Abstract
Introduction: Long non-coding RNAs (lncRNAs) have emerged recently as major players in tumor biology and may be used for cancer diagnosis, prognosis, and potential therapeutic targets. Although down-regulation of lncRNA LET in several cancers has been studied, its role in gastric cancer remains unknown. The aim of our study was to investigate the expression, and clinical significance of lncRNA LET in gastric cancer. Methods: The expression of lncRNA LET was detected by quantitative real-time PCR (qRT-PCR) in pairs of tumor tissues and adjacent non-tumor tissues of 93 gastric cancer patients. Then, we analyzed the potential relationship between lncRNA LET expression levels in tumor tissues and clinicopathological features of gastric cancer, and clinical outcome. Results: We found that lncRNA LET expression was markedly down-regulated in tumor tissues compared with adjacent non-tumor tissues, and associated with depth of invasion, lymph node metastasis, distant metastasis, and TNM stage. Kaplan-Meier analysis showed that patients with low lncRNA LET expression had a poor overall survival than those with high lncRNA LET expression. Moreover, univariate and multivariate analyses showed that low lncRNA LET expression was an independent poor prognostic factor for gastric cancer patients. Conclusions: Our data provided the first evidence that lncRNA LET might be a novel prognostic indicator in gastric cancer and might be a potential target for diagnosis and gene therapy.
Keywords: Gastric cancer, long non coding RNA, LET, prognosis
Introduction
Gastric cancer is the second leading cause of cancer death, and is the most common gastrointestinal malignancy in China [1,2]. Although the majority of the patients at an early stage of gastric carcinoma can be cured by surgery, more than half of those at an advanced stage of the disease die of carcinoma recurrence, even after undergoing curative gastrectomy [3]. Thus, the overall 5-year survival rate is about 40%, and that of the patients with distant metastasis is less than 5% [4,5]. Therefore, new markers with high sensitivity and specificity for gastric cancer detection and better progression control targets are urgent needed.
It is well known that protein coding genes account for only 2% of the total genome, whereas the vast majority of the human genome can be transcripted into non coding RNAs (ncRNA) [6]. Among them are long non coding RNAs (lncRNAs), which are more than 200 nucleotides in length and unable to be translated into proteins [7]. LncRNAs are known to play important roles in cellular development, differentiation, and many other biological processes [8-10]. Multiple lines of evidence have revealed the contribution of lncRNAs as having oncogenic and tumor suppressor roles in tumorigenesis [11].
Recent studies indicated that lncRNA LET was down-regulated in several cancers such as hepatocellular carcinomas, colorectal cancer, and gallbladder cancer, which play key roles in tumor development and progression [12-14]. However, the relationship between expression of lncRNA LET and gastric cancer development and/or progression remains unclear.
In our current study, we examined the expression level of lncRNA LET in gastric cancer tissues and adjacent non-tumor tissues. The relationships between its expression and clinicopathological features were then analyzed so as to evaluate whether lncRNA LET expression could be a useful biomarker for prognosis in gastric cancer patients.
Materials and methods
Patients and specimens
A total of 93 gastric cancer samples were obtained from patients who had underwent surgery at The First Affiliated Hospital of Xinxiang Medical University between 2007 and 2009, and were diagnosed with gastric cancer according AJCC cancer staging manual based on histopathological evaluation [15]. Clinical pathology information was available for all samples (Table 1). No local or systemic treatment was conducted in these patients before the operation. All specimens were immediately frozen in liquid nitrogen until use. The study was approved by the Research Ethics Committee of Xinxiang Medical University. Informed consents were obtained from all patients.
Table 1.
Association of lncRNA LET with clinicopathological features
| Clinicopathological features | Total | LncRNA LET expression | P value | |
|---|---|---|---|---|
|
| ||||
| Low | High | |||
| Age (years) | 0.559 | |||
| < 60 | 41 | 18 | 23 | |
| ≥ 60 | 52 | 26 | 26 | |
| Gender | 0.141 | |||
| Male | 60 | 25 | 35 | |
| Female | 33 | 19 | 14 | |
| Tumor size (cm) | 0.537 | |||
| < 5 | 35 | 18 | 17 | |
| ≥ 5 | 58 | 26 | 32 | |
| Differentiation | 0.723 | |||
| Well | 27 | 12 | 15 | |
| Moderate + Poor | 66 | 32 | 34 | |
| Depth of invasion | 0.000 | |||
| T1 + T2 | 36 | 8 | 28 | |
| T3 + T4 | 57 | 36 | 21 | |
| Lymph node metastasis | 0.000 | |||
| No | 51 | 12 | 39 | |
| Yes | 42 | 32 | 10 | |
| Distant metastasis | 0.000 | |||
| No | 48 | 13 | 35 | |
| Yes | 45 | 31 | 14 | |
| TNM stage | 0.001 | |||
| I + II | 47 | 14 | 33 | |
| III + IV | 46 | 30 | 16 | |
RNA extraction and qRT-PCR analyses
Total RNA was extracted from tissues using TRIzol reagent (Invitrogen). For qRT-PCR, RNA was reverse transcribed to cDNA by using a Reverse Transcription Kit (Takara). Real-time PCR analyses were performed with Power SYBR Green (Takara). Results were normalized to the expression of GAPDH. The PCR primers for lncRNA LET or GAPDH were as follows: lncRNA LET sense, 5’-CCTTCCTGACAGCCAGTGTG-3’ and reverse, 5’-CAGAATGGAAATACTGGAGCAAG-3’; GAPDH sense, 5’-GTCAACGGATTTGGTCTGTATT-3’ and reverse, 5’-AGTCTTCTGGGTGGCAGTGAT-3’. qRT-PCR and data collection were performed on ABI 7900. The relative expression of lncRNA LET was calculated and normalized using the 2−ΔΔCt method relative to GAPDH.
Statistical analysis
All statistical analyses were performed using SPSS 18.0 software (IBM). The statistical significance between groups was determined using the Student’s t test. Association between expression level of lncRNA LET and each clinicopathologic parameter was evaluated using Pearson’s Chi-square test. Patient survival was evaluated using the Kaplan-Meier method and compared using log-rank test. Univariate and multivariate Cox regression analyses were performed to analyze the survival data. The data are shown as the mean ± SD from at least three independent experiments. A two-sided P value of less than 0.05 was considered to statistically significant.
Results
Expression of lncRNA LET is downregulated in human gastric cancer tissues
We firstly examined lncRNA LET expression level in 93 paired gastric cancer samples and adjacent non-tumor tissues by qRT-PCR, and normalized to GAPDH. Figure 1 showed that the lncRNA LET level was significantly down-regulated in gastric cancer tissues compared with corresponding adjacent non-tumor tissues (P < 0.05). The data indicate that abnormal lncRNA LET expression may be related to gastric cancer pathogenesis.
Figure 1.
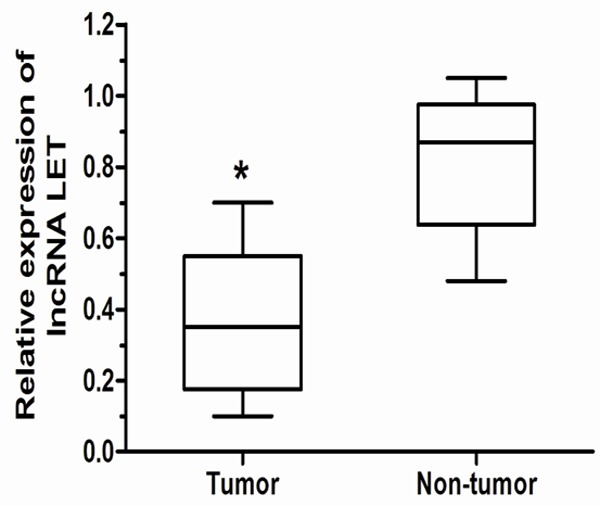
qRT-PCR analysis of lncRNA LET expression in gastric cancer. lncRNA LET was significantly down-regulated in gastric cancer tissues compared to the adjacent non-tumor tissues. ΔCt method was used to measure the relative lncRNA LET expression, which was normalized by the GAPDH expression level. *P < 0.05.
lncRNA LET expression and clinicopathologic factors in gastric cancer
To assess the correlation of lncRNA LET expression with clinicopathologic data, the expression levels of lncRNA LET in tumor tissues were categorized as low or high in relation to the mean value. Clinicopathologic factors were analyzed in the high and low lncRNA LET expression groups (Table 1). The low lncRNA LET group was correlated with deeper depth of invasion, more lymph node metastasis, more distant metastasis, and higher TNM stage (P < 0.05) than the high lncRNA LET expression group. However, lncRNA LET expression level was not associated with other clinicopathologic factors such as age, gender, tumor size, and differentiation (P > 0.05) (Table 1).
Correlation between lncRNA LET expression and prognosis of gastric cancer patients
We further examined whether lncRNA LET expression level correlated with outcome of gastric cancer patients after gastrectomy. Overall survival (OS) curves were plotted according to lncRNA LET expression level by the Kaplan-Meier analysis and log-rank test, the results were presented in Figure 2, patients with low lncRNA LET expression level had poorer overall survival than the high lncRNA LET expression group (P < 0.05). Our data demonstrated that down-regulated expression of lncRNA LET in gastric cancer was significantly correlated with patients’ survival time.
Figure 2.
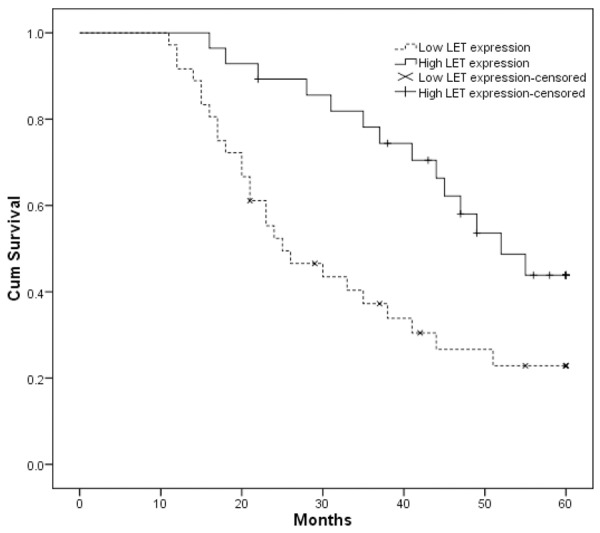
Kaplan-Meier survival analysis of association between lncRNA LET expression level and overall survival of gastric cancer patients. Patients with low expression of lncRNA LET showed decreased overall survival compared with patients with high level of lncRNA LET expression. Survival curves were compared using log-rank test.
In order to estimate the clinical significance of various prognostic factors that might influence survival in the study population, univariate analyses was performed for OS in 93 patients with gastric cancer, respectively. As shown in Table 2, depth of invasion, lymph node metastasis, distant metastasis, TNM stage, and lncRNA LET expression were statistically significant risk factors affecting OS of gastric cancer patients. The other clinicopathological features, such as age, gender, tumor size and differentiation were not statistically significant prognosis factors (P > 0.05). Low intratumoral lncRNA LET expression is a significant negative predictor for OS (P < 0.05). To evaluate the robustness of the prognostic value of intratumoral lncRNA LET expression, variables with a value of P < 0.05 were selected for multivariate analysis. As shown in Table 2, multivariate analysis revealed that lncRNA LET expression depth of invasion, lymph node metastasis, distant metastasis, and TNM stage were independent prognostic markers for gastric cancer. Taken together, these data indicated that low lncRNA LET expression level was an independent risk factor for gastric cancer patients.
Table 2.
Univariate and multivariate logistic regression analysis of factors associated with clinicopathological features
| Clinicopathological feature | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| Risk ratio | 95% CI | P | Risk ratio | 95% CI | P | |
| Age (years) | ||||||
| ≥ 60 vs. < 60 | 1.362 | 0.614-2.325 | 0.406 | |||
| Gender | ||||||
| Male vs. Female | 0.894 | 0.493-1.524 | 0.275 | |||
| Tumor size | ||||||
| ≥ 5 cm vs. < 5 cm | 1.571 | 0.847-2.386 | 0.337 | |||
| Differentiation | ||||||
| Moderate + Poor vs. Well | 2.1745 | 0.674-3.738 | 0.397 | |||
| Depth of invasion | ||||||
| T3 + T4 vs. T1 + T2 | 1.974 | 0.813-3.426 | 0.029 | 1.663 | 0.697-3.215 | 0.017 |
| Lymph node metastasis | ||||||
| Yes vs. No | 2.607 | 0.718-5.326 | 0.013 | 2.402 | 0631-5.174 | 0.009 |
| Distant metastasis | ||||||
| Yes vs. No | 3.017 | 1.429-6.617 | 0.009 | 2.826 | 1.374-6.036 | 0.011 |
| TNM stage | ||||||
| III + IV vs. I + II | 3.618 | 1.495-7.214 | 0.012 | 3.177 | 1.318-6.915 | 0.008 |
| LncRNA LET | ||||||
| Low vs. High | 2.513 | 1.414-5.847 | < 0.001 | 2.275 | 1.301-5.176 | 0.007 |
Discussion
Accurate prediction of the prognosis for the individual gastric cancer patient is of great importance, and molecular biomarkers that could be served as prognostic factors would be useful in determining an individualized treatment plan for a gastric cancer patient [16]. However, the biomarkers used in this tumor group today are not satisfactory [17], and it is needed to exploit additional markers to fine-tune this process.
Previous investigations indicated that lncRNAs were involved in multiple cellular functions including proliferation, apoptosis and differentiation, thus, lncRNAs have been implemented in diverse physiological and pathological processes ranging from development to cancer. For example, Rajnish’s data showed that lncRNA HOTAIR was increased in expression in primary breast tumors and metastases, and HOTAIR expression level in primary tumors was a powerful predictor of eventual metastasis and death, in vitro assay, they demonstrated that HOTAIR exerts its oncogenic functions via binding the PRC2 (polycomb repressive complex 2), which methylates histone H3 on K27 to promote gene repression [18]. Zhang found that lncRNA ANRIL was up-regulated and served as an independent predictor for overall survival of human gastric cancer, Further experiments revealed that E2F1 could induce ANRIL and ANRIL-mediated growth promotion is in part due to epigenetic repression of miR-99a/miR-449a in Trans by binding to PRC2 [19]. Liu et al. showed that lncRNA GAS5 expression was downregulated in bladder cancer; furthermore, gain-of-function and loss-of-function studies showed that GAS5 could inhibit bladder cancer cell proliferation [20]. Qin demonstrated that lncRNA TSLC1-AS1 expression was down-regulated in glioma tumor, over-expression of TSLC1-AS1 resulted in inhibition of cell proliferation, migration and invasion in U8 cells, which indicated that lncRNA TSLC1-AS1 may serve as a potential biomarker and therapeutic target for glioma [21]. However, the relationship between lncRNA and cancer patient prognosis remains largely unknown.
LncRNA LET was previously shown to be down-regulated and function as a tumor suppressor in primary hepatocellular carcinoma, they also demonstrated that the down-regulation of lncRNA LET was vital in the stabilization of nuclear factor 90 protein, which leads to hypoxia-induced cancer cell invasion [12], Ma revealed that lncRNA LET was significantly down-regulated in gallbladder cancer compared to their adjacent normal tissues. Meanwhile, patients with low expression of lncRNA LET have significantly poorer prognosis than those with high expression [13]. This common characteristic thus strengthened the clinical application value of lncRNA LET. Therefore, we hypothesized that lncRNA LET expression was also decreased in gastric cancer tissues and decreased of this lncRNA could predict the prognosis of gastric cancer patients.
To test this hypothesis, tissue samples from 93 patients with gastric cancer were selected. The qRT-PCR showed that lncRNA LET was significantly down-regulated in gastric cancer tissues compared to the adjacent non-tumor tissues. We then used the mean expression level of lncRNA LET as a cutoff to divide the 93 patients into the lncRNA LET low group and lncRNA LET high group to further investigate the association between lncRNA LET expression and clinicopathological characteristics. Patients with low expression of lncRNA LET showed deeper depth of invasion, more lymph node metastasis, more distant metastases and higher TNM stage than the lncRNA LET high group. These results indicated that down-regulation of lncRNA LET might play key roles in gastric cancer progression and development.
As lncRNA LET expression was found to be associated with gastric cancer invasion and metastasis, considering the invasion of cancer to nearby tissues and metastasis to distal tissues are crucial factors affecting the prognosis of patients, lncRNA LET might be a potential prognostic marker for patients with gastric cancer. In order to investigate the prognostic role of lncRNA LET on gastric cancer, we performed Kaplan-Meier analysis of overall survival. Results showed that patients with gastric cancer of low lncRNA LET expression tend to have worse overall survival in comparison to patients with high expression. To further evaluate the prognostic value of lncRNA LET in gastric cancer, we performed Cox proportional hazards model, our results proved that decreased lncRNA LET expression was an independent prognostic marker for predicting the poor prognosis of gastric cancer patients. Thus, lncRNA LET expression could be used as a molecular prognostic factor to identify patients who are more likely to have higher risk of death.
In conclusion, we have proved that lncRNA LET expression was decreased in gastric cancer and associated with tumor progression. The present study also demonstrated for the first time that lncRNA LET expression was an independent prognostic factor of patients with gastric cancer. Therefore, it is possible that lncRNA LET may play an important role in invasiveness and metastasis of gastric cancer. It is also possible that lncRNA LET serves as prognostic marker in clinical practice and a new therapeutic method for the treatment of gastric cancer.
Disclosure of conflict of interest
None.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Jing JJ, Liu HY, Hao JK, Wang LN, Wang YP, Sun LH, Yuan Y. Gastric cancer incidence and mortality in Zhuanghe, China, between 2005 and 2010. World J Gastroenterol. 2012;18:1262–9. doi: 10.3748/wjg.v18.i11.1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakajima T. Gastric cancer treatment guidelines in Japan. Gastric Cancer. 2002;5:1–5. doi: 10.1007/s101200200000. [DOI] [PubMed] [Google Scholar]
- 4.Crew KD, Neugut AI. Epidemiology of gastric cancer. World J Gastroenterol. 2006;12:354–362. doi: 10.3748/wjg.v12.i3.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang L. Incidence and mortality of gastric cancer in China. World J Gastroenterol. 2006;12:17–20. doi: 10.3748/wjg.v12.i1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 7.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 8.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 11.Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang F, Huo XS, Yuan SX, Zhang L, Zhou WP, Wang F, Sun SH. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol Cell. 2013;49:1083–1096. doi: 10.1016/j.molcel.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 13.Ma MZ, Kong X, Weng MZ, Zhang MD, Qin YY, Gong W, Zhang WJ, Quan ZW. Long noncoding RNALET is a positive prognostic factor and exhibits-tumorsuppressive activity in gallbladder cancer. Mol Carcinog. 2014 doi: 10.1002/mc.22215. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 14.He Y, Meng XM, Huang C, Wu BM, Zhang L, Lv XW, Li J. Long noncoding RNAs: Novel insights into hepatocelluar carcinoma. Cancer Lett. 2014;344:20–27. doi: 10.1016/j.canlet.2013.10.021. [DOI] [PubMed] [Google Scholar]
- 15.Egner JR. AJCC cancer staging manual. JAMA. 2010;304:1726–1727. [Google Scholar]
- 16.Ueda T, Volinia S, Okumura H, Shimizu M, Taccioli C, Rossi S, Alder H, Liu CG, Oue N, Yasui W. Relation between microRNA expression and progression and prognosis of gastric cancer: a microRNA expression analysis. Lancet Oncol. 2010;11:136–146. doi: 10.1016/S1470-2045(09)70343-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pietrantonio F, De Braud F, Da Prat V, Perrone F, Pierotti MA, Gariboldi M, Fanetti G, Biondani P, Pellegrinelli A, Bossi I, Di Bartolomeo M. A review on biomarkers for prediction of treatment outcome in gastric cancer. Anticancer Res. 2013;33:1257–1266. [PubMed] [Google Scholar]
- 18.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang EB, Kong R, Yin DD, You LH, Sun M, Han L, Xu TP, Xia R, Yang JS, De W, Chen J. Long noncoding RNA ANRIL indicates a poor prognosis of gastric cancer and promotes tumor growth by epigenetically silencing of miR-99a/miR-449a. Oncotarget. 2014;5:2276–2292. doi: 10.18632/oncotarget.1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Z, Wang W, Jiang J, Bao E, Xu D, Zeng Y, Tao L, Qiu J. Downregulation of GAS5 promotes bladder cancer cell proliferation, partly by regulating CDK6. PLoS One. 2013;8:e73991. doi: 10.1371/journal.pone.0073991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qin X, Yao J, Geng P, Fu X, Xue J, Zhang Z. LncRNA TSLC1-AS1 is a novel tumor suppressor in glioma. Int J Clin Exp Pathol. 2014;7:3065. [PMC free article] [PubMed] [Google Scholar]


