Abstract
Objectives: The aim of this study was to investigate the expression of ECT2 in gastric cancer and its clinical significance. Methods and results: We investigated the differentially expressed genes between gastric cancer tissues and normal gastric mucosa by cDNA microarray, and then we found ECT2 was up-regulated in gastric cancer. What is more, we verified ECT2 expression level by quantitative real-time reverse transcription-polymerase chain reaction (qRT-PCR) and measured its protein level by immunohistochemistry (IHC). qRT-PCR analysis indicated ECT2 was significantly up-regulated in gastric cancer and Immunohistochemistry confirmed the percentage of ECT2-positive specimens was significantly higher in gastric carcinoma than in non-tumor tissues. Up-regulation of ECT2 is associated with the degree of histological differentiation (P = 0.007), invasion depth (P = 0.047), lymph node metastasis (P = 0.016), distant metastasis (P = 0.021) and TNM stage (P = 0.016), patients with up-regulated ECT2 had a lower overall survival rate (P = 0.000). Cox regression analysis revealed that up-regulation of ECT2 is an independent prognostic factor in gastric cancer patients (P = 0.012). Conclusion: Up-regulation of ECT2 might contribute to the progression of gastric carcinogenesis and may be a useful prognostic indicator in gastric cancer.
Keywords: ECT2, microarray, gastric cancer, prognosis
Introduction
Gastric cancer (GC) is the fourth most common cancer and second leading cause of cancer-related death worldwide [1]. In spite of its declining incidence, GC still remains among the leading causes of death from cancer in China [2]. The development and progression of gastric cancer involves a number of genetic and epigenetic alterations of tumor suppressor and tumor-related genes [3], until now we know little about the molecular mechanism of gastric carcinogenesis. Owing to the absence of specific symptoms in the early stages, gastric cancer is usually detected at an advanced stage in many patients with lymph node invasion and metastasis, the total prognosis of gastric cancer is poor [4]. In addition, the TNM stage which depend on the depth of invasion of gastric wall (T), the involvement of lymph nodes (N) and the presence of distant metastasis (M) is the most important prognostic factor for GC, however, prognosis often varies among patients in the same stage [5]. So it is critically important to understand the molecular mechanism of gastric carcinogenesis and find specific and sensitive molecular targets for early diagnosis, effective treatment and prediction of prognosis of GC.
ECT2 (epithelial cell transforming sequence 2) was identified as a proto-oncogene capable of transforming NIH/3T3 fibroblasts [6], The C-terminus of ECT2 protein is the catalytic core, and consists of a Dbl-homology (DH) and a pleckstrinhomology (PH) domain which confer guanine nucleotide exchange activity toward Rho GTPases [7], the N-terminal contains tandem repeats of the BRCT domain which is highly conserved in proteins related to cell cycle check point and DNA damage response [8], ECT2 protein is reported to interact with the Rho GTPases family leading to cell malignant transformation [9], triggers cytokinesis [8] and regulates epithelial cell polarity [10]; up to now, ECT2 is considered as an oncogene in human cancer [11], and it has been showed up-regulation in a variety of human tumors including glioma [12], lung cancer [13], esophageal cancer [13], pancreatic cancer [14], et al. However few studies have analyzed the role of ECT2 expression in GC. In the present study, we examined the expression of ECT2 in gastric carcinoma tissues and evaluated its clinicopathological significance and prognostic value to determine the possible use of ECT2 as a prognostic molecular marker in gastric cancer.
Materials and methods
Tissue specimens for gene chip and quantitative real-time PCR
Fresh specimens were obtained from 52 patients who underwent curative surgery for gastric cancer at Zhejiang Provincial People’s Hospital from January to December in 2012. The cancer tissues and their matched normal mucosa (obtained at greater than 5 cm apart from the edge of primary tumor focus) were immediately frozen in liquid nitrogen after resection and stored at -80°C, among these specimens, 20 couples were for Gene Chip Array, another 32 couples were for Quantitative real-time PCR.
Tissue samples for immunohistochemistry
A total of 112 patients who had curative surgery for gastric cancer at Zhejiang Provincial People’s Hospital from 2007 to 2009 were enrolled in this study. Resected tumor samples were immediately collected after surgical resection and fixed in formaldehyde solution, then embed in paraffin. The group was comprised of 80 males and 32 females with a median age of 59.2 years (range from 30 to 82 years). Pathological data including age, gender, tumor size and depth, differentiation status, growth pattern, lymph node metastasis were obtained, all these patients were classified by TNM stage as follows: TNM stage I (n = 13), TNM stage II (n = 26), TNM stage III (n = 56) and TNM stage IV (n = 17); diagnose was confirmed by two pathologists, tumor grade and stage classifications were assigned according to the 7th edition of the AJCC staging manual [15]. None of the patients had received radiotherapy or chemotherapy before surgery. The last follow-up was December 2012. Eighty normal gastric tissue samples from patients without malignant tumor were obtained by endoscopy as controls.
The use of all above specimens in this study was approved by the ethics committee of Zhejiang Provincial People’s Hospital after obtaining informed consent from all patients.
RNA extraction
We extracted total RNA from 20 couples of fresh gastric cancer tissues and matched normal gastric tissues using TRIZOL Reagent (Cat#15596-018, life technologies, Carlsbad, CA, US) according to the manufacturer’s instruction, then inspected RNA integrity by an Agilent Bioanalyzer 2100 (Agilent technologies, Santa Clara, CA, US), total RNA was qualified when its 2100 RIN ≥ 7.0 and 28 s/18 s ≥ 0.7.
RNA amplification and labeling
Qualified total RNA was amplified, labeled and purified by using GeneChip 3’IVT Express Kit (Cat#901229, Affymetrix, Santa Clara, CA, US) followed the manufacturer’s instruction to obtain biotin labeled cRNA.
Array hybridization
We performed Array hybridization and wash by using GeneChip® Hybridization, Wash and Stain Kit (Cat#900720, Affymetrix, Santa Clara, CA, US) in Hybridization Oven 645 (Cat#00-0331-220V, Affymetrix, Santa Clara, CA, US) and Fluidics Station 450 (Cat#00-0079, Affymetrix, Santa Clara, CA, US).
Data acquisition
Slides were scanned by Gene Chip scanner 3000 (Cat#00-00212 Affymetrix, Santa Clara, CA, US) and Command Console Software 3.1 (Affymetrix, Santa Clara, CA, US) with default settings, then MAS 5.0 algorithm, Gene Spring Software 11.0 (Agilent technologies, Santa Clara, CA, US) was used to normalize the raw data. The value of T vs. N signal log ratio ≥ 2 or ≤ 0.5 was regarded as differentially expressed.
Quantitative real-time PCR
Total RNA was isolated according to the manufacturer’s instructions (Life Technologies, CA, USA). RNA was reverse-transcribed into cDNA (PrimeScript RT-PCR kit; Takara Bio). GAPDH was used as an internal control, qRT-PCR was performed in a total volume of 20 μl reaction mixture. Quantitative ECT2 mRNA levels were assessed by real-time PCR with ABI PRISM 7700 Sequence Detection System (Applied Biosystems, Hangzhou, Zhejiang, China) using SYBR Premix Ex Taq (Applied Takara Bio). The sequences for qRT-PCR primers were as follows: human ECT2 forward 5’-CGGCTCAGTGGAGGGAAG-3’ and reverse 5’-GGAAGGCTGACAAGGGAGG-3’; GAPDH forward were 5’-TGAAGGTCGGAGTCAACGG-3’ and reverse were 5’-CTGGAAGATGGTGATGGGATT-3’. PCR was run as follows: 95°C for 4 min, amplification for 40 cycles with denaturation at 95°C for 10 seconds, annealing at 57°C for 30 seconds, and extension at 72°C for 30 seconds. Each qRT-PCRs was repeated 3 times, the average was calculated, the ECT2 expression level was expressed as 2-ΔΔCt, ΔCt = Ct(ECT2)-Ct(GAPDH).
Immunohistochemistry
The sections of formalin-fixed paraffin-embedded tissues were treated with routine deparaffinization and hydration followed by antigen retrieval which was performed in 10 mM citrate buffer (pH 6.0) at 120°C for 3 min. Then treated with 3% hydrogen peroxide for 15 min at room temperature, after rinsing with phosphate-buffered saline (PBS), non-specific binding was blocked with 10% goat serum. The sections were incubated at 4°C for overnight using the primary antibodies at a dilution of 1:300 against ECT2 (Santa Cruz Biotechnology, Santa Cruz, CA, USA), then incubated for 15 min with biotin-labeled secondary antibody at room temperature, stained with 3,3-diaminobenzidine (DAB), counterstained with hematoxylin. Negative controls were stained without primary antibody.
Immunohistochemical evaluation
All samples were observed under Nikon light microscope (Nikon Corporation, Tokyo, Japan) independently by two experienced pathologists without the knowledge of patients. Immunoreactivity for ECT2 was evaluated using a combined scoring system according to intensity and extent, the staining intensity was categorized as follows: (0 for no staining, 1+ for weak immunoreactivity, 2+ for moderate immunoreactivity and 3+ for strong immunoreactivity). The percentage of cells demonstrating positive ECT2 staining were scored as follows: 0, < 5% positive cells, 1+, 5-25% positive cells, 2+, 26-50% positive cells, 3+, 51-75% positive cells, 4+, 76-100% positive cells. The intensity and proportion scores were then multiplied to obtain a total score. Total score ranged from 0 to 12, scores of 0-3 were considered as negative and ≥ 4 were considered as positive.
Statistical analysis
Statistical analyses were performed using SPSS for Windows, version 15.0 (Chicago, IL, USA), χ2 test or Fisher’s exact test was used to examine the relationship between ECT2 expression and clinicopathological variables; Wilcoxon signed Ranks Test was used to detect ECT2 mRNA levels in paired samples of gastric cancer. Kaplan-Meier survival curves were generated and survival data were analyzed with the log-rank test, univariate and multivariate Cox regression analysis was used to identify independent prognostic factors. P < 0.05 was considered to have statistical significance.
Results
ECT2 gene expression in gene chip
We analyzed the difference of gene expression profiles between 20 gastric cancer tissues and normal gastric mucosa by cDNA microarray, many genes were abnormally expressed in gastric cancer, these genes were correlated with cell adhesion, angiogenesis, transcription, cell proliferation and apoptosis, cell cycle regulate, DNA repair et al. ECT2 was one of the up-regulated genes in GC tissues which showed more than 3 fold differential expression (P = 0.00-0412) (Table 1). We selected it as a newly recognized biomarker to investigate its role in GC.
Table 1.
Up-regulation genes in microarray
| Gene_ Symbol | Probe_ Set_ ID | Fold change | P values | Entrez_Gene | Chromosomal_ Location | Chromosome_Number. Avadis | Archival_UniGene_Cluster |
|---|---|---|---|---|---|---|---|
| ECT2 | 219787_s_at | 3.165477 | 0.000412 | 1894 | chr3q26.1- q26.2 | chr3 | Hs.122579 |
| COL3A1 | 201852_x_at | 2.764341 | 0.000479 | 1281 | chr2q31 | chr2 | Hs.119571 |
| CHEK1 | 205394_at | 2.763041 | 0.007799 | 1111 | chr11q24.2 | chr11 | Hs.20295 |
| VEGFC | 209946_at | 2.762513 | 0.000531 | 7424 | chr4q34.3 | chr4 | Hs.79141 |
| KIF14 | 206364_at | 2.757725 | 0.003571 | 9928 | chr1q32.1 | chr1 | Hs.116649 |
| AMIGO2 | 222108_at | 2.755118 | 0.000495 | 347902 | chr12q13.11 | chr12 | Hs.121520 |
| EDNRA | 204464_s_at | 2.748538 | 0.003231 | 1909 | chr4q31.22 | chr4 | Hs.76252 |
| MSRB3 | 225782_at | 2.747015 | 0.007808 | 253827 | chr12q14.3 | chr12 | Hs.35092 |
| ASPM | 232238_at | 2.742532 | 0.00228 | 259266 | chr1q31 | chr1 | Hs.145479 |
| EPPK1 | 232165_at | 2.737158 | 0.000959 | 83481 | chr8q24.3 | chr8 | Hs.200412 |
| ANTXR1 | 220092_s_at | 2.646845 | 0.015279 | 84168 | chr2p13.1 | chr2 | Hs.8966 |
| ATP10A | 214255_at | 2.283599 | 0.001352 | 57194 | chr15q11.2 | chr15 | Hs.44697 |
| ENO1 | 201231_s_at | 2.281446 | 0.000294 | 2023 | chr1p36.2 | chr1 | Hs.254105 |
| SGOL2 | 230165_at | 2.281425 | 0.005322 | 151246 | chr2q33.1 | chr2 | Hs.44269 |
ECT2 mRNA expression in GC tissue and non- cancerous mucosa
ECT2 mRNA expression was analyzed in 32 paired GC samples and surrounding non-cancerous tissues. We observed that ECT2 mRNA expression level was higher in gastric cancer tissues compared with paired normal tissues in 24 cases (75%), (Wilcoxon signed Ranks Test, Z = -3.216, P = 0.001). The mean expression of ECT2 was significantly higher in GC (Figure 1A). The relative ECT2 expression levels of 32 paired tissues are shown in (Figure 1B).
Figure 1.
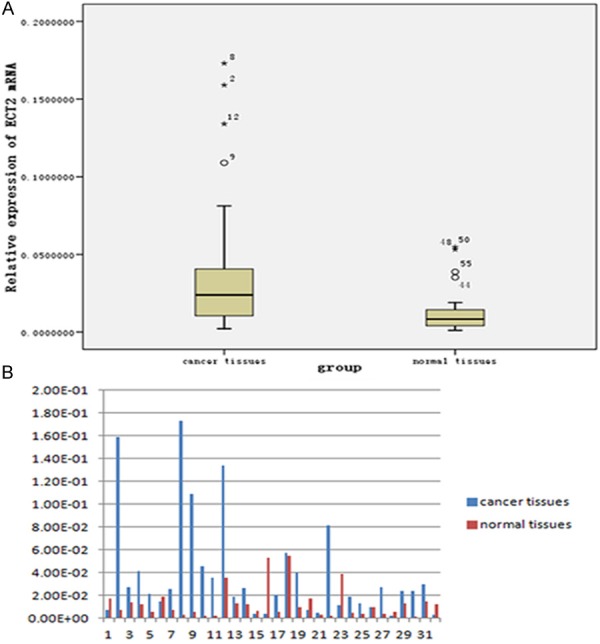
Quantitative real-time PCR results of GC and normal tissues. A. Expression of ECT2 was significantly higher in gastric cancer tissues than in normal tissues (P = 0.001). B. Relative ECT2 expression in 32 paired specimens.
ECT2 protein levels in GC tissue and non-cancerous mucosa
The ECT2 protein expression levels were examined in 112 gastric cancer specimens, we found ECT2 protein was over-expressed in 79 (70.5%) GC cases, and its staining was detected in both cytoplasm and nucleus; and ECT2 protein was detected in 13 of 80 (16.3%) human non-tumor mucosa, all these normal samples showed ECT2 expression at low levels. (Figure 2)
Figure 2.
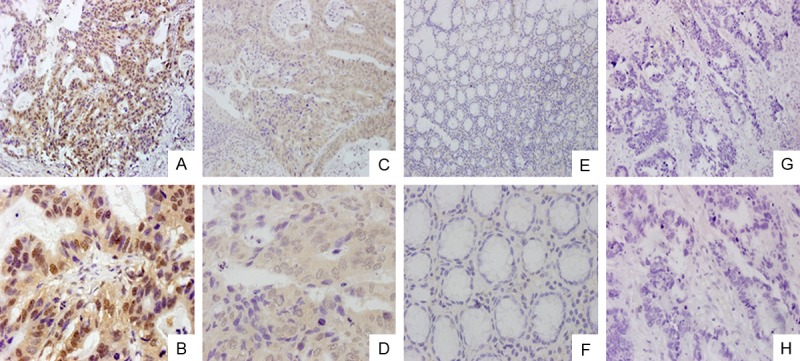
Immunohistochemical analysis of ECT2. A, B. ECT2 was strongly expressed in gastric cancer tissues; magnifications were × 200 and × 600, respectively. C, D. ECT2 was weakly expressed in gastric cancer tissues; magnifications were × 200 and × 600, respectively. E, F. ECT2 negative in normal gastric tissues; magnifications were × 200 and × 600, respectively. G, H. negative sample for immunostaining of ECT2, with phosphate-buffered saline replacing primary antibody against ECT2; magnifications were × 200 and × 600.
Relationship between ECT2 expression and clinicopathological factors
The correlation between ECT2 expression and clinicopathological features was analyzed. ECT2 expression was significantly associated with the degree of histological differentiation (P = 0.007), invasive depth (P = 0.047), lymph node metastasis (P = 0.016), distant metastasis (P = 0.021) and TNM stage (P = 0.016), there were no correlations between ECT2 protein expression and gender (P = 0.238), age (P = 0.105), tumor size (P = 0.669), tumor location (P = 0.354) (Table 2). The mean survival time of ECT2-positive group was significantly shorter than that of ECT2-negative group (42.255 ± 2.453 months vs. 50.427 ± 3.357 months, P = 0.000) (Figure 3). The 5-year survival rate of ECT2-positive group was significantly worse than that of ECT2-negative group (24.1%, 19/79 vs. 72.7%, 24/33, P = 0.000). Cox regression analysis showed that ECT2 protein expression (P = 0.012), TNM stage (P = 0.000), distant metastasis (P = 0.036), local invasion (P = 0.006), lymph node metastasis (P = 0.004) were independent prognostic factors for GC patients (Table 3).
Table 2.
Association between ECT2 expression and clinicopathological parameters
| Clinicopathologic parameter | Cases (n) | ECT2 expression | χ2 | P values | |
|---|---|---|---|---|---|
|
| |||||
| Positive | Negative | ||||
| Gender | 1.392 | 0.238 | |||
| Males | 80 | 59 | 21 | ||
| Females | 32 | 20 | 12 | ||
| Age (years) | 2.632 | 0.105 | |||
| < 60 | 58 | 37 | 21 | ||
| ≥ 60 | 54 | 42 | 12 | ||
| Tumor location | 2.077 | 0.354 | |||
| Fundus | 33 | 26 | 7 | ||
| Body | 38 | 24 | 14 | ||
| Antrum | 41 | 29 | 12 | ||
| Tumor size (cm) | 0.183 | 0.669 | |||
| < 5 | 61 | 42 | 19 | ||
| ≥ 5 | 51 | 37 | 14 | ||
| Tumor classification | 7.279 | 0.007* | |||
| Well or moderate | 43 | 24 | 19 | ||
| Poor | 69 | 55 | 14 | ||
| Local invasion | 3.938 | 0.047* | |||
| T1-2 | 24 | 13 | 11 | ||
| T3-4 | 88 | 66 | 22 | ||
| Lymph node metastasis | 5.756 | 0.016* | |||
| Yes | 79 | 61 | 18 | ||
| No | 33 | 18 | 5 | ||
| Distant metastasis | 5.363 | 0.021* | |||
| Yes | 17 | 16 | 1 | ||
| No | 95 | 63 | 32 | ||
| TNM stages | 10.313 | 0.016* | |||
| I | 13 | 7 | 6 | ||
| II | 26 | 14 | 12 | ||
| III | 56 | 42 | 14 | ||
| IV | 17 | 16 | 1 | ||
Significant difference.
Figure 3.
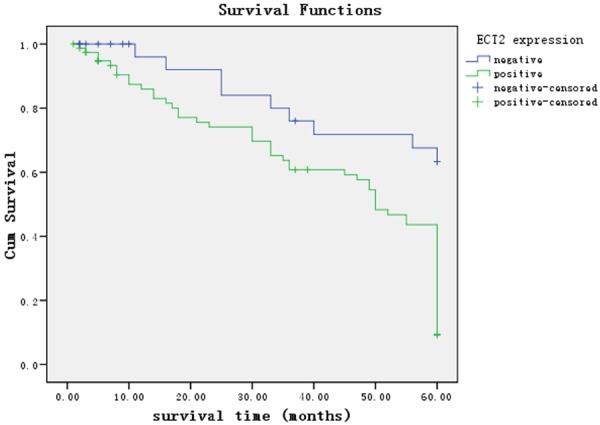
Kaplan-Meier survival curves of patients positive and negative for ECT2 expression. Patients in the ECT2 positive group had a significantly poorer prognosis than these in the ECT2 negative group (Mantel-Cox, P = 0.000).
Table 3.
Univariate and Multivariate Cox regression survival analysis of clinicopathological parameters and ECT2 expression in gastric cancer patients
| Variable | n | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Relative risk | 95% CI | P | Relative risk | 95% CI | P | ||
| Gender (M/F) | 80/32 | 1.052 | 0.620-1.786 | 0.850 | NR | ||
| Age (< 60 Y/≥ 60 Y) | 58/54 | 1.314 | 0.816-2.116 | 0.261 | NR | ||
| Tumor location (Fundus/Body/Antrum) | 33/38/41 | 0.916 | 0.681-1.233 | 0.564 | NR | ||
| Tumor size (< 5/≥ 5 cm) | 61/51 | 1.796 | 1.114-2.895 | 0.016* | 1.460 | 0.885-2.407 | 0.139 |
| Tumor classification (W+M/P) | 43/69 | 1.086 | 0.658-1.793 | 0.746 | NR | ||
| Local invasion (T 1-2/T 3-4) | 24/88 | 2.452 | 1.171-5.135 | 0.017* | 0.184 | 0.054-0.621 | 0.006* |
| Lymph node metastasis (+/-) | 79/33 | 2.592 | 1.414-4.751 | 0.002* | 0.134 | 0.034-0.522 | 0.004* |
| Distant metastasis (+/-) | 17/95 | 3.447 | 1.900-6.253 | 0.000* | 0.211 | 0.049-0.902 | 0.036* |
| TNM Stage (I/II/III/IV) | 13/26/56/17 | 2.231 | 1.597-3.117 | 0.000* | 9.892 | 2.979-32.845 | 0.000* |
| ECT2 expression (+/-) | 79/33 | 3.264 | 1.619-6.582 | 0.001* | 2.705 | 1.250-5.853 | 0.012* |
Abbreviations: CI: confidence interval
Significant difference.
Discussion
Gastric cancer is usually diagnosed at advanced stages in China, at this time, treatment options are limited and prognosis for long-term survival is poor [16]; the treatment of gastric cancer includes a combination of surgery, chemotherapy, and radiation therapy [17]; however, the overall 5-year survival rate is low at 40% [18], thus, in order to improve the management and treatment of GC patients, it is essential to investigate novel tumor-specific markers for early diagnosis, targeted therapy and prediction of prognosis. Microarray technology makes it possible to measure the expression levels of thousands of genes, and identifying meaningful and useful molecular targets from these large data. In this study, we selected ECT2 gene for further validation which is showed more than 3-fold higher expression in gastric cancer tissues in gene expression microarray.
The Rho family of small GTPases, represented by RhoA, Rac1, and Cdc42, act as molecular switches in diverse signaling pathways that regulate actin cytoskeleton modeling, cell motility, cell adhesion, cell cycle progression and gene transcription [19]. The Rho GTPases cycle between a GTP bound active state and a GDP bound inactive state, and have been implicated in the malignant phenotype of many human cancers as a result of their participation in aberrant signaling in tumor cells and overexpression in human tumors [20]. ECT2 is a member of the Dbl family of guanine nucleotide exchange factors (GEFs), which catalyze the exchange of GDP for GTP, thereby activate the Rho GTPases in signal transduction [21]. ECT2 is expressed in many tissues including kidney, liver, spleen, lung, bladder, ovary and brain [22], it exhibits nuclear localization in interphase, In this stage, ECT2 may be in an inactive state, where the N-terminal domain interacts with the catalytic domain to inhibit its exchange activity [23], and it is released to the cytoplasm in prometaphase after nuclear membrane breakdown and subsequent activate Rho GTPases to trigger cytokinesis [24]. If ECT2 becomes mislocalized to the cytoplasm it will untimely activate the Rho signaling pathways and lead to malignant transformation [9]. In addition, recent study identified another mechanism that ECT2 can act as a GEF from within the nucleus to activate Rac and then leads to malignant transformation [25]. Up-expression of ECT2 has been reported in many types of cancers, such as glioma [12], lung and esophageal cancer [13], pancreatic cancer [14], ovarian cancer [25], osteosarcoma [26], and has been associated with poor patient survival prognosis, but the role of ECT2 in gastric cancer remains unknown, it is the first study to investigate the expression of ECT2 in gastric cancer and its clinical significance in the development and progression of GC patients.
ECT2 gene showed more than 3-fold up-regulation in cancerous tissues in gene chip, and we verified the differential expression by qRT-PCR and immunohistochemistry. Our results confirmed that ECT2 mRNA and protein levels were significantly higher in tumor tissue samples than adjacent non-tumor tissues. Furthermore, ECT2 expression was correlated with tumor differentiation, TNM stage, lymph node metastasis and distant metastasis in GC patients, patients with positive ECT2 expression had a lower overall survival rate than that of patients with negative ECT2 expression. Furthermore, multivariate analysis showed that ECT2 expression level was an independent significant dangerous factor for survival after resection. Our study suggests that up-regulation of ECT2 is a common feature in gastric cancer and could be a valuable prognostic marker. However, considering the small number of samples in this study, further investigations including larger samples and experiments in vitro and vivo are needed in the future; in addition, how ECT2 functions in these processes is not entirely known, further studies are needed to elucidate the molecular mechanisms by which ECT2 participates in the development and progression of GC.
In summary, ECT2 is up-regulated in GC tissues and associated with tumor classification, invasion depth, lymph node metastasis, distant metastasis, TNM stage, and poor prognosis in gastric cancer patients; our results suggest that ECT2 may be used as a valuable prognostic indicator in patients with gastric cancer.
Acknowledgements
This study was supported by Zhejiang Provincial Natural Science Foundation of China (Grant No. LZ12H16004).
Disclosure of conflict of interest
None.
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Yang L. Incidence and mortality of gastric cancer in China. World J Gastroenterol. 2006;12:17–20. doi: 10.3748/wjg.v12.i1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tamura G. Alterations of tumor suppressor and tumor-related genes in the development and progression of gastric cancer. World J Gastroenterol. 2006;12:192–198. doi: 10.3748/wjg.v12.i2.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Q, Jin XS, Yang ZY, Wei M, Liu BY, Gu QL. Upregulated Hoxc6 expression is associated with poor survival in gastric cancer patients. Neoplasma. 2013;60:439–445. doi: 10.4149/neo_2013_057. [DOI] [PubMed] [Google Scholar]
- 5.Gravalos C, Jimeno A. HER2 in gastric cancer: a new prognostic factor and a novel therapeutic target. Ann Oncol. 2008;19:1523–1529. doi: 10.1093/annonc/mdn169. [DOI] [PubMed] [Google Scholar]
- 6.Miki T, Smith CL, Long JE, Eva A, Fleming TP. Oncogene ect2 is related to regulators of small GTP-binding proteins. Nature. 1993;362:462–465. doi: 10.1038/362462a0. [DOI] [PubMed] [Google Scholar]
- 7.Solski PA, Wilder RS, Rossman KL, Sondek J, Cox AD, Campbell SL, Der CJ. Requirement for C-terminal sequences in regulation of Ect2 guanine nucleotide exchange specificity and transformation. J Biol Chem. 2004;279:25226–25233. doi: 10.1074/jbc.M313792200. [DOI] [PubMed] [Google Scholar]
- 8.Kim JE, Billadeau DD, Chen J. The tandem BRCT domains of Ect2 are required for both negative and positive regulation of Ect2 in cytokinesis. J Biol Chem. 2005;280:5733–5739. doi: 10.1074/jbc.M409298200. [DOI] [PubMed] [Google Scholar]
- 9.Saito S, Liu XF, Kamijo K, Raziuddin R, Tatsumoto T, Okamoto I, Chen X, Lee CC, Lorenzi MV, Ohara N, Miki T. Deregulation and mislocalization of the cytokinesis regulator ECT2 activate the Rho signaling pathways leading to malignant transformation. J Biol Chem. 2004;279:7169–7179. doi: 10.1074/jbc.M306725200. [DOI] [PubMed] [Google Scholar]
- 10.Liu XF, Ohno S, Miki T. Nucleotide exchange factor ECT2 regulates epithelial cell polarity. Cell Signal. 2006;18:1604–1615. doi: 10.1016/j.cellsig.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 11.Fields AP, Justilien V. The guanine nucleotide exchange factor (GEF) Ect2 is an oncogene in human cancer. Adv Enzyme Regul. 2010;50:190–200. doi: 10.1016/j.advenzreg.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sano M, Genkai N, Yajima N, Tsuchiya N, Homma J, Tanaka R, Miki T, Yamanaka R. Expression level of ECT2 proto-oncogene correlates with prognosis in glioma patients. Oncol Rep. 2006;16:1093–1098. [PubMed] [Google Scholar]
- 13.Hirata D, Yamabuki T, Miki D, Ito T, Tsuchiya E, Fujita M, Hosokawa M, Chayama K, Nakamura Y, Daigo Y. Involvement of epithelial cell transforming sequence-2 oncoantigen in lung and esophageal cancer progression. Clin Cancer Res. 2009;15:256–266. doi: 10.1158/1078-0432.CCR-08-1672. [DOI] [PubMed] [Google Scholar]
- 14.Zhang ML, Lu S, Zhou L, Zheng SS. Correlation between ECT2 gene expression and methylation change of ECT2 promoter region in pancreatic cancer. Hepatobiliary Pancreat Dis Int. 2008;7:533–538. [PubMed] [Google Scholar]
- 15.Edge SB, Byrd DR, Compton CC, Fritz AG, Greene FL, Trotti A. AJCC cancer staging manual. 7th edition. New York, NY: Springer; 2010. [Google Scholar]
- 16.Wang WF, Li J, Du LT, Wang LL, Yang YM, Liu YM, Liu H, Zhang X, Dong ZG, Zheng GX, Wang CX. Kruppel-like factor 8 overexpression is correlated with angiogenesis and poor prognosis in gastric cancer. World J Gastroenterol. 2013;19:4309–4315. doi: 10.3748/wjg.v19.i27.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang CY, Zhao JJ, Lv L, Chen YB, Li YF, Jiang SS, Wang W, Pan K, Zheng Y, Zhao BW, Wang DD, Chen YM, Yang L, Zhou ZW, Xia JC. Decreased expression of AZGP1 is associated with poor prognosis in primary gastric cancer. PLoS One. 2013;8:e69155. doi: 10.1371/journal.pone.0069155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang YY, Ye ZY, Zhao ZS, Tao HQ, Li SG. Systems biology approach to identification of biomarkers for metastatic progression in gastric cancer. J Cancer Res Clin Oncol. 2010;136:135–141. doi: 10.1007/s00432-009-0644-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Das B, Shu X, Day GJ, Han J, Krishna UM, Falck JR, Broek D. Control of intramolecular interactions between the pleckstrin homology and Dbl homology domains of Vav and Sos1 regulates Rac binding. J Biol Chem. 2000;275:15074–15081. doi: 10.1074/jbc.M907269199. [DOI] [PubMed] [Google Scholar]
- 20.Gomez del Pulgar T, Benitah SA, Valeron PF, Espina C, Lacal JC. Rho GTPase expression in tumourigenesis: evidence for a significant link. Bioessays. 2005;27:602–613. doi: 10.1002/bies.20238. [DOI] [PubMed] [Google Scholar]
- 21.Rossman KL, Der CJ, Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol. 2005;6:167–180. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- 22.Saito S, Tatsumoto T, Lorenzi MV, Chedid M, Kapoor V, Sakata H, Rubin J, Miki T. Rho exchange factor ECT2 is induced by growth factors and regulates cytokinesis through the N-terminal cell cycle regulator-related domains. J Cell Biochem. 2003;90:819–836. doi: 10.1002/jcb.10688. [DOI] [PubMed] [Google Scholar]
- 23.Justilien V, Jameison L, Der CJ, Rossman KL, Fields AP. Oncogenic activity of Ect2 is regulated through protein kinase C iota-mediated phosphorylation. J Biol Chem. 2011;286:8149–8157. doi: 10.1074/jbc.M110.196113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hara T, Abe M, Inoue H, Yu LR, Veenstra TD, Kang YH, Lee KS, Miki T. Cytokinesis regulator ECT2 changes its conformation through phosphorylation at Thr-341 in G2/M phase. Oncogene. 2006;25:566–578. doi: 10.1038/sj.onc.1209078. [DOI] [PubMed] [Google Scholar]
- 25.Huff LP, Decristo MJ, Trembath D, Kuan PF, Yim M, Liu J, Cook DR, Miller CR, Der CJ, Cox AD. The Role of Ect2 Nuclear RhoGEF Activity in Ovarian Cancer Cell Transformation. Genes Cancer. 2013;4:460–475. doi: 10.1177/1947601913514851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang H, Yin Z, Ning K, Wang L, Guo R, Ji Z. Prognostic value of microRNA-223/epithelial cell transforming sequence 2 signaling in patients with osteosarcoma. Hum Pathol. 2014;45:1430–6. doi: 10.1016/j.humpath.2014.02.018. [DOI] [PubMed] [Google Scholar]


