.29 |
| At5g58330 | cpNADP-MDH | 1287.07 | 601.46 | 2.14 | 1.10 |
| At1g79750 | NADP-ME | 814.71 | 87.96 | 9.26 | 3.21 |
| At2g19900 | NADP-ME | 2480.24 | 194.89 | 12.73 | 3.67 |
| At5g11670 | NADP-ME | 1161.41 | 90.10 | 12.89 | 3.69 |
| At1g17290 | AlaAT | 393.42 | 103.14 | 3.81 | 1.93 |
| At4g31990 | AspAT | 748.92 | 87.97 | 8.51 | 3.09 |
| At5g19550 | AspAT | 59.18 | 80.65 | 0.73 | -0.45 |
| At5g11520 | AspAT | 79.59 | 112.71 | 0.71 | -0.50 |
| At2g22250 | AspAT | 37.61 | 28.50 | 1.32 | 0.40 |
| At1g53240 | mtNAD-MDH | 190.23 | 129.15 | 1.47 | 0.56 |
| At4g00570 | NAD-ME | 42.94 | 27.82 | 1.54 | 0.63 |
| At4g37870 | PEP-CK | 125.01 | 168.60 | 0.74 | -0.43 |
| At3g47520 | cpNAD-MDH | 90.47 | 34.47 | 2.62 | 1.39 |
| At1g08650 | PEPC-K | 17.44 | 1.59 | 10.98 | 3.46 |
| At5g47840 | AMK2 | 489.96 | 230.02 | 2.13 | 1.09 |
| At5g35170 | adenylate kinase family protein | 141.21 | 216.67 | 0.65 | -0.62 |
| At5g09650 | PPA6 | 1042.96 | 739.28 | 1.41 | 0.50 |
| At2g26900 | BASS 2 | 1083.92 | 249.28 | 4.35 | 2.12 |
| At3g56160 | BASS 4 | 39.89 | 6.88 | 5.80 | 2.54 |
| At5g33320 | PPT1 | 225.80 | 39.88 | 5.66 | 2.50 |
| At3g01550 | PPT2 | 25.57 | 4.82 | 5.30 | 2.41 |
| At5g46110 | TPT | 1386.73 | 2111.57 | 0.66 | -0.61 |
| At5g12860 | Dit1 | 390.82 | 259.62 | 1.51 | 0.59 |
| At5g64280 | Dit2 | 91.71 | 24.54 | 3.74 | 1.90 |
Ha-AS = Haloxylon ammodendron assimilating shoots, Ha-C = Haloxylon ammodendron cotyledons.
All genes required for the NAD-ME type of C4 photosynthesis were also up-regulated in assimilating shoots compared with cotyledons (Table 1). The transcripts encoding NAD-ME (At4g00570) were up-regulated 1.5-fold, and those encoding AlaAT (At1g17290) and mtNAD-MDH (At1g53240) were up-regulated 3.8- and 1.5-fold, respectively.
Transcripts encoding the regulatory factors PEPC kinase (PEPC-K) increased 11-fold, which was significant (Table 1). Additionally, the transport proteins required for C4 photosynthesis were significantly up-regulated (see Table 1), such as pyruvate sodium symporter (BASS2), phosphoenolpyruvate/phosphate translocators (PPT1 and PPT2) and chloroplast dicarboxylate transporters (DiT1 and DiT2).
Photorespiratory Genes Were Down-regulated in Assimilating Shoots
A major advantage of the C4 pathway is the reduction in photorespiration because high CO2 concentration around Rubisco in bundle sheath cells effectively suppresses photorespiration. Detailed analysis of gene expression of all photorespiration genes showed that the transcripts of nearly all genes related to photorespiration were lower in abundance in assimilating shoots than in cotyledons (Table 2). The genes AtAGT1 (At2g13360), AtGGT1 (At1g23310), AtGLDT1 (At1g11860), AtSHM1 (At4g37930) and AtGLYK (At1g80380), all of which play major roles in photorespiration, were significantly down-regulated (log2 (Fold change) ≤-1).
Table 2. Transcript abundance of photorespiration genes.
| Enzyme | Gene name | Gene ID | Ha-AS(RPKM) | Ha-C(RPKM) | log2(Ha-AS/Ha-C) |
|---|---|---|---|---|---|
| 2PG phosphatase | AtPGLP1 | At5g36790* | 243.47 | 228.19 | 0.09 |
| Glycolate oxidase | AtGOX1 | At3g14420 | 911.87 | 3690.36 | -2.02 |
| AtGOX2 | At3g14415 | 330.80 | 1326.77 | -2.00 | |
| AtGOX3 | At4g18360 | 151.29 | 679.40 | -2.17 | |
| AtHAOX1 | At3g14130 | 8.32 | 12.51 | -0.59 | |
| AtHAOX2 | At3g14150 | 9.20 | 16.56 | -0.85 | |
| Ser:glyoxylate aminotransferase | AtAGT1 | At2g13360 | 362.28 | 2052.76 | -2.50 |
| Glu:glyoxylate aminotransferase | AtGGT1 | At1g23310 | 670.03 | 1607.05 | -1.26 |
| AtGGT2 | At1g70580 | 527.48 | 1194.60 | -1.18 | |
| Gly decarboxylase P-protein | AtGLDP1 | At4g33010 | 503.01 | 1461.69 | -1.54 |
| AtGLDP2 | At2g26080 | 266.08 | 770.75 | -1.53 | |
| Gly decarboxylase H-protein | AtGLDH1 | At2g35370 | 153.54 | 644.93 | -2.07 |
| AtGLDH2 | At2g35120 | 84.97 | 54.76 | 0.63 | |
| AtGLDH3 | At1g32470 | 168.58 | 721.63 | -2.10 | |
| Gly decarboxylase T-protein | AtGLDT1 | At1g11860 | 546.86 | 1701.11 | -1.64 |
| Gly decarboxylase L-protein | AtmLPD1 | At3g17240 | 164.91 | 271.07 | -0.72 |
| AtmLPD2 | At1g48030 | 156.18 | 277.42 | -0.83 | |
| Ser hydroxymethyltransferase | AtSHM1 | At4g37930 | 696.67 | 1457.01 | -1.06 |
| AtSHM2 | At5g26780 | 378.13 | 774.57 | -1.03 | |
| Hydroxypyruvate reductases | AtHPR1 | At1g68010 | 647.42 | 1269.02 | -0.97 |
| AtHPR2 | At1g79870* | 115.77 | 104.57 | 0.15 | |
| Glycerate kinase | AtGLYK | At1g80380 | 46.14 | 115.70 | -1.33 |
Ha-AS = Haloxylon ammodendron assimilating shoots, Ha-C = Haloxylon ammodendron cotyledons.
*no significant differences in the expression of these genes between cotyledons and assimilating shoots (p>0.01).
Genes Controlling Vein Density Were Up-regulated in Assimilating Shoots
Kranz anatomy is accompanied by high vein density [16], so we assessed the transcript abundance of known genes controlling vein density. All of these genes were up-regulated in assimilating shoots (Table 3). Among them, the transcription factors MONOPTEROS/AUXIN RESPONSE FACTOR5 (MP/ARF5) MP and ATHB8, which are involved in the auxin signal transduction pathway controlling leaf vascular development, were up-regulated 3.9- and 1.9-fold, respectively.
Table 3. Transcription abundance of known genes related to vein density.
| Gene ID | Name | Ha-AS(RPKM) | Ha-C (RPKM) | Fold Change | log2(Ha-AS/Ha-C) |
|---|---|---|---|---|---|
| At4g32880 | ATHB8 | 13.79 | 3.53 | 3.91 | 1.97 |
| At1g19850 | MP/ARF5 | 6.05 | 3.26 | 1.86 | 0.89 |
| At1g13980 | GNOM/EMB30 | 22.25 | 18.68 | 1.19 | 0.25 |
| At2g36120 | DOT1 | 248.82 | 157.52 | 1.58 | 0.66 |
| At1g13290 | DOT5 | 3.16 | 1.86 | 1.70 | 0.77 |
| At5g60690 | REVOLUTA (REV) | 15.66 | 5.91 | 2.65 | 1.41 |
| At5g13300 | SFC | 14.41 | 9.46 | 1.52 | 0.61 |
| At1g73590 | PIN-FORMED1 (PIN1) | 10.80 | 4.00 | 2.70 | 1.43 |
| At1g20330 | CVP1/SMT2 | 37.70 | 20.13 | 1.87 | 0.91 |
| At1g05470 | CVP2 | 2.95 | 1.28 | 2.31 | 1.20 |
| At5g55540 | LOP1(now tornado1–2) | 1.09 | 0.19 | 5.72 | 2.52 |
| At1g65620 | AS2 | 0.53 | 0.00 | 19.02 | |
| At5g60200 | DOF5.3 | 1.94 | 0.92 | 2.10 | 1.07 |
Ha-AS = Haloxylon ammodendron assimilating shoots, Ha-C = Haloxylon ammodendron cotyledons.
Differentially Expressed Transcription Factors Encoding Genes
A total of 248 transcription factor-encoding genes were identified, showing differential expression [FDR≤0.001 and abs (|log2(Fold change)|≥1)] in assimilating shoots compared with cotyledons (Table 4). In total, 107 genes were up-regulated, and 141 genes were repressed in assimilating shoots. To avoid capturing variation in transcript abundance associated with differences between the different developmental stages that do not relate to C4 photosynthesis, we also sequenced the transcriptomes of Arabidopsis cotyledons and leaves in parallel to minimize the influence of developmental stage-specific effects. Excluding the genes that had the same developmental expression pattern in H. ammodendron and Arabidopsis, there were 96 putative positive regulators and 130 putative negative regulators. We tested the likelihood that SCARECROW/SHORTROOT components [47] of the C4 regulatory network were among these identified genes (see Table 5). Scarecrow plays a role in establishing Kranz anatomy in maize leaves, and its mutation results in abnormal Kranz anatomy [48]. SHR was included among the identified genes, and SCR was up-regulated by 1.3-fold in assimilating shoots, although it was not identified.
Table 4. Differentially expressed transcription factor-encoding genes.
| Putative positive regulators | |||||
| At1g01250 | AP2-EREBP family | At3g57670 | C2H2 family | At2g34710 | Homeobox family |
| At1g68550* | AP2-EREBP family | At4g27240 | C2H2 family | At3g18010 | Homeobox family |
| At1g79700 | AP2-EREBP family | At5g03740* | C2H2 family | At4g08150 | Homeobox family |
| At4g16750 | AP2-EREBP family | At5g39550 | C2H2 family | At4g32880 | Homeobox family |
| At4g23750* | AP2-EREBP family | At5g54630 | C2H2 family | At5g46880 | Homeobox family |
| At4g37750 | AP2-EREBP family | At5g57520 | C2H2 family | At5g60690 | Homeobox family |
| At5g11190 | AP2-EREBP family | At1g68200 | C3H family | At1g24260 | MADS family |
| At5g17430 | AP2-EREBP family | At2g44580 | C3H family | At2g03710 | MADS family |
| At5g57390 | AP2-EREBP family | At3g63530 | C3H family | At2g45660 | MADS family |
| At2g46530 | ARF family | At5g45290 | C3H family | At3g30260 | MADS family |
| At1g27660 | bHLH family | At1g54160 | CCAAT-HAP2 family | At4g22950 | MADS family |
| At1g61660 | bHLH family | At3g14020* | CCAAT-HAP2 family | At4g24540* | MADS family |
| At1g68810 | bHLH family | At5g12840 | CCAAT-HAP2 family | At5g15800 | MADS family |
| At2g27230 | bHLH family | At5g06510* | CCAAT-HAP2 family | At5g60910 | MADS family |
| At2g41130 | bHLH family | At1g08970 | CCAAT-HAP5 family | At2g47460 | MYB family |
| At3g06120 | bHLH family | At5g43250 | CCAAT-HAP5 family | At3g61250* | MYB family |
| At3g20640 | bHLH family | At3g04850 | CPP family | At4g32730 | MYB family |
| At5g10570 | bHLH family | At3g22760 | CPP family | At5g15310 | MYB family |
| At5g57150 | bHLH family | At3g22780 | CPP family | At5g49330 | MYB family |
| At5g65640 | bHLH family | At4g14770 | CPP family | At1g54330 | NAC family |
| At2g18160 | bZIP family | At3g01330 | E2F-DP family | At1g62700 | NAC family |
| At1g28050 | C2C2-CO-like family | At3g48160 | E2F-DP family | At3g57150 | NAC family |
| At2g33500 | C2C3-CO-like family | At3g10760 | G2-like | At4g28500 | NAC family |
| At3g50410 | C2C2-Dof family | At3g46640 | G2-like | At2g24630 | REM family |
| At1g08000 | C2C2-Gata family | At5g42630 | G2-like | At1g02065 | SBP family |
| At3g06740 | C2C3-Gata family | At1g50420* | GRAS family | At1g69170 | SBP family |
| At4g16141 | C2C4-Gata family | At1g63100 | GRAS family | At5g43270 | SBP family |
| At5g26930 | C2C5-Gata family | At4g37650 | GRAS family | At4g37490 | TCP family |
| At5g49300 | C2C6-Gata family | At5g41920 | GRAS family | At1g16070 | TUB family |
| At2g45190 | C2C2-YABBY family | At2g22840 | GRF family | At4g39410 | WRKY family |
| At4g00180 | C2C2-YABBY family | At1g30490 | Homeobox family | At1g75240 | ZF-HD family |
| At1g75710 | C2H2 family | At1g46480 | Homeobox family | At2g02540 | ZF-HD family |
| At2g29660 | C2H2 family | At1g52150* | Homeobox family | At2g18350 | ZF-HD family |
| At3g12270 | C2H2 family | At1g62990 | Homeobox family | At4g24660 | ZF-HD family |
| At3g14740 | C2H2 family | At1g79840 | Homeobox family | At5g65410 | ZF-HD family |
| At3g44750* | C2H2 family | At2g27990 | Homeobox family | ||
| Putative negative regulators | |||||
| At1g01030 | ABI3VP1 family | At5g50915 | bHLH family | At2g46680 | Homeobox family |
| At1g25560 | ABI3VP1 family | At5g62610 | bHLH family | At3g01220 | Homeobox family |
| At1g03800 | AP2-EREBP family | At5g67060 | bHLH family | At3g61890 | Homeobox family |
| At1g12610* | AP2-EREBP family | At1g08320 | bZIP family | At5g15150 | Homeobox family |
| At1g19210 | AP2-EREBP family | At3g10800 | bZIP family | At5g47370 | Homeobox family |
| At1g46768 | AP2-EREBP family | At1g25440 | C2C2-CO-like family | At4g26170 | HRT family |
| At1g50640 | AP2-EREBP family | At1g49130 | C2C2-CO-like family | At4g36990 | HSF family |
| At1g64380 | AP2-EREBP family | At1g68520 | C2C2-CO-like family | At5g62020 | HSF family |
| At1g74930 | AP2-EREBP family | At1g73870 | C2C2-CO-like family | At1g68320 | MYB family |
| At2g28550 | AP2-EREBP family | At2g24790 | C2C2-CO-like family | At1g52890 | NAC family |
| At2g40340 | AP2-EREBP family | At3g02380 | C2C2-CO-like family | At1g01720 | NAC family |
| At3g11020 | AP2-EREBP family | At4g39070 | C2C2-CO-like family | At1g52880 | NAC family |
| At3g15210 | AP2-EREBP family | At5g15840 | C2C2-CO-like family | At1g69490 | NAC family |
| At3g20310 | AP2-EREBP family | At5g15850 | C2C2-CO-like family | At1g77450 | NAC family |
| At3g54990* | AP2-EREBP family | At5g57660 | C2C2-CO-like family | At2g17040 | NAC family |
| At4g17490 | AP2-EREBP family | At1g29160 | C2C2-Dof family | At4g17980 | NAC family |
| At4g25470 | AP2-EREBP family | At1g51700 | C2C2-Dof family | At4g27410 | NAC family |
| At4g25490 | AP2-EREBP family | At1g64620 | C2C2-Dof family | At4g28530 | NAC family |
| At4g34410* | AP2-EREBP family | At1g69570 | C2C2-Dof family | At5g08790 | NAC family |
| At4g36900 | AP2-EREBP family | At3g21270 | C2C2-Dof family | At5g18270 | NAC family |
| At5g05410 | AP2-EREBP family | At1g27730 | C2H2 family | At5g39610* | NAC family |
| At5g07310 | AP2-EREBP family | At3g19580 | C2H2 family | At5g61430 | NAC family |
| At5g13330 | AP2-EREBP family | At3g49930 | C2H2 family | At5g63790 | NAC family |
| At5g25190* | AP2-EREBP family | At5g12850 | C2H2 family | At1g64530 | NLP family |
| At5g44210 | AP2-EREBP family | At5g04340 | C2H2 family | At1g13260 | RAV family |
| At5g47230 | AP2-EREBP family | At5g67450 | C2H2 family | At1g68840* | RAV family |
| At5g50080 | AP2-EREBP family | At1g26800 | C3H family | At2g36080 | RAV family |
| At5g51190 | AP2-EREBP family | At2g15580 | C3H family | At2g46870* | RAV family |
| At5g51990 | AP2-EREBP family | At3g10910 | C3H family | At3g11580 | RAV family |
| At5g61600 | AP2-EREBP family | At3g58720 | C3H family | At3g25730 | RAV family |
| At5g61890 | AP2-EREBP family | At4g13100 | C3H family | At5g06250 | RAV family |
| At5g64750* | AP2-EREBP family | At1g67910 | CAMTA family | At1g53230 | TCP family |
| At5g20730 | ARF family | At2g13570 | CCAAT-HAP3 family | At2g38250 | Trihelix family |
| At1g76110 | ARID family | At1g68670 | G2-like family | At5g01380 | Trihelix family |
| At1g04880 | ARID family | At2g03500 | G2-like family | At1g29860 | WRKY family |
| At3g48100 | ARR-B family | At3g04030 | G2-like family | At1g62300 | WRKY family |
| At1g09530 | bHLH family | At5g18240 | G2-like family | At1g69310 | WRKY family |
| At1g18400 | bHLH family | At1g07520 | GRAS family | At1g80840* | WRKY family |
| At1g22380 | bHLH family | At1g07530 | GRAS family | At2g23320 | WRKY family |
| At1g26260 | bHLH family | At2g29060 | GRAS family | At2g38470 | WRKY family |
| At1g73830* | bHLH family | At2g37650 | GRAS family | At2g47260 | WRKY family |
| At2g18300 | bHLH family | At4g17230 | GRAS family | At4g04450 | WRKY family |
| At2g20180 | bHLH family | At4g24150 | GRF family | At4g18170 | WRKY family |
| At3g07340 | bHLH family | At5g53660 | GRF family | At4g22070 | WRKY family |
| At3g21330 | bHLH family | At1g26960 | Homeobox family | At4g31800 | WRKY family |
| At4g34530* | bHLH family | At1g69780 | Homeobox family | At5g26170 | WRKY family |
| At4g36540 | bHLH family | At2g44910 | Homeobox family | At5g46350* | WRKY family |
Ha-AS = Haloxylon ammodendron assimilating shoots, Ha-C = Haloxylon ammodendron cotyledons.
*these genes had the same developmental expression pattern in H. ammodendron and Arabidopsis.
Table 5. SCARECROW/SHORTROOT regulatory network.
| Maize Gene ID | Arabidopsis Gene ID | Arabidopsis ortholog | Hal-AS (RPKM) | Hal-C (RPKM) | log2(Ha-AS/Ha-C) |
|---|---|---|---|---|---|
| GRMZM2G131516 | AT3G54220 | SCR | 27.26 | 20.31 | 0.42 |
| GRMZM2G132794 | AT4G37650 | SHR | 12.63 | 0.90 | 3.81 |
| GRMZM2G172657 | |||||
| GRMZM2G150011 | AT1G13290 | DOT5 | 3.16 | 1.86 | 0.77 |
Ha-AS = Haloxylon ammodendron assimilating shoots, Ha-C = Haloxylon ammodendron cotyledons.
Discussion
The Haloxylon genus comprises three closely related species, including H. ammodendron (saxaul), H. aphyllum (black saxaul) and H. persicum (white saxaul). H. aphyllum and H. persicum were reported to have different types of photosynthesis in assimilating shoots and cotyledons. Similarly, H. ammodendron assimilating shoots had Salsoloid-type Kranz anatomy (Fig. 1B), indicative of C4 photosynthesis, while the structure of cotyledons was of the non-Kranz type (Fig. 1A). The developmental transition from a C3 pathway to a two-celled C4 pathway in Haloxylon species in nature is a good system with which to study the genetic regulatory network of C4 syndrome.
Stable carbon isotope analysis is used as a screening method to determine the photosynthetic pathway when it is unknown in a species [46]. We measured the δ13C values of cotyledons and assimilating shoots. The H. ammodendron cotyledons had a δ13C value of-15.58±0.72 ‰ (mean ± SE, n = 3), falling into the range of typical values for C4 plants of-6 to-19 ‰ [49]. Although this finding is contrary to the anatomical results, it is consistent with a previous report of Haloxylon that proposed that these C4-type values are a result of old C4 assimilates stored in the cotyledons during seed formation [29]. Interestingly, the assimilating shoots exhibited a δ13C value of-21.89±1.05 ‰ (mean ± SE, n = 3) and were closer to that of C3 plants (-23 ‰ to-32 ‰). Because Haloxylon seeds have no endosperm, cotyledon photosynthesis provides C3 assimilates to support early plant development [29]. H. ammodendron has two large and long-lived cotyledons; therefore, the δ13C values of young assimilating shoots (10 days of age) are more negative and closer to that of C3 due to a mixture of assimilates from C3 (cotyledons) and C4 photosynthesis (assimilating shoots).
In this study, we performed deep mRNA-Seq using the Illumina HiSeq 2000 platform to analyze the transcriptomes of H. ammodendron assimilating shoots and cotyledons, which exhibit different types of photosynthesis. Using Arabidopsis as the reference genome, 2959 differentially expressed genes [FDR≤0.001 and abs (|log2(Fold change)|≥1)] were identified, with 1852 and 1107 transcripts being more abundant in assimilating shoots and cotyledons, respectively (see Table A in S1 File).
It was reported that the assimilating shoots of H. aphyllum and H. persicum mainly perform NADP-ME-type C4 shuttling and a small fraction of NAD-ME-type C4 shuttling, as shown by photosynthetic enzyme activity analysis and immunoblot analysis [29]. The mRNA-Seq analysis presented here confirmed this discovery and further showed that up-regulation occurs at the level of transcript abundance. All genes necessary for the core C4 cycle of NADP-ME type plants were significantly up-regulated, and all genes required for the NAD-ME type of C4 photosynthesis were also up-regulated, but to a lesser extent, in assimilating shoots compared with cotyledons (Table 1). This suggests that NADP-ME-type C4 photosynthesis is predominant, and NAD-ME-type C4 photosynthesis makes a small contribution to photosynthetic CO2 fixation. Likewise, nearly all genes encoding photorespiratory proteins had lower steady-state transcriptional levels in assimilating shoots (Table 2).
We identified a list of positive and negative regulators of C4 syndrome (Table 4). Although not all of these transcription factors are known to be components of the C4 regulatory network, our observations suggest that at least a subset of these factors are very likely involved in vein density and the development of Kranz anatomy. Within the list, the transcription factor ATHB8 (Table 4) and the upstream gene MP (Table 3) in the auxin signal transduction pathway controlling leaf vascular development were both up-regulated. This evidence is supportive of a role for at least a subset cohort in vein density. Very recently, Wang et al. (2013) performed a genome-wide comparative analysis of developmental trajectories in Kranz (foliar leaf blade) and non-Kranz (husk leaf sheath) leaves of the C4 plant maize to look for regulators of Kranz anatomy and identified 48 putative positive regulators, of which, 40 genes were assigned to 38 Arabidopsis orthologous genes [47]. Although maize has a Panicoid-type (classical NADP-ME type) Kranz anatomy and Haloxylon species have a Salsoloid-type Kranz anatomy, there remains high overlap between the list of positive regulators in each study. Within our list of putative positive regulators of C4 syndrome, 11 of 38 Arabidopsis transcription factor-encoding genes were also significantly up-regulated in H. ammodendron assimilating shoots. Another two genes, AT3G54220 and AT3G13960, were increased by 1.3- and 1.9-fold (Table 6). This high overlap ratio confirms that the methods we used identified Kranz anatomy regulators.
Table 6. High overlap between putative positive Kranz regulators identified by Wang et al. (2013) and this study.
| Maize Gene ID | Annotation | Arabidopsis Gene ID | Arabidopsis ortholog | Ha-AS (RPKM) | Ha-C (RPKM) | log2(Ha-AS/Ha-C) |
|---|---|---|---|---|---|---|
| GRMZM2G132794 | GRAS (SHR) | AT4G37650 | SHR | 12.63 | 0.90 | 3.81 |
| GRMZM2G172657 | GRAS (SHR) | |||||
| GRMZM2G163975 | bHLH family | AT1G27660 | 5.00 | 2.43 | 1.04 | |
| GRMZM2G039074 | Myb family | AT5G42630 | ATS | 1.04 | 0.06 | 4.03 |
| GRMZM2G178182 | bHLH family | AT5G50915 | 7.44 | 15.34 | -1.04 | |
| GRMZM2G472945 | TLP-family | AT1G16070 | TLP8 | 1.68 | 0.17 | 3.34 |
| GRMZM2G178102 | HD-ZIP III (PHV-like) | AT1G30490 | PHV | 2.63 | 0.90 | 1.55 |
| GRMZM2G131516 | GRAS (SCR) | AT3G54220 | SCR | 27.26 | 20.31 | 0.42 |
| GRMZM2G136494 | MRPI-like ZnF | AT1G75710 | ANT | 13.51 | 4.30 | 1.65 |
| GRMZM2G028046 | MRPI-like ZnF | |||||
| GRMZM2G021573 | AP2-EREBP (ANT-like) | AT4G37750 | 7.82 | 0.93 | 3.07 | |
| GRMZM2G146688 | AP2-EREBP (ANT-like) | |||||
| GRMZM2G040924 | MIXTA-like Myb | AT3G61250 | 1.95 | 0.51 | 1.95 | |
| GRMZM2G111045 | MIXTA-like Myb | |||||
| GRMZM2G171365 | ZmMADS1 | AT2G45660 | AGL20 | 19.94 | 0.93 | 4.43 |
| GRMZM2G425236 | ZnF-HD family | AT4G24660 | ATHB22 | 28.69 | 7.66 | 1.91 |
| GRMZM2G097275 | SBP family | AT5G43270 | SPL2 | 0.77 | 0.21 | 1.91 |
| GRMZM5G850129 | GRF | AT3G13960 | GRF5 | 6.71 | 3.50 | 0.94 |
Ha-AS = Haloxylon ammodendron assimilating shoots, Ha-C = Haloxylon ammodendron cotyledons. Wang et al. (2013) identified 48 putative positive Kranz regulators through comparative analysis of Kranz (foliar leaf blade) and non-Kranz (husk leaf sheath) leaves of maize. Forty genes were assigned to 38 Arabidopsis orthologous genes [47]. Fourteen of 38 Arabidopsis transcription factor-encoding genes were also identified in this study, and 11 transcription factors were significantly up-regulated (log2 (Fold change) >1) in H. ammodendron assimilating shoots.
The differences in transcript abundance between the different photosynthetic organs of H. ammodendron, cotyledons and assimilating shoots, reflect the development of C4 syndrome. Previous studies suggested C4 cycle genes, trans-factors and even cis-elements were recruited from ancestral C3 plants [50–53], but the mechanism behind is not clear. With the same genomic context of the cotyledons and assimilating shoots, this natural and developmental transition from C3 to C4 would be an ideal model system to study the molecular mechanism of recruitment, if whole genome sequence and genetic transformation are available.
Supporting Information
(TIF)
Table A. 2959 differentially expressed genes [FDR≤0.001 and abs (|log2(Fold change)|≥1)] Table B. Up-regulated GO categories. Table C. Down-regulated GO categories.
(XLSX)
Acknowledgments
We would like to thank Yutao Wang (Kashgar Teachers College, Kashgar, China) for providing the images of Haloxylon ammodendron in its natural habitat.
Data Availability
RNAseq data produced in this study have been submitted to the NCBI/SRA database under accession number SRP049928.
Funding Statement
This work was supported by the China Postdoctoral Science Foundation (Grant No. 2013T60468) and the National Basic Research Program of China (Grant No. 2012CB114204). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Cerling TE, Harris JM, MacFadden BJ, Leakey MG, Quade J, et al. (1997) Global vegetation change through the Miocene/Pliocene boundary. Nature 389: 153–158. [Google Scholar]
- 2. Christin PA, Besnard G, Samaritani E, Duvall MR, Hodkinson TR, et al. (2008) Oligocene CO2 decline promoted C4 photosynthesis in grasses. Curr Biol 18: 37–43. [DOI] [PubMed] [Google Scholar]
- 3. Edwards EJ, Osborne CP, Stromberg CAE, Smith SA, Bond WJ, et al. (2010) The origins of C4 grasslands: integrating evolutionary and ecosystem science. Science 328: 587–591. 10.1126/science.1177216 [DOI] [PubMed] [Google Scholar]
- 4. Osborne CP, Beerling DJ (2006) Nature's green revolution: the remarkable evolutionary rise of C4 plants. Phil Trans R Soc B 361: 173–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pagani M, Zachos JC, Freeman KH, Tipple B, Bohaty S (2005) Marked decline in atmospheric carbon dioxide concentrations during the Paleogene. Science 309: 600–603. [DOI] [PubMed] [Google Scholar]
- 6. Li Y, Xu J, Haq NU, Zhang H, Zhu X-G (2014) Was low CO2 a driving force of C4 evolution: Arabidopsis responses to long-term low CO2 stress. J Exp Bot 65: 3657–3667. 10.1093/jxb/eru193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sage RF (2004) The evolution of C4 photosynthesis. New Phytol 161: 341–370. [DOI] [PubMed] [Google Scholar]
- 8. Sage RF, Christin PA, Edwards EJ (2011) The C4 plant lineages of planet Earth. J Exp Bot 62: 3155–3169. 10.1093/jxb/err048 [DOI] [PubMed] [Google Scholar]
- 9. Sage RF, Sage TL, Kocacinar F (2012) Photorespiration and the evolution of C4 photosynthesis. Annu Rev Plant Biol 63: 19–47. 10.1146/annurev-arplant-042811-105511 [DOI] [PubMed] [Google Scholar]
- 10. Lloyd J, Farquhar GD (1994) 13C discrimination during CO2 assimilation by the terrestrial biosphere. Oecologia 99: 201–215. [DOI] [PubMed] [Google Scholar]
- 11. Still CJ, Berry JA, Collatz GJ, DeFries RS (2003) Global distribution of C3 and C4 vegetation: Carbon cycle implications. Global Biogeochem Cycles 17: 1006. [Google Scholar]
- 12. Hatch MD, Osmond CB (1976) Compartmentation and transport in C4 photosynthesis In: Stocking CR, Heber U, editors. Transport in Plants III: Springer; Berlin Heidelberg: pp. 144–184. [Google Scholar]
- 13. Laetsch WM (1974) The C4 syndrome: a structural analysis. Annu Rev Plant Physiol 25: 27–52. [Google Scholar]
- 14. Aubry S, Brown NJ, Hibberd JM (2011) The role of proteins in C3 plants prior to their recruitment into the C4 pathway. J Exp Bot 62: 3049–3059. 10.1093/jxb/err012 [DOI] [PubMed] [Google Scholar]
- 15. Hibberd JM, Sheehy JE, Langdale JA (2008) Using C4 photosynthesis to increase the yield of rice—rationale and feasibility. Curr Opin Plant Biol 11: 228–231. 10.1016/j.pbi.2007.11.002 [DOI] [PubMed] [Google Scholar]
- 16. Westhoff P, Gowik U (2010) Evolution of C4 photosynthesis—looking for the master switch. Plant Physiol 154: 598–601. 10.1104/pp.110.161729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Björkman O, Gauhl E, Nobs MA (1969) Comparative studies of Atriplex species with and without B-carboxylation photosynthesis and their first-generation hybrid. Year B Carnegie Inst Wash 68: 620–633. [Google Scholar]
- 18. Björkman O, Pearcy RW, Nobs MA (1970) Hybrids between Atriplex species with and without B-carboxylation photosynthesis. Photosynthetic characteristics. Year B Carnegie Inst Wash 69: 640–648. [Google Scholar]
- 19. Boynton JE, Nobs MA, Björkman O, Pearcy RW (1970) Hybrids between Atriplex species with and without B-carboxylation photosynthesis. Leaf anatomy and ultrastructure. Year B Carnegie Inst Wash 69: 629–632. [Google Scholar]
- 20. Nobs MA, Björkman O, Pearcy RW (1970) Hybrids between Atriplex species with and without B-carboxylation photosynthesis. Cytogenetic and morphological characteristics. Year B Carnegie Inst Wash 69: 625–629 [Google Scholar]
- 21. Pearcy RW, Björkman O (1970) Hybrids between Atriplex species with and without B-carboxylation photosynthesis. Biochemical characteristics. Year B Carnegie Inst Wash 69: 632–640. [Google Scholar]
- 22. Björkman O, Nobs MA, Berry JA (1971) Further studies on hybrids between C3 and C4 species of Atriplex . Year B Carnegie Inst Wash 70: 507–511. [Google Scholar]
- 23. Brown RH, Bouton JH (1993) Physiology and genetics of interspecific hybrids between photosynthetic types. Annu Rev Plant Physiol Plant Mol Biol 44: 435–456. [Google Scholar]
- 24. Tolley BJ, Sage TL, Langdale JA, Hibberd JM (2012) Individual maize chromosomes in the C3 plant oat can increase bundle sheath cell size and vein density. Plant Physiol 159: 1418–1427. 10.1104/pp.112.200584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. von Caemmerer S, Quick WP, Furbank RT (2012) The development of C4 rice: current progress and future challenges. Science 336: 1671–1672. 10.1126/science.1220177 [DOI] [PubMed] [Google Scholar]
- 26. Ueno O, Samejima M, Muto S, Miyachi S (1988) Photosynthetic characteristics of an amphibious plant, Eleocharis vivipara: Expression of C4 and C3 modes in contrasting environments. Proc Natl Acad Sci USA 85: 6733–6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ueno O (2001) Environmental regulation of C3 and C4 differentiation in the amphibious sedge Eleocharis vivipara . Plant Physiol 127: 1524–1532. [PMC free article] [PubMed] [Google Scholar]
- 28. Ueno O (1998) Induction of Kranz anatomy and C4-like biochemical characteristics in a submerged amphibious plant by abscisic acid. Plant Cell 10: 571–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pyankov VI, Black CC, Artyusheva EG, Voznesenskaya EV, Ku MSB, et al. (1999) Features of photosynthesis in Haloxylon species of Chenopodiaceae that are dominant plants in central Asian deserts. Plant Cell Physiol 40: 125–134. [Google Scholar]
- 30. Pyankov VI, Voznesenskaya EV, Kuz'min AN, Ku MSB, Ganko E, et al. (2000) Occurrence of C3 and C4 photosynthesis in cotyledons and leaves of Salsola species (Chenopodiaceae). Photosynth Res 63: 69–84. [DOI] [PubMed] [Google Scholar]
- 31. Sawers RJH, Liu P, Anufrikova K, Hwang JTG, Brutnell TP (2007) A multi-treatment experimental system to examine photosynthetic differentiation in the maize leaf. BMC Genomics 8: 12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li P, Ponnala L, Gandotra N, Wang L, Si Y, et al. (2010) The developmental dynamics of the maize leaf transcriptome. Nat Genet 42: 1060–1067. 10.1038/ng.703 [DOI] [PubMed] [Google Scholar]
- 33. Majeran W, Friso G, Ponnala L, Connolly B, Huang M, et al. (2010) Structural and metabolic transitions of C4 leaf development and differentiation defined by microscopy and quantitative proteomics in maize. Plant Cell 22: 3509–3542. 10.1105/tpc.110.079764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. John CR, Smith-Unna RD, Woodfield H, Covshoff S, Hibberd JM (2014) Evolutionary convergence of cell-specific gene expression in independent lineages of C4 grasses. Plant Physiol 165: 62–75. 10.1104/pp.114.238667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gowik U, Bräutigam A, Weber KL, Weber APM, Westhoff P (2011) Evolution of C4 photosynthesis in the genus Flaveria: how many and which genes does it take to make C4? Plant Cell 23: 2087–2105. 10.1105/tpc.111.086264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bräutigam A, Kajala K, Wullenweber J, Sommer M, Gagneul D, et al. (2011) An mRNA blueprint for C4 photosynthesis derived from comparative transcriptomics of closely related C3 and C4 species. Plant Physiol 155: 142–156. 10.1104/pp.110.159442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xu J, Li Y, Ma X, Ding J, Wang K, et al. (2013) Whole transcriptome analysis using next-generation sequencing of model species Setaria viridis to support C4 photosynthesis research. Plant Mol Biol 83: 77–87. 10.1007/s11103-013-0025-4 [DOI] [PubMed] [Google Scholar]
- 38. Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628. 10.1038/nmeth.1226 [DOI] [PubMed] [Google Scholar]
- 40. Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, et al. (2008) The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320: 1344–1349. 10.1126/science.1158441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kent WJ (2002) BLAT—the BLAST-like alignment tool. Genome Res 12: 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Carbon S, Ireland A, Mungall CJ, Shu S, Marshall B, et al. (2009) AmiGO: online access to ontology and annotation data. Bioinformatics 25: 288–289. 10.1093/bioinformatics/btn615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bender MM (1968) Mass spectrometric studies of carbon 13 variations in corn and other grasses. Radiocarbon 10: 468–472. [Google Scholar]
- 45. Bender MM (1971) Variations in the 13C/12C ratios of plants in relation to the pathway of photosynthetic carbon dioxide fixation. Phytochemistry 10: 1239–1244. [Google Scholar]
- 46. Farquhar GD, Ehleringer JR, Hubick KT (1989) Carbon isotope discrimination and photosynthesis. Annu Rev Plant Physiol Plant Mol Biol 40: 503–537. [Google Scholar]
- 47. Wang P, Kelly S, Fouracre JP, Langdale JA (2013) Genome-wide transcript analysis of early maize leaf development reveals gene cohorts associated with the differentiation of C4 Kranz anatomy. Plant J 75: 656–670. 10.1111/tpj.12229 [DOI] [PubMed] [Google Scholar]
- 48. Slewinski TL, Anderson AA, Zhang C, Turgeon R (2012) Scarecrow plays a role in establishing Kranz anatomy in maize leaves. Plant Cell Physiol 53: 2030–2037. 10.1093/pcp/pcs147 [DOI] [PubMed] [Google Scholar]
- 49. Su P (2010) Photosynthesis of C4 desert plants In: Ramawat KG, editor. Desert Plants: Springer; Berlin Heidelberg: pp. 243–259. [Google Scholar]
- 50. Brown NJ, Newell CA, Stanley S, Chen JE, Perrin AJ, et al. (2011) Independent and parallel recruitment of preexisting mechanisms underlying C4 photosynthesis. Science 331: 1436–1439. 10.1126/science.1201248 [DOI] [PubMed] [Google Scholar]
- 51. Kajala K, Brown NJ, Williams BP, Borrill P, Taylor LE, et al. (2012) Multiple Arabidopsis genes primed for recruitment into C4 photosynthesis. Plant J 69: 47–56. 10.1111/j.1365-313X.2011.04769.x [DOI] [PubMed] [Google Scholar]
- 52. Aubry S, Kelly S, Kümpers BMC, Smith-Unna RD, Hibberd JM (2014) Deep evolutionary comparison of gene expression identifies parallel recruitment of trans-factors in two independent origins of C4 photosynthesis. PLoS Genet 10: e1004365 10.1371/journal.pgen.1004365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Külahoglu C, Denton AK, Sommer M, Maß J, Schliesky S, et al. (2014) Comparative transcriptome atlases reveal altered gene expression modules between two Cleomaceae C3 and C4 plant species. Plant Cell 26: 3243–3260. 10.1105/tpc.114.123752 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
Table A. 2959 differentially expressed genes [FDR≤0.001 and abs (|log2(Fold change)|≥1)] Table B. Up-regulated GO categories. Table C. Down-regulated GO categories.
(XLSX)
Data Availability Statement
RNAseq data produced in this study have been submitted to the NCBI/SRA database under accession number SRP049928.