Abstract
The identification of cancer-associated long non-coding RNAs and the investigation of their molecular and biological functions are vital for understanding the molecular biology and progression of cancer. The CARLo-5, a newly identified long non-coding RNA, was found to be upregulated in colon cancer. However, little is known about its role in gastric cancer. In the present study, a great upregulation of CARLo-5 was observed in gastric cancer compared to paired adjacent normal tissues. Knockdown of CARLo-5 in gastric cancer cell lines significantly inhibited the cell proliferation via inducing G0/G1 cell-cycle arrest and apoptosis. Furthermore, ERK/MAPK pathway was found to be inactivated in the gastric cells after CARLo-5 knockdown. These results indicated that CARLo-5 might serve as a pro-oncogenic lncRNA that promotes proliferation of gastric cancer and activates the ERK/MAPK pathway.
Keywords: CARLo-5, gastric cancer, lncRNA, proliferation, ERK, MAPK
Introduction
Gastric cancer (GC) is the fourth most frequent cancer and second most frequent cause of cancer-related deaths worldwide [1]. Despite recent advances in diagnostic techniques and in treatment including target therapy, there are still large numbers of GC patients with poor prognosis [2-4]. The pathogenic mechanism contributing to the aggressive biological feature in this cancer remains to be clarified.
The genome sequencing projects revealed that the human genome is comprised of less than 2% protein coding genes, and more than 90% of the genome is transcribed as noncoding RNAs (ncRNA) [5]. Long non-coding RNAs (lncRNAs) are a class of non-coding RNA transcripts longer than 200 nucleotides and are implicated in a number of important events, such as various cellular processes, development, and human diseases [6]. Increasing evidence demonstrate that lncRNAs exhibit unique profiles in various human cancers, which reflect disease progression and serve as a prognostic marker [7-9].
LncRNA CARLo-5, a recently identified long noncoding mapped to chromosome 8q24, was found to be generally upregulated in colon cancer tissues compared to their neighboring normal tissues [10]. CARLo-5 expression was significantly correlated with the rs6983267 allele, which was associated with increased cancer susceptibility [10]. Kim et al [10] demonstrated that MYC enhancer region physically interacts with the active regulatory region of the CARLo-5 promoter, suggesting that the cancer-associated variant rs6983267 in MYC enhancer could regulate CARLo-5 expression through long-range interaction with the active regulatory region of its promoter. It also reveals that CARLo-5 has a role in cell-cycle regulation and development of colon cancer [10]. However, the prognostic role of CARLo-5 in cancer is elusive and few studies have examined in detail its molecular mechanism in gastric cancer.
In the present study, we determined CARLo-5 expression pattern and its correlation with clinicopathological factors in gastric cancer patients. The oncogenic activity of CARLo-5 was investigated in gastric cancer cell lines.
Materials and methods
Human tissue specimens
All patients gave written informed consent to the study, which was approved by the Ethics Committee of Yijishan Hospital at Wannan Medical University (Anhui, China). The study methodologies conformed to the standards set by the declaration of Helsinki. Fifty-one paired GC and adjacent non-tumor gastic tissues (≥ 3 cm away from tumor) were obtained from patients who underwent resection of the primary gastric cancer at Yijishan Hospital between 2012 and 2013 and were diagnosed with GC based on histopathological evaluation. Each sample was snap-frozen in liquid nitrogen and stored at -80°C prior to RNA isolation and qRT-PCR analysis. No anti-cancer treatments were given before biopsy collection. Complete clinicopathological data of the patients from which the specimens were collected were available. No selection bias was introduced in GC samples collection for this study.
Cell lines
Three gastric cancer cell lines (SGC7901, BGC823, MGC803), and a normal gastric epithelium cell line (GES-1) were purchased from the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China). The cell lines were cultured in DMEM or RPMI 1640 (Gibco BRL), supplemented with 10% fetal bovine serum (FBS, HyClone) as well as 100 U/ml penicillin and 100 μg/ml streptomycin (Invitrogen). Cells were maintained in a humidified incubator at 37°C in the presence of 5% CO2. All cell lines have been passaged for fewer than 6 months.
RNA extraction and quantitative real-time PCR
Total RNA from tissues and cells was extracted using Trizol reagent (Invitrogen, CA) according to the manufacturer’s protocol. RNA was reverse transcribed to cDNA by a Reverse Transcription Kit (Takara, Dalian, China). The cDNA template was amplified by real-time RT-PCR using the SYBR® Premix Dimmer Eraser kit (TaKaRa, Dalian, China). The quantitative real-time polymerase chain reaction (qRT-PCR) was performed on ABI 7500 system (Applied Biosystems, CA, USA) according to the manufacturer’s instructions. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was measured as an internal control for cell lines and β-actin was measured as an internal control for paired tumor and normal tissues. The relative expression fold change of mRNAs was calculated by the 2-ΔΔCt method. The primer sequences were as follows: β-actin: 5’-GAAATCGTGCGTGACATTAA-3’ (forward), 5’-AAGGAAGGCTGGAAGAGTG-3’ (reverse); GADPH: 5’-GTCAACGGATTTGGTCTGTATT-3’ (forward), 5’-AGTCTTCTGGGTGGCAGTGAT-3’ (reverse); CARLo-5: 5’-GCCACAAATCAACAACAACAACAACAA-3’ (forward), 5’-AGAGTGATGCCAAGGCTGTTATTGTCAA-3’ (reverse). The qRT-PCR reaction was conducted under the following conditions: 95°C for 30 s, 40 cycles of 95°C for 5 s and 60°C for 60 s. For cell expression and tumor samples, each sample was run in triplicate. qRT-PCR results were analyzed and expressed relative to CT (threshold cycle) values, and then converted to fold changes.
Transfection of gastric cancer cells
The nucleotide sequences of siRNA for CARLo-5 were as follows: CARLo-5 siRNA-1 (siCARLo5-1): GGAGGGUGCUUGACAAUAAUU; CARLo-5 siRNA-2 (siCARLo5-2): GAGAAGACCAUAAGAAGAU [10]. Negative control siRNA (si-NC) were purchased from Invitrogen (Invitrogen, USA). Cells were grown on six-well plates to 75% confluence and transfected with siRNA oligonucleotides using Lipofectamine 2000 (Invitrogen, USA) according to the manufacturer’s protocol, and siRNAs were used at 50 nM final concentration. Forty-eight hours after transfection, cells were harvested for qRT-PCR or western blot analyses.
Cell viability assay
The transfected cells were seeded into 96-well plates at a density of 1 × 104 cells/well. Cell proliferation was measured using MTT assay. The cells were incubated in 0.1 mg/ml MTT at 37°C for 4 h and lysed in dimethyl sulfoxide (DMSO) at room temperature for 10 min. The absorbance in each well was measured by a microplate reader (Bio-Rad, Hercules, CA, USA). Cell proliferation was documented every 24 h.
Colony formation assay
1000 of transfected cells were placed in each well of 6-well plates and maintained in proper media containing 10% FBS for two weeks, during which the medium was replaced every 4 days. Colonies were then fixed with methanol and stained with 0.1% crystal violet (Sigma) in PBS for 15 minutes. Colony formation was determined by counting the number of stained colonies.
Flow cytometric analysis
Transfected Gastric cells were plated in 6-well plates. After 48 h incubation, the cultures were incubated with propidium iodide (PI) for 30 min in the dark. Cultures were collected and analyzed for cell cycle using a flow cytometer (FACSCalibur, BD Biosciences) after PI staining. The cultures were also stained with annexin V-fluorescein isothiocyanate, and the cell apoptosis was analyzed using a flow cytometer.
Nuclear stain
Transfected cells were allowed to adhere on slide glasses overnight. Then, they were washed with PBS three times and fixed with ethanol for 10 min. After being air dried, cells were washed with PBS and stained with Hoechst 33258 (1 mg/mL) for 10 min. The chromatin structures of the cells were examined by fluorescence microscopy.
Western blot analysis
To determine the expressed protein levels, transfected cells were harvested from 6-well culture plates, lysed in RIPA lysis buffer (Cell Signaling Technology) supplemented with protease inhibitors (Roche) on ice. The lysates were then collected and subjected to ultrasonication and centrifugation at 12000 rpm for 10 min. The supernatants were collected, and protein content was determined by Bradford assay. Total proteins (30-50 μg) were resolved using 10% SDS-PAGE separating gel (Bio-Rad Laboratories, CA) and blotted onto a PVDF Immobilon-P membrane (Millipore). The PVDF membranes were then blocked with 10% non-fat powdered milk in Tris-buffered saline Solution with Tween (TBS-T) at room temperature for 2 h and incubated overnight with primary antibody at 4°C overnight: anti-p16 (1:1000), anti-p21 (1:1000), anti-p27 (1:1000), anti-Bax (1:1000), anti-Bcl-2 (1:1000), anti-caspase-3 (1:1000), anti-p-ERK (1:1000), anti-ERK (1:1000), anti-p-p38 (1:1000), and anti-tubulin (1:2000). All the primary antibodies were purchased from Abcam. After three 5-min washes in TBS-T, the membranes were incubated with horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG antibody (1:2000; Abcam) for 4 h at room temperature and then washed again in TBS-T and visualized with BeyoECL Plus kit. All experiments were performed in triplicate.
Statistical analysis
The statistical analysis was performed with SPSS 17.0 software. The data are presented as the means ± standard deviation (S.D.). Continuous variables were compared by Student’s t test or the ANOVA test. If the test result of the variance homogeneity between the groups was significant, the Mann-Whitney test was appropriately adopted. In samples with small size (n < 30) and with non-normal distribution and/or elevated dispersion, we also used non-parametric statistics. Two-tailed P-values less than 0.05 were considered statistically significant.
Results
Expression levels of CARLo-5 in GC cell lines
To explore whether CARLo-5 was upregulated in GC, we first determined its expression in diverse GC cell lines with RT-PCR and normalized to GADPH. When normalized to normal gastric epithelium cell line (GES-1), three cell lines (BGC-823, MGC-803, and SGC-7901) expressed higher levels of CARLo-5 (Figure 1A).
Figure 1.
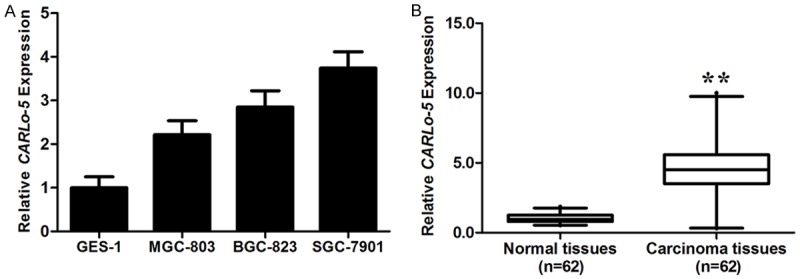
CARLo-5 expression in GC cell lines, cancer tissues and its clinical significance. A. qRT-PCR analysis of CARLo-5 expression levels in gastric cancer cell lines (BGC-823, MGC-803, and SGC-7901) compared with the normal gastric epithelium cell line (GES-1). Data represent the mean ± S.D. from three independent experiments. B. Difference in expression levels of CARLo-5 expression levels between gastric cancer tissues and matched non-tumor tissues. The expression of CARLo-5 was normalized to β-actin. The statistical differences between samples were analyzed with paired samples t-test (n = 51, P < 0.0001). Horizontal lines in the box plots represent the medians, the boxes represent the interquartile range, and the whiskers represent the 2.5th and 97.5th percentiles. The relative expression fold change of mRNAs was calculated by the 2-ΔΔCt method. *, P < 0.05; **, P < 0.01.
Expression of CARLo-5 is upregulated in GC tissues
The expression level of CARLo-5 was detected in 51 paired GC tissues and adjacent normal tissues by RT-PCR, and normalized to β-actin. CARLo-5 was significantly upregulated in cancer tissues compared with normal counterparts (P < 0.01, Figure 1B).
In order to further understand the significance of CARLo-5 overexpression in GC, we determined the potential associations between CARLo-5 expression and patients’ clinicopathological features. The detailed relationships between CARLo-5 expression status and clinicopathological variables of 51 patients were summarized in Table 1. It is worth noting that, high CARLo-5 expression was significantly correlated with tumor size (P = 0.004) and advanced TNM stage (P = 0.012). However, CARLo-5 expression level was not correlated with other parameters such as patient’s gender and tumor position (Table 1).
Table 1.
Correlation between CARLo-5 expression and clinicopathological characteristics of gastric cancer
| Clinical parameters | Number of patients (%) | Average Fold changea | P-valueb |
|---|---|---|---|
| Age (years) | |||
| < 50 | 24 (47.06%) | 4.81 | 0.276 |
| > 50 | 27 (52.94%) | 4.35 | |
| Gender | |||
| Male | 26 (50.98%) | 5.08 | 0.297 |
| Female | 25 (49.02%) | 4.52 | |
| Size | |||
| > 5 cm | 23 (45.10%) | 6.74 | 0.004 |
| < 5 cm | 28 (54.90%) | 3.78 | |
| Histologic differentiation | |||
| well/moderately | 35 (68.63%) | 4.33 | 0.134 |
| poor | 16 (31.37%) | 4.97 | |
| TNM stage | |||
| I/II | 30 (58.82%) | 4.03 | 0.012 |
| III/IV | 21 (41.18%) | 5.25 | |
| Invasion depth | |||
| T1/T2 | 24 (47.06%) | 4.26 | 0.121 |
| T3/T4 | 27 (52.94%) | 4.89 | |
| Lymph node metastasis | |||
| Yes | 25 (49.02%) | 4.91 | 0.567 |
| No | 26 (50.98%) | 4.54 | |
| Distant metastasis | |||
| Yes | 7 (13.73%) | 4.53 | 0.786 |
| No | 44 (86.27%) | 4.75 |
Median of relative expression;
P < 0.05 was considered significant (Mann-Whitney U test).
Manipulation of CARLo-5 expression via small-interference RNA (siRNA) in GC cells
To further explore the molecular mechanism of CARLo-5 in GC, we inhibited the expression level of CARLo-5 in GC cell lines. As illustrated in Figure 1A, SGC-7901 cells harbored higher expression levels of CARLo-5. Then, CARLo-5 siRNAs were transfected into SGC-7901 cells. To avoid off-target effects, we designed two siRNAs targeting different regions of CARLo-5. RT-PCR revealed that the expression level of CARLo-5 was significantly reduced in SGC-7901 cells (Figure 2A). As shown in Figure 2A, CARLo-5 was most efficiently inhibited by siCARLo-2, the most effective siRNA. Thus, siCARLo-2 was used in subsequent experiments.
Figure 2.
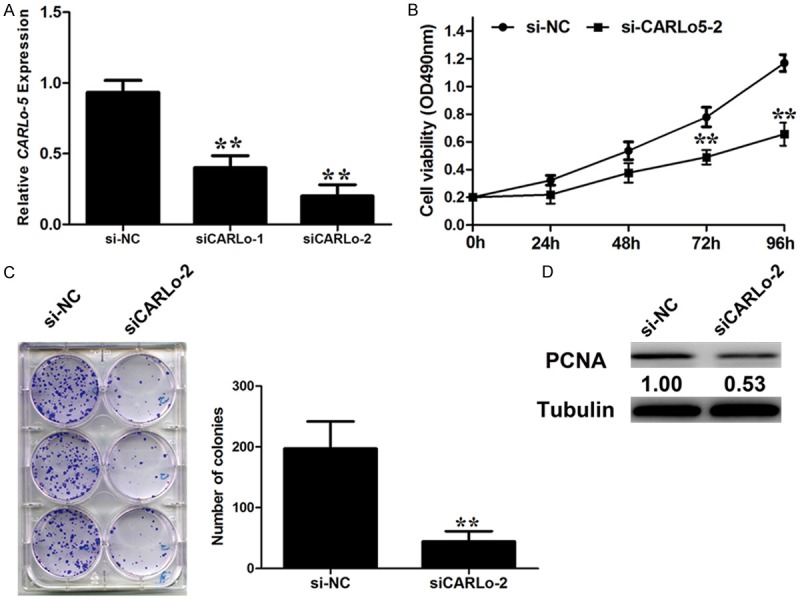
Effect of CARLo-5 on cell proliferation. A. qRT-PCR analysis of CARLo-5 expression following treatment of SGC-7901 cells with two specific siRNAs targeting CARLo-5. B. SGC-7901 cells were transfected with si-CARLo-2 or si-NC. MTT assays were performed to determine the proliferation of SGC-7901 cells. C. Changes in the colony-formation ability were shown after si-CARLo-2 transfection. D. Changes in the proliferation marker, PCNA, were shown by western blotting analysis and normalized to Tubulin after si-CARLo-2 transfection. Data represent the mean ± S.D. from three independent experiments. *, P < 0.05; **, P < 0.01.
Knockdown of CARLo-5 inhibits of GC cell proliferation in vitro
Kim et al [10] demonstrated that CARLo-5 promoted colon cancer cells proliferation. Then, we explored effects of CARLo-5 on the biological behaviors on GC cells. MTT assay showed that knockdown of CARLo-5 expression significantly inhibited the cell proliferation in SGC-7901 cells compared to the control cells (Figure 2B). Similarly, colony formation assay revealed inhibition of CARLo-5 markedly decreased the clonogenic survival in SGC-7901 cells (Figure 2C). Consistent with the proliferation promotion effects of CARLo-5 on GC cells, western blot analysis showed the expression of proliferating cell nuclear antigen (PCNA) was greatly attenuated in SGC-7901 transfected with siCARLo-2 compared to cells transfected with si-NC (Figure 2D). These data suggest that CARLo-5 may play a role in the cell proliferation.
Knockdown of CARLo-5 inhibits GC cell proliferation via inducing G0/G1 arrest
Flow cytometric analysis was performed to further explore whether the effect of CARLo-5 on proliferation of GC cells was via altering cell-cycle progression. The data showed that the cell-cycle progression of siCARLo-2 transfected cells was significantly stalled at G0-G1 phase compared to cells transfected with si-NC (Figure 3A). To explore the mechanism of CARLo-5’s role in G0/G1 arrest, we determined the alteration in the expression levels of G0/G1 arrest markers, p27, p21 and p16 by western blot. The results showed that p27, p21 and p16 protein expression levels were increased with inhibition of CARLo-5 expression (Figure 3B).
Figure 3.
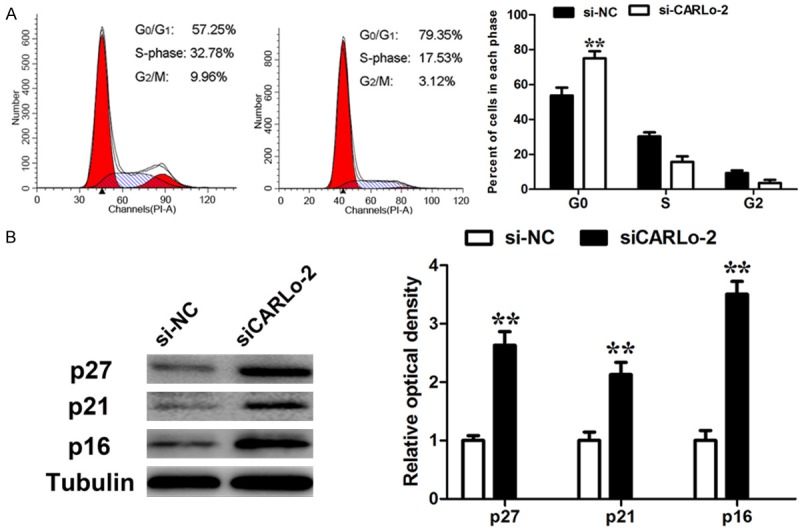
Effect of CARLo-5 on cell cycle. A. Cell cycle analysis determined the relative cell numbers in each cell-cycle phase after propidium iodide staining of CARLo-5-downregulated SGC-7901 cells. Numbers inside bars represent percentages of cells in each phase. Data represent the mean ± S.D. from three independent experiments. B. SGC-7901 cells, treated as described in Figure 3A, were collected for western blotting analysis of the G0/G1 arrest markers, p16, p21, and p27. Relative protein expression was identified (n = 3). *, P < 0.05; **, P < 0.01.
Knockdown of CARLo-5 inhibits GC cell proliferation through inducing apoptosis
Flow cytometric analysis was performed to examine whether the anti-proliferative effects of CARLo-5 knockdown could possibly be through inducing GC cell apoptosis. Knockdown of CARLo-5 expression could obviously induce cell apoptosis (Figure 4A). In addition, nuclear staining with the Hoechst 33258 stain revealed characterized features of late apoptosis, including condensation and fragmentation of nuclei in the siCARLo-2 transfected cells (Figure 4B). To explore the underlying mechanism of CARLo-5’s role in cell apoptosis, we determined the expression of pro-apoptotic factor, Bax, Bcl and caspase-3 by western blot. CARLo-5 knockdown increased the expression levels of caspase-3 (Figure 5A) and Bax/Bcl-2 ratio (Figure 5B). These results demonstrated that CARLo-5 knockdown may inhibit the proliferation of GC cells through inducing cell G0/G1 arrest and apoptosis.
Figure 4.
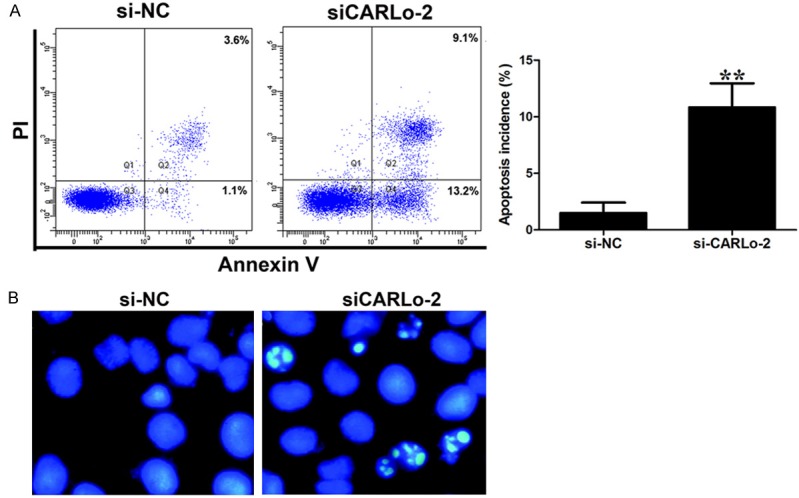
Effect of CARLo-5 on cell apoptosis. A. Annexin V/PI staining and flow cytometry analysis assessing apoptosis in SGC-7901 cells after si-CARLo-2 transfection. Data represent the mean ± S.D. from three independent experiments. *, P < 0.05; **, P < 0.01. B. Nuclear staining with the Hoechst 33258 stain revealed characterized features of late apoptosis, including condensation and fragmentation of nuclei in the siCARLo-2 transfected cells.
Figure 5.
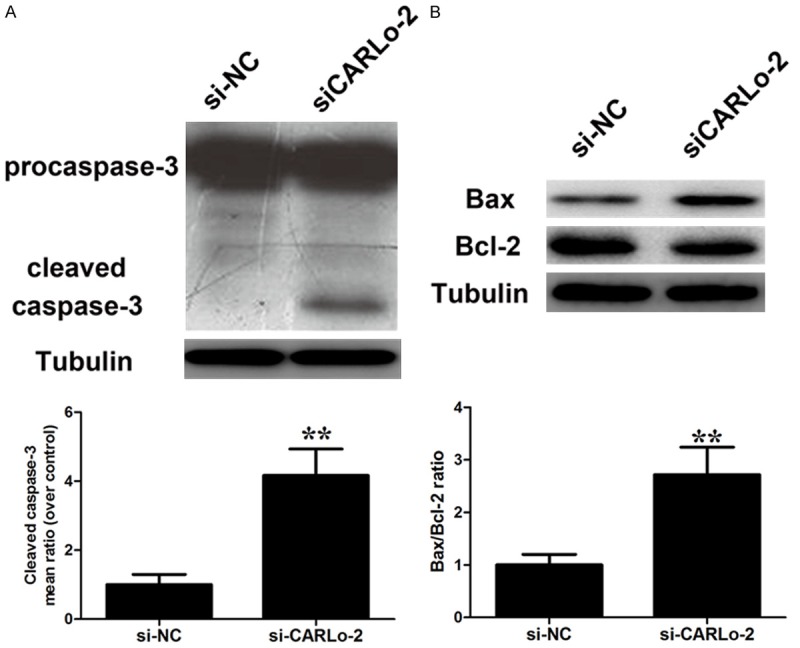
Effect of CARLo-5 on cell apoptosis. Changes in pro-apoptotic factor caspase-3 activation (A) and Bax/Bcl-2 ratio (B) were shown by western blotting analysis and normalized to Tubulin after si-CARLo-2 transfection. Data represent the mean ± S.D. from three independent experiments. *, P < 0.05; **, P < 0.01.
CARLo-5 downregulation may exert its anti-proliferative effect through inactivation of ERK/MAPK pathway
To further define the underlying molecular mechanism modulated by CARLo-5’s in the growth and apoptosis of GC cells, we focused on the ERK/MAPK pathway. Knockdown of CARLo-5 resulted in the decrease of phosphorylation of ERK and MARK (p38) (Figure 6A).
Figure 6.
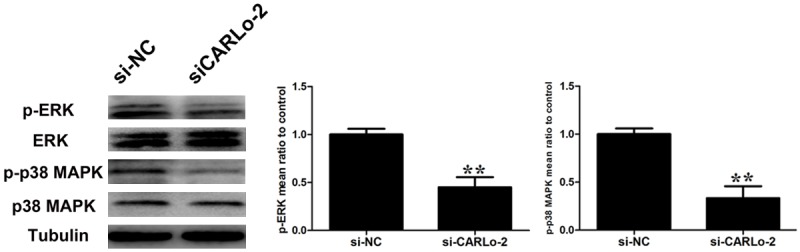
The effects of CARLo-5 downregulation on ERK/MAPK pathway. Changes in ERK and p38 MAPK activation were shown by western blotting analysis after si-CARLo-2 transfection. Data represent the mean ± S.D. from three independent experiments. *, P < 0.05; **, P < 0.01.
Discussion
Recent advances in high-throughput gene sequencing analysis have improved our understanding that < 2% of human genome can be transcribed, yielding many short or long noncoding RNAs with limited or no protein-coding capacity [5]. MicroRNAs (miRNAs) are a well-described class of non-coding RNAs. They are ~22-nucleotides long and act as negative regulators of gene expression by inhibiting mRNA translation or promoting mRNA degradation [11,12]. miRNAs are found to play a vital role in gastric cancer [13-15]. Long non-coding RNAs (lncRNAs) are a class of non-coding RNA transcripts longer than 200 nucleotides and are implicated in a number of important events, such as various cellular processes, development, and human diseases [6]. LncRNAs regulate the expression of genes at the epigenetic, transcriptional and post-transcriptional levels, and play an important role in physiological processes [16]. Recent studies have demonstrated that lncRNAs are important players in the development of gastric cancer [17-19]. Yet only a quite small number of lncRNAs have been characterized and their role in cancer remains to be explored.
CARLo-5 was first identified as an upregulated lncRNA in colon cancer, wherein CARLo-5 promotes tumor proliferation [10]. As CARLo-5 plays an important role in colon cancer progression, we investigated the biological role of CARLo-5 in GC development and studied its clinical significance. In our present study, we found that the average levels of CARLo-5 were markedly higher in GC tissues compared to those in paired non-tumor tissues. The high expression level of CARLo-5 in gastric cancer was correlated with tumor size and advanced TNM stage. It suggests that CARLo-5 might exhibit oncogenic activity and play an important part in GC development.
We demonstrated that CARLo-5 knockdown resulted in a significant reduction in GC cell proliferation. To explore the molecular mechanism modulated by CARLo-5 in the proliferation of GC cells, we examined the effects of CARLo-5 on the cell-cycle progression and apoptosis. Flow cytometric analysis revealed that CARLo-5 knockdown induced G0/G1 cell-cycle arrest and cell apoptosis. Western blot analysis of G0/G1 arrest markers and pro-apoptotic factors confirmed the results. It suggests that CARLo-5 knockdown may inhibit GC cell growth through inducing G0/G1 arrest and apoptosis.
The MAPK pathway coordinately regulates diverse cellular processes, such as proliferation, apoptosis and differentiation [20]. In this study, we showed that CARLo-5 knockdown resulted in significant decrease in the expression of p-ERK and MAPK (p38). However, CARLo-5 downregulation almost had no effect on the activation of AKT.
In summary, we demonstrate that the expression of CARLo-5 was significantly upregulated in GC tissues. We also showed that CARLo-5 promoted the proliferation of GC cells, suggesting that CARLo-5 may play a vital role in GC development. Our study may add our understanding to the molecular mechanisms through which CARLo-5 contributes to the tumor progression, which may facilitate the development of lncRNA-directed diagnostics and therapeutics against cancers.
Disclosure of conflict of interest
None.
References
- 1.Shah MA, Kelsen DP. Gastric cancer: a primer on the epidemiology and biology of the disease and an overview of the medical management of advanced disease. J Natl Compr Canc Netw. 2010;8:437–447. doi: 10.6004/jnccn.2010.0033. [DOI] [PubMed] [Google Scholar]
- 2.Allum WH, Blazeby JM, Griffin SM, Cunningham D, Jankowski JA, Wong R Association of Upper Gastrointestinal Surgeons of Great Britain and Ireland, the British Society of Gastroenterology and the British Association of Surgical Oncology. Guidelines for the management of oesophageal and gastric cancer. Gut. 2011;60:1449–1472. doi: 10.1136/gut.2010.228254. [DOI] [PubMed] [Google Scholar]
- 3.Saka M, Morita S, Fukagawa T, Katai H. Present and future status of gastric cancer surgery. Jpn J Clin Oncol. 2011;41:307–313. doi: 10.1093/jjco/hyq240. [DOI] [PubMed] [Google Scholar]
- 4.Tan HT, Low J, Lim SG, Chung MC. Serum autoantibodies as biomarkers for early cancer detection. FEBS J. 2009;276:6880–6904. doi: 10.1111/j.1742-4658.2009.07396.x. [DOI] [PubMed] [Google Scholar]
- 5.Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339:159–166. doi: 10.1016/j.canlet.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 6.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 7.Ge XS, Ma HJ, Zheng XH, Ruan HL, Liao XY, Xue WQ, Chen YB, Zhang Y, Jia WH. HOTAIR, a prognostic factor in esophageal squamous cell carcinoma, inhibits WIF-1 expression and activates Wnt pathway. Cancer Sci. 2013;104:1675–1682. doi: 10.1111/cas.12296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang JF, Guo YJ, Zhao CX, Yuan SX, Wang Y, Tang GN, Zhou WP, Sun SH. Hepatitis B virus X protein (HBx)-related long noncoding RNA (lncRNA) down-regulated expression by HBx (Dreh) inhibits hepatocellular carcinoma metastasis by targeting the intermediate filament protein vimentin. Hepatology. 2013;57:1882–1892. doi: 10.1002/hep.26195. [DOI] [PubMed] [Google Scholar]
- 9.Yang F, Zhang L, Huo XS, Yuan JH, Xu D, Yuan SX, Zhu N, Zhou WP, Yang GS, Wang YZ, Shang JL, Gao CF, Zhang FR, Wang F, Sun SH. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology. 2011;54:1679–1689. doi: 10.1002/hep.24563. [DOI] [PubMed] [Google Scholar]
- 10.Kim T, Cui R, Jeon YJ, Lee JH, Lee JH, Sim H, Park JK, Fadda P, Tili E, Nakanishi H, Huh MI, Kim SH, Cho JH, Sung BH, Peng Y, Lee TJ, Luo Z, Sun HL, Wei H, Alder H, Oh JS, Shim KS, Ko SB, Croce CM. Long-range interaction and correlation between MYC enhancer and oncogenic long noncoding RNA CARLo-5. Proc Natl Acad Sci U S A. 2014;111:4173–8. doi: 10.1073/pnas.1400350111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Palanichamy JK, Rao DS. miRNA dysregulation in cancer: towards a mechanistic understanding. Front Genet. 2014;5:54. doi: 10.3389/fgene.2014.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bouyssou JM, Manier S, Huynh D, Issa S, Roccaro AM, Ghobrial IM. Regulation of microRNAs in cancer metastasis. Biochim Biophys Acta. 2014;1845:255–265. doi: 10.1016/j.bbcan.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu Q, Yang Z, An Y, Hu H, Yin J, Zhang P, Nie Y, Wu K, Shi Y, Fan D. MiR-19a/b modulate the metastasis of gastric cancer cells by targeting the tumour suppressor MXD1. Cell Death Dis. 2014;5:e1144. doi: 10.1038/cddis.2014.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma J, Liu J, Wang Z, Gu X, Fan Y, Zhang W, Xu L, Zhang J, Cai D. NF-kappaB-dependent microRNA-425 upregulation promotes gastric cancer cell growth by targeting PTEN upon IL-1β induction. Mol Cancer. 2014;13:40. doi: 10.1186/1476-4598-13-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He XP, Shao Y, Li XL, Xu W, Chen GS, Sun HH, Xu HC, Xu X, Tang D, Zheng XF, Xue YP, Huang GC, Sun WH. Downregulation of miR-101 in gastric cancer correlates with cyclooxygenase-2 overexpression and tumor growth. FEBS J. 2012;279:4201–4212. doi: 10.1111/febs.12013. [DOI] [PubMed] [Google Scholar]
- 16.Krishnan J, Mishra RK. Emerging trends of long non-coding RNAs in gene activation. FEBS J. 2014;281:34–45. doi: 10.1111/febs.12578. [DOI] [PubMed] [Google Scholar]
- 17.Yang F, Xue X, Zheng L, Bi J, Zhou Y, Zhi K, Gu Y, Fang G. Long non-coding RNA GHET1 promotes gastric carcinoma cell proliferation by increasing c-Myc mRNA stability. FEBS J. 2014;281:802–813. doi: 10.1111/febs.12625. [DOI] [PubMed] [Google Scholar]
- 18.Yang F, Bi J, Xue X, Zheng L, Zhi K, Hua J, Fang G. Up-regulated long non-coding RNA H19 contributes to proliferation of gastric cancer cells. FEBS J. 2012;279:3159–3165. doi: 10.1111/j.1742-4658.2012.08694.x. [DOI] [PubMed] [Google Scholar]
- 19.Sun M, Xia R, Jin F, Xu T, Liu Z, De W, Liu X. Downregulated long noncoding RNA MEG3 is associated with poor prognosis and promotes cell proliferation in gastric cancer. Tumour Biol. 2014;35:1065–1073. doi: 10.1007/s13277-013-1142-z. [DOI] [PubMed] [Google Scholar]
- 20.Koul HK, Pal M, Koul S. Role of p38 MAP Kinase Signal Transduction in Solid Tumors. Genes Cancer. 2013;4:342–359. doi: 10.1177/1947601913507951. [DOI] [PMC free article] [PubMed] [Google Scholar]


