Abstract
MicroRNAs (miRNAs) are small noncoding RNA molecules that regulate gene expression at the post transcriptional level. Compelling evidence shows that there are causative links between miRNAs deregulation and cancer development and progression. In this study, we demonstrated that miR-584 was downregulated in human glioma and could suppress growth of the human glioma cell line U87-MG and U251-MG. Bioinformatics analysis indicated that PTTG1IP was a putative target of miR-584. In a Luciferase reporter system, we confirmed that PTTG1IP was a direct target gene of miR-584. These findings indicate that miR-584 suppresses glioma cell growth by negatively regulating the expression of PTTG1IP, suggesting that miR-584 has a tumor suppressive role in human glioma pathogenesis.
Keywords: Glioma, microRNA, miR-584, PTTG1IP
Introduction
Gliomas are the most common malignant primary brain tumors in adults and exhibit a spectrum of aberrantly aggressive phenotype. A combination of surgery, radiotherapy and chemotherapy is widely used to treat gliomas, particularly malignant gliomas. However, the prognosis of the disease remains poor, with a median survival in the range of 15-17 months [1-4]. Therefore, it is crucial to investigate the mechanism involved in the development and progression of glioma and to find new therapeutic targets.
MicroRNAs (miRNAs) are a class of small (~22 nucleotides) non-coding RNAs that function as negative regulators of gene expression at the post-transcriptional level by binding to complimentary sequences in, mainly, the 3’-untranslated regions (3’-UTRs) of specific mRNAs [5-7]. Abnormal expression and the loss of the dynamic balance between oncogenes and tumor suppressor genes typically lead to tumor formation and the development of cancer [8-12]. miRNAs have been shown as important regulators in diverse biological processes of cancer, such as cell proliferation [13], angiogenesis [12], cell differentiation [14], cell apoptosis [15], adhesion, and metastasis [16]. These data emphasize the importance of miRNAs in cancer development and provide new insights into understanding the molecular mechanism of tumorigenesis. In gliomas, various miRNAs such as miR-10b [17], miR-23a [18], miR-92b [19]and miR-17 [20], have been associated with the initiation and progression of glioblastoma and with their invasive nature. In contrast, miR-214 [15], miR-16-1 [21], miR-204 [22] and miR-145 [23] have been implicated as tumor suppressor miRNAs in these tumors. However, the exact role of miR-584 in gliomas has not been revealed yet.
In this study, we focused on the downregulated miR-584 in glial tumors and glioma cell lines, U87-MG and U251-MG. Further investigation revealed that in glioma cell lines miRNA-584 functioned as a tumor suppressor and overexpression of miRNA-584 reduced cell proliferation, decreased cell invasive capacity and increased apoptosis. The PTTG1IP gene predicted by bioinformatics analysis as a target gene of the miR-584, was validated by fluorescent reporter assay. These findings suggested that miR-584 could act as a biomarker in glioma and its restoration might be a possible therapeutic approach.
Materials and methods
Clinical specimen and RNA isolation
Tissue specimens and clinical information were obtained as part of an approved study by the Institutional Review Board at the Tianjin Medical University, China. Thirteen human glioma tissues were collected with patient consent at the time of operation, grading of tumors was carried out with WHO criteria (World Health Organization, 2007). The clinical information of patients was shown in Table 1. The matched normal tissue was taken from the distal end of the operative excisions, far from the tumor. Immediately after surgery, samples were snap-frozen and stored in liquid nitrogen. Large and small RNAs were isolated from tissue samples with the mirVana miRNA Isolation Kit (Ambion, USA), according to the manufacturer’s instructions.
Table 1.
The Information of Clinical Patients
| Index | Gender | Age | Clinical diagnosis (WHO) |
|---|---|---|---|
| 1 | Male | 51 | I |
| 2 | Female | 43 | I |
| 3 | Female | 46 | I |
| 4 | Male | 67 | I |
| 5 | Female | 58 | I |
| 6 | Male | 67 | II |
| 7 | Male | 73 | II |
| 8 | Female | 51 | II |
| 9 | Female | 87 | II |
| 10 | Male | 55 | III |
| 11 | Male | 49 | III |
| 12 | Female | 67 | III |
| 13 | Female | 61 | IV |
Cell culture and transfection
Two human glioma cell line U87-MG and U251-MG were maintained in Dulbecco’s modified Eagle’s medium (Gibco, USA), and supplemented with 10% fetal bovine serum, 100 IU•mL-1 penicillin, and 100 μg•mL-1 streptomycin. Cells were incubated at 37°C in a humidified chamber supplemented with 5% CO2. The transfection was performed using the Lipofectamine 2000 Reagent (Invitrogen, USA) following the manufacturer’s instructions.
All the RNA oligonucleotides were purchased from GenePharme (Shanghai, China).
Cell viability and proliferative capacity assay
To determine cell viability and proliferative capacity, cells were examined using the 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) and colony formation assays as described previously. U87-MG and U251-MG cells were seeded in 96-well plates at a density of 5000 cells per well, and then transfected with miR-584 mimics or miR-control on the next day. The MTT assay was used to determine relative cell viability at 0, 12, 24 and 48 h. Ten microliters of MTT (at a final concentration of 0.5 mg/ml) solution was added to 100 μl of culture medium, and incubated for 4 h at 37°C; the absorbance at 570 nm (A570) was then measured using an uQuant Universal Microplate Spectrophotometer (Bio-Tek Instruments, USA).
For the colony formation assay, the number of viable cell colonies was determined after 15 days after inoculation of 150 cells/well in triplicate in 12-well plates. The cells were stained with crystal violet. The rate of colony formation was calculated with the equation: colony formation rate = (number of colonies/number of seeded cells) ×100%
Cell apoptosis assay
The apoptotic ratios of cells were determined with the Annexin V-7-ADD apoptosis detection kit (Roche, Switzerland). Briefly, 48 hours after transfection, the cells were collected and washed twice with cold PBS buffer, resuspended in 200 μl of binding buffer, incubated with 20 μl of Annexin-V-R-PE for 20 minutes in an dark ice bath, and then 7-AAD 10 μl before analyzing by flow cytometry. Cells treated with DMSO were used as the negative control.
Western blot analysis
Total cellular extracts were extracted using RIPA buffer. Proteins were separated by 10% SDS denatured polyacrylamide gel, and then transferred onto a nitrocellulose membrane. Membranes were incubated with an antibody against PTTG1IP or an antibody against glyceraldehyde-3-phosphate dehydrogenase (GAPDH) overnight at 4°C. The membranes were washed and incubated with horseradish peroxidase-conjugated secondary antibody. Protein expression was assessed by enhanced chemiluminescence and exposure to chemiluminescent film. Lab works image acquisition and analysis software was used to quantify band intensities. Antibodies were purchased from Abcam (Cambridge, UK).
Target prediction and Luciferase reporter assays
Based on bioinformatic prediction (TargetScan, RNA22 and microrna. org), PTTG1IP was selected as candidate target of miR-584. The 3’UTR segments of PTTG1IP containing putative binding sites for miR-584 were obtained by PCR and inserted into pmirGLO vector. The wild-type reporter construct pmirGLO/PTTG1IP-3’UTR and the mutant reporter construct pmirGLO/PTTG1IP-3’UTR mut, in which the site of perfect complementarity to miR-584 was mutated using site-directed mutagenesis PCR, were used for miRNA functional analysis. Wild-type and mutant insertions were confirmed by DNA sequencing. All primer information is available in Table 2. For Luciferase reporter experiments, U87-MG cells were co-transfected with the miR-584 mimics in a 48-well plate followed by the pmirGLO/PTTG1IP-3’UTR reporter vector or the pmirGLO/PTTG1IP-3’UTR mut. Firefly luciferase and Renilla luciferase levels were measured at 48 h after transfection. Each experiment was repeated at least three times.
Table 2.
The primers used in this study
| Name | Sequence |
|---|---|
| U6 RT | 5’-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAAATATGGAAC-3’ |
| U6 forward | 5’-TGCGGGTGCTCGCTTCGGCAGC-3’ |
| miR-584 mimics | 5’-UUAUGGUUUGCCUGGGACUGAG-3’ |
| miR-584 control | 5’-AAACGGUUUAAAUAGGCUCCCCG-3’ |
| GAPDH forward | 5’-CGTGACATTAAGGAGAAGCTG-3’ |
| GAPDH reverse | 5’-CTAGAAGCATTTGCGGTGGAC-3’ |
Real-time quantitative PCR
Small RNA (5 μg) was reverse transcribed into cDNA using M-MLV reverse transcriptase (Promega, USA) with the specific primers. The cDNA was used as template to amplify either mature miR-584 or an endogenous control U6 snRNA by PCR. The PCR was performed as follows: 94°C for 3 min, followed by 40 cycles of 94°C for 30 s, 50°C for 30 s and 72°C for 30 s. The real-time PCR was performed using SYBR Premix Ex Taq (TaKaRa, Japan) on the iQ5 Real-Time PCR Detection system (Bio-Rad). The relative expression of miR-584 was defined as follows: quantity of miR-424/quantity of U6 within the same sample. Briefly, a cDNA library was generated through reverse transcription using M-MLV reverse transcriptase (Promega) with large RNA (5 μg). The cDNA was used to amplify the genes PTTG1IP and the β-actin gene, which served as an endogenous control. The PCR was performed as follows: 94°C for 3 min, followed by 40 cycles of 94°C for 30 s, 58°C for 30 s and 72°C for 30 s. Real-time PCR was performed as described above, and the relative gene expression level was defined as follows: quantity of gene/quantity of β-actin within the same sample.
Statistical analysis
Data are expressed as the means ± standard deviation (SD), and P ≤ 0.05 is considered to be statistically significant using the Students-Newman-Keuls test.
Results
Depressed expression of miR-584 in glial tumors, glioma cell lines
To determine the expression of miR-584 in human glioma tissues and adjacent normal tissues, we used quantitative real time RT-PCR to detect 13 pairs of glioma samples (Figure 1A). Furthermore, miR-584 expression was significantly low in glioma cell line U87-MG and U251-MG by real-time RT-PCR (Figure 1B). It was shown that miR-584 expression level was generally and significantly downregulated in glioma.
Figure 1.
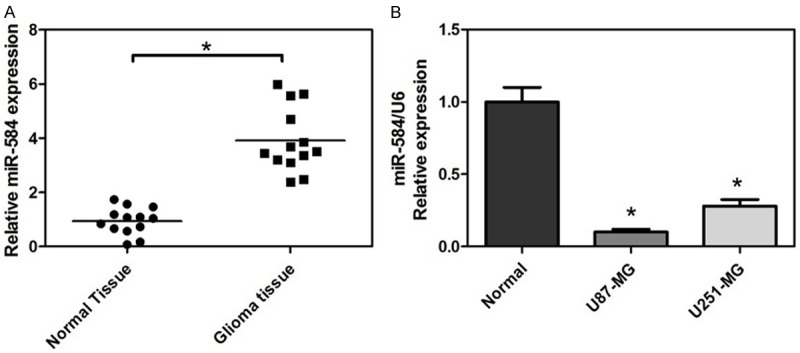
Depressed expression of miR-584 in glial tumors, glioma cell lines. A. Endogenous level of miR-584 in glioma samples and normal brain tissues by real-time qRT-PCR by qRT-PCR. B. The relative abundance of miR-584 in glioma cell lines, U87-MG and U251-MG, and the normal glial cell line. The expression of miR-584 is normalized to U6 small nuclear RNA (*P < 0.05).
Overexpression of miR-584 suppresses cell growth and promotes apoptosis of the glioma cell lines
First, we transfected either miR-584 mimics or an miR-control into U87-MG and U251-MG cells, and detected miR-584 levels by real time RT-PCR. Expression of miR-584 was increased 12 fold in the U87-MG cells and 13 fold in U251-MG cells transfected with miR-584 mimics as compared with controls (Figure 2A). Cell viability of glioma cells transfected with miR-584 mimics was evaluated with the 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyl-tetrazolium bromide (MTT) assay; miR-584 mimics reduced cell viability at 12, 24, 48 or 72 h after transfection (Figure 2B and 2C). In parallel, we analyzed colony formation and cellular proliferation to assess the effect of miR-584 on the proliferative capacity of glioma cells. The colony formation rate of U87-MG and U251-MG cells transfected with miR-584 mimics, meanwhile, were significantly lower than that of the control group (Figure 2D and 2E). The fluorescence activated cell sorting (FACS) analysis showed that the forced expression of miR-584 lead to glioma cell apoptosis. The percentage of total apoptotic cells (early apoptotic + late apoptotic) significantly increased in response to miR-584 overexpression compared with miR-control overexpression in U87-MG and U251-MG cells (Figure 2F and 2G). These results indicated that miR-584 suppressed the ability to proliferate and promoted apoptosis in glioma cells.
Figure 2.
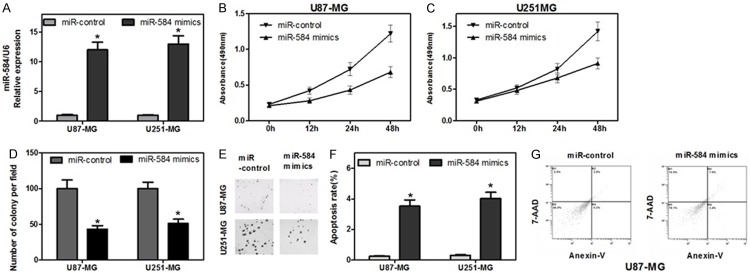
Overexpression of miR-584 suppresses cell growth and Promotes Apoptosis of the glioma cell lines. A. Measurement of miR-584 expression levels by real-time RT-PCR. Small RNA was extracted from U87-MG and U251-MG cells transfected with miR-584 mimics or miR-control, and U6 snRNA served as an endogenous normalize. The relative miR-584 expression level (mean ± SD) is shown (*P < 0.05). B and C. Cell viability was detected through MTT assay. After U87-MG and U251-MG cells were transfected with the miR-584 mimics or miR-control, the MTT assay was used to determine the relative cell growth activity at 0, 12, 24 h and 48 h post-transfection (*P < 0.05). The relative cell growth activity was normalized to the growth activity of U87-MG and U251-MG cells in the control groups. D and E. Cell proliferation ability was evaluated by a colony formation assay. U87-MG and U251-MG cells transfected with miR-584 mimics or miR-control were seeded in 12-well plates. On the 8th day after seeding, the number of colonies was counted (*P < 0.05). F and G. Apoptosis of U87-MG and U251-MG cells following miR-584 mimics transfection was analyzed by flow cytometry. The cells were stained with annexin V-fluorescein isothiocyanate and counterstained with 7-ADD (*P < 0.05).
MiR-584 decreased the Invasion and migration abilities of glioma cells
Cell migration and invasion are two essential processes of cancer metastasis. Thus, we first examined the invasive ability of glioma cells transfected with miR-584 and found that cells transfected with miR-584 presented less invasion ability than those with control miRNA (Figure 3A and 3B). There was clear difference in the migration ability between glioma cells transfected with miR-584 and those with miR-control, revealing that glioma cells transfected with miR-584 may close the wound more slowly when compared with those with miR-control (Figure 3C-E). These data demonstrated that miR-584 might inhibit migration and invasion in glioma.
Figure 3.
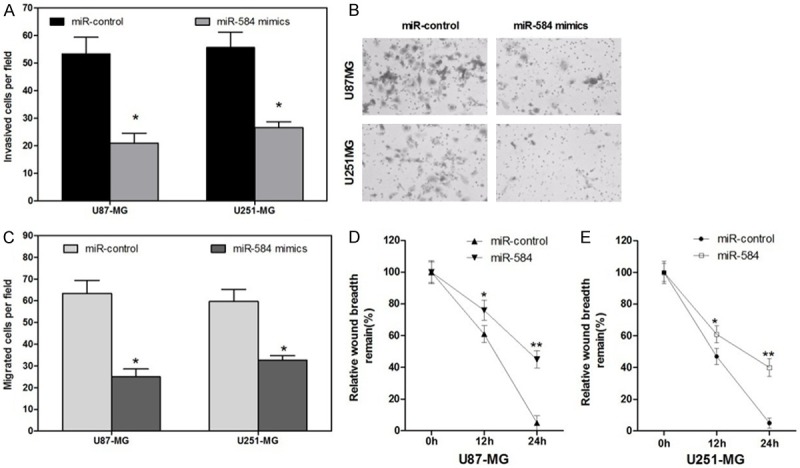
MiR-584 decreased the Invasion and Migration Abilities of Glioma Cells. A and B. Transwell assays showed that cells transfected with miR-584 mimics presented less invasion ability than those with miR-control (*P < 0.05). C and D. Wound healing assays showed that glioma cells transfected with miR-584 mimics may close the wound more slowly when compared with those with miR-control (*P < 0.05).
MiR-584 targets PTTG1IP and negatively regulates its expression
MicroRNAs regulate a variety of cellular activities through regulation of the expression of target genes. To determine the mechanism of miR-584-mediated cell cycle dysregulation in glioma cells, we next identified target genes that could be responsible for the effect of miR-584. Thus, bioinformatic analyses (TargetScan, RNA22 and microrna.org) were used to identify potential target genes of miR-584. To further confirm that miR-584 directly targets PTTG1IP, we performed luciferase reporter assays to examine whether miR-584 interacts directly with its target PTTG1IP. We constructed a series of 3’UTR fragments, including the full-length wild-type PTTG1IP 3’UTR and a binding site mutant (Figure 4A). These fragments were then inserted into the pmirGLO luciferase reporter plasmid. In U87-MG cells, we found that the cotransfection of miR-584 and the wildtype PTTG1IP 3’UTR caused a significant decrease in luciferase units compared with the controls. However, the cotransfection of the mutant PTTG1IP 3’UTR and miR-584 mimics failed to alter the luciferase intensity (Figure 4B). Furthermore, overexpression of miR-584 reduced PTTG1IP mRNA and protein expression in U87-MG and U251-MG cells (Figure 4C-E). Taken together, these results suggest that miR-584 binds directly to the 3’UTR of PTTG1IP, thereby repressing gene expression.
Figure 4.
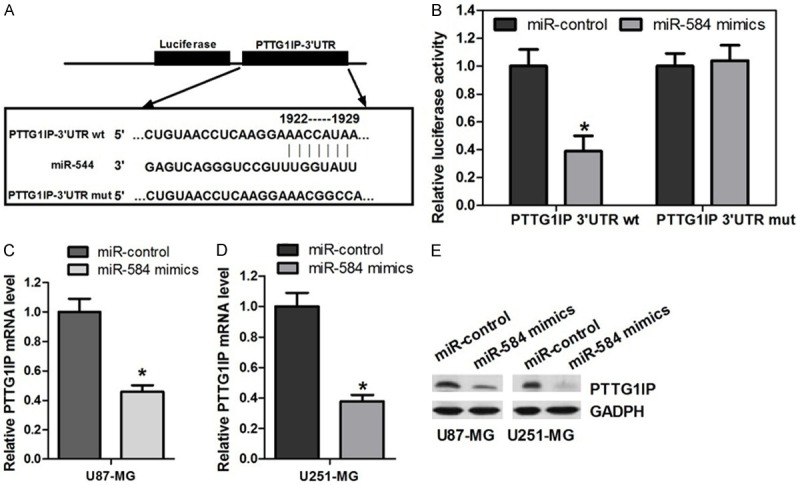
MiR-584 targets PTTG1IP and negatively regulates its expression. A. The predicted miR-584 binding site on the PTTG1IP mRNA 3’-UTR and the deletion mutation at the miR-584 “seed region” binding site, on the PTTG1IP mRNA 3’-UTR are shown. B. U87-MG cells were transfected with the wild type or mutated version of the luciferase-PTTG1IP 3’-UTR reporter vector as well as the miR-584 mimics or miR-control. The miR-584 mimics reduced the intensity the luciferase-PTTG1IP 3’-UTR reporter vector, while the mutant luciferase-PTTG1IP 3’-UTR failed to alter the luciferase intensity (*P < 0.05). C and D. Measurement of PTTG1IP mRNA expression levels by qRT-PCR. RNA was extracted from U87-MG and U251-MG cells transfected with the miR-584 mimics or miR-control. The endogenous expression levels of the β-actin mRNA were used for normalization, and the relative PTTG1IP expression levels are shown (*P < 0.05). E. Measurement of PTTG1IP expression levels by Western blot analysis. Protein was extracted from U87-MG and U251-MG cells transfected with the miR-584 mimics or miR-control. The endogenous expression levels of the GAPDH protein were used for normalization, and the relative PTTG1IP protein expression levels are shown (*P < 0.05).
Quantitative analysis of PTTG1IP expression in glioma tissue
To determine the expression of PTTG1IP in glioma and adjacent normal tissue, real-time RT-PCR for PTTG1IP was performed on thirteen tissue pairs, each consisting of an glioma and adjacent normal tissue. In general, PTTG1IP expression levels were significantly higher in glioma tissues than in the matched normal tissues (Figure 5). In contrast, miR-584 expression levels were predominantly downregulated in glioma tissue (Figure 1A).
Figure 5.
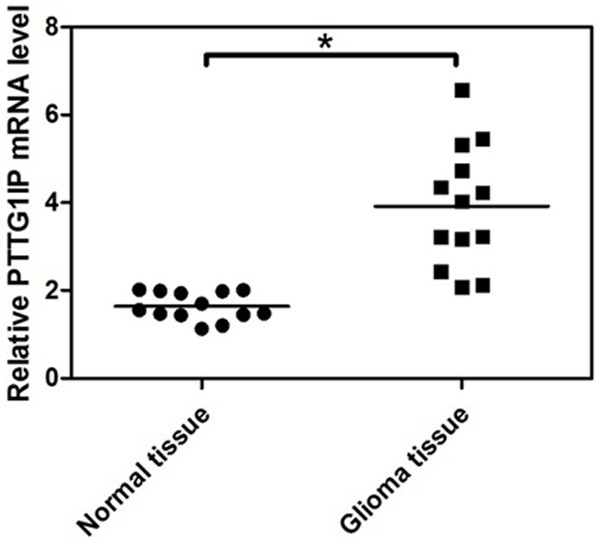
Quantitative analysis of PTTG1IP expression in glioma tissue. To test the expression of PTTG1IP in glioma tissues and adjacent normal tissues, a real-time RT-PCR assay was performed on the thirteen pairs of glioma tissue samples. The expression level of PTTG1IP is normalized to β-actin (*P < 0.05).
Discussion
There are a lot of studies focusing on the relationship between cancer and deregulated miRNA expression. Several miRNAs have been identified as oncogenes and tumor suppressors that are involved in glioma development. For instance, miR-21 [24], and miR-27b [25], have been shown to be expressed in glioma and to increase cell growth and invasion. In contrast, decreased expression of miR-205 [26], miR-34c [27], miR-451 [28] and miR-218 [29] may inhibit glioma cell proliferation and invasion. For miR-584, our findings are consistent with studies on other types of malignancy, such as breast [30], kidney [31] and colon [32]. Fils-Aimé N et al reported that TGF-β silences the expression of miR-584, resulting in enhanced PHACTR1 expression, and further leading to actin rearrangement and breast cancer cell migration [30]. Ueno K et al found that Expression of miR-584 in RCC (A-498 and 769-P) cells was downregulated compared with HK-2 cells. It demonstated that miR-584 is a new tumour suppressor miR in ccRCC (clear cell renal cell carcinoma) and inhibits cell motility through downregulation of ROCK-1 [31].
PTTG Binding Factor (PBF or PTTG1IP) is a little characterised proto-oncogene and is identified through its ability to interact with PTTG1, the human securin [33,34]. PBF is widely expressed in normal human tissues, including normal thyroid [33,35]. Whilst expression is low in normal breast tissue, immunohistochemical analysis demonstrated that PBF was strongly expressed in epithelial cells of all types and grades of breast tumour assessed [36]. MiR-584 inhibits cancer growth by blocking the expression and activity of PTTG1IP. The loss of miR-584 leads to the upregulation of PTTG1IP, consequently resulting in malignant transformation.
The current study identified that miR-584 were significantly down-regulated in glioma samples compared with normal samples. To further realize the role of miR-584 in glioma, we enhanced miR-584 expression in U87-MG and U251-MG cells using the miR-424 mimics, and found that overexpressed miR-584 suppressed the growth of glioma cells and induced cell apoptosis. We used the Dual-Luciferase reporter assay to confirm the target gene of miR-584 were PTTG1IP. In this study, we confirmed that miR-584-PTTG1IP signalling pathway represents a functional mechanism by which miR-584 suppresses glioma. However, miRNAs usually work in the regulation of multiple targets and we could not tell that there is no other signalling pathway working by miR-584 in glioma.
In summary, we demonstrated that miR-584 expression was markedly decreased in human glioma cell lines, and for the first time, we described the roles of miR-584 in cellular proliferation, migration, and invasion abilities in glioma cells. In addition, this study suggested that PTTG1IP is one of putative target genes of miR-584.
Acknowledgements
This work was support by Fund of scientific research in department of education of Yunnan (No. 2014Y174).
Disclosure of conflict of interest
None.
References
- 1.Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, Chin L, DePinho RA, Cavenee WK. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 2.Hottinger AF, Stupp R, Homicsko K. Standards of care and novel approaches in the management of glioblastoma multiforme. Chin J Cancer. 2014;33:32–39. doi: 10.5732/cjc.013.10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Posti JP, Bori M, Kauko T, Sankinen M, Nordberg J, Rahi M, Frantzen J, Vuorinen V, Sipila JO. Presenting symptoms of glioma in adults. Acta Neurol Scand. 2014 doi: 10.1111/ane.12285. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 4.Noda SE, El-Jawahri A, Patel D, Lautenschlaeger T, Siedow M, Chakravarti A. Molecular advances of brain tumors in radiation oncology. Semin Radiat Oncol. 2009;19:171–178. doi: 10.1016/j.semradonc.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 5.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 6.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 7.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berger F, Reiser MF. Micro-RNAs as potential new molecular biomarkers in oncology: have they reached relevance for the clinical imaging sciences? Theranostics. 2013;3:943–952. doi: 10.7150/thno.7445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu T, Tang H, Lang Y, Liu M, Li X. MicroRNA-27a functions as an oncogene in gastric adenocarcinoma by targeting prohibitin. Cancer Lett. 2009;273:233–242. doi: 10.1016/j.canlet.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 10.Wei Q, Li YX, Liu M, Li X, Tang H. MiR-17-5p targets TP53INP1 and regulates cell proliferation and apoptosis of cervical cancer cells. IUBMB Life. 2012;64:697–704. doi: 10.1002/iub.1051. [DOI] [PubMed] [Google Scholar]
- 11.Xu XM, Wang XB, Chen MM, Liu T, Li YX, Jia WH, Liu M, Li X, Tang H. MicroRNA-19a and -19b regulate cervical carcinoma cell proliferation and invasion by targeting CUL5. Cancer Lett. 2012;322:148–158. doi: 10.1016/j.canlet.2012.02.038. [DOI] [PubMed] [Google Scholar]
- 12.Dews M, Homayouni A, Yu D, Murphy D, Sevignani C, Wentzel E, Furth EE, Lee WM, Enders GH, Mendell JT, Thomas-Tikhonenko A. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet. 2006;38:1060–1065. doi: 10.1038/ng1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Felli N, Fontana L, Pelosi E, Botta R, Bonci D, Facchiano F, Liuzzi F, Lulli V, Morsilli O, Santoro S, Valtieri M, Calin GA, Liu CG, Sorrentino A, Croce CM, Peschle C. MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proc Natl Acad Sci U S A. 2005;102:18081–18086. doi: 10.1073/pnas.0506216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen H, Huang Q, Dong J, Zhai DZ, Wang AD, Lan Q. Overexpression of CDC2/CyclinB1 in gliomas, and CDC2 depletion inhibits proliferation of human glioma cells in vitro and in vivo. BMC Cancer. 2008;8:29. doi: 10.1186/1471-2407-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao Z, Tan X, Zhao A, Zhu L, Yin B, Yuan J, Qiang B, Peng X. microRNA-214-mediated UBC9 expression in glioma. BMB Rep. 2012;45:641–646. doi: 10.5483/BMBRep.2012.45.11.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma L, Young J, Prabhala H, Pan E, Mestdagh P, Muth D, Teruya-Feldstein J, Reinhardt F, Onder TT, Valastyan S, Westermann F, Speleman F, Vandesompele J, Weinberg RA. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol. 2010;12:247–256. doi: 10.1038/ncb2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guessous F, Alvarado-Velez M, Marcinkiewicz L, Zhang Y, Kim J, Heister S, Kefas B, Godlewski J, Schiff D, Purow B, Abounader R. Oncogenic effects of miR-10b in glioblastoma stem cells. J Neurooncol. 2013;112:153–163. doi: 10.1007/s11060-013-1047-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu X, Chen D, Cui Y, Li Z, Huang J. Targeting microRNA-23a to inhibit glioma cell invasion via HOXD10. Sci Rep. 2013;3:3423. doi: 10.1038/srep03423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang K, Wang X, Zou J, Zhang A, Wan Y, Pu P, Song Z, Qian C, Chen Y, Yang S, Wang Y. miR-92b controls glioma proliferation and invasion through regulating Wnt/beta-catenin signaling via Nemo-like kinase. Neuro Oncol. 2013;15:578–588. doi: 10.1093/neuonc/not004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu S, Wang S, Geng S, Ma S, Liang Z, Jiao B. Increased expression of microRNA-17 predicts poor prognosis in human glioma. J Biomed Biotechnol. 2012;2012:970761. doi: 10.1155/2012/970761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li X, Ling N, Bai Y, Dong W, Hui GZ, Liu D, Zhao J, Hu J. MiR-16-1 plays a role in reducing migration and invasion of glioma cells. Anat Rec (Hoboken) 2013;296:427–432. doi: 10.1002/ar.22626. [DOI] [PubMed] [Google Scholar]
- 22.Ying Z, Li Y, Wu J, Zhu X, Yang Y, Tian H, Li W, Hu B, Cheng SY, Li M. Loss of miR-204 expression enhances glioma migration and stem cell-like phenotype. Cancer Res. 2013;73:990–999. doi: 10.1158/0008-5472.CAN-12-2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee HK, Bier A, Cazacu S, Finniss S, Xiang C, Twito H, Poisson LM, Mikkelsen T, Slavin S, Jacoby E, Yalon M, Toren A, Rempel SA, Brodie C. MicroRNA-145 is downregulated in glial tumors and regulates glioma cell migration by targeting connective tissue growth factor. PLoS One. 2013;8:e54652. doi: 10.1371/journal.pone.0054652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corsten MF, Miranda R, Kasmieh R, Krichevsky AM, Weissleder R, Shah K. MicroRNA-21 knockdown disrupts glioma growth in vivo and displays synergistic cytotoxicity with neural precursor cell delivered S-TRAIL in human gliomas. Cancer Res. 2007;67:8994–9000. doi: 10.1158/0008-5472.CAN-07-1045. [DOI] [PubMed] [Google Scholar]
- 25.Lee JJ, Drakaki A, Iliopoulos D, Struhl K. MiR-27b targets PPARgamma to inhibit growth, tumor progression and the inflammatory response in neuroblastoma cells. Oncogene. 2012;31:3818–3825. doi: 10.1038/onc.2011.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yue X, Wang P, Xu J, Zhu Y, Sun G, Pang Q, Tao R. MicroRNA-205 functions as a tumor suppressor in human glioblastoma cells by targeting VEGF-A. Oncol Rep. 2012;27:1200–1206. doi: 10.3892/or.2011.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu Z, Wu Y, Tian Y, Sun X, Liu J, Ren H, Liang C, Song L, Hu H, Wang L, Jiao B. Differential effects of miR-34c-3p and miR-34c-5p on the proliferation, apoptosis and invasion of glioma cells. Oncol Lett. 2013;6:1447–1452. doi: 10.3892/ol.2013.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tian Y, Nan Y, Han L, Zhang A, Wang G, Jia Z, Hao J, Pu P, Zhong Y, Kang C. MicroRNA miR-451 downregulates the PI3K/AKT pathway through CAB39 in human glioma. Int J Oncol. 2012;40:1105–1112. doi: 10.3892/ijo.2011.1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xia H, Yan Y, Hu M, Wang Y, Wang Y, Dai Y, Chen J, Di G, Chen X, Jiang X. MiR-218 sensitizes glioma cells to apoptosis and inhibits tumorigenicity by regulating ECOP-mediated suppression of NF-kappaB activity. Neuro Oncol. 2013;15:413–422. doi: 10.1093/neuonc/nos296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fils-Aime N, Dai M, Guo J, El-Mousawi M, Kahramangil B, Neel JC, Lebrun JJ. MicroRNA-584 and the protein phosphatase and actin regulator 1 (PHACTR1), a new signaling route through which transforming growth factor-beta Mediates the migration and actin dynamics of breast cancer cells. J Biol Chem. 2013;288:11807–11823. doi: 10.1074/jbc.M112.430934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ueno K, Hirata H, Shahryari V, Chen Y, Zaman MS, Singh K, Tabatabai ZL, Hinoda Y, Dahiya R. Tumour suppressor microRNA-584 directly targets oncogene Rock-1 and decreases invasion ability in human clear cell renal cell carcinoma. Br J Cancer. 2011;104:308–315. doi: 10.1038/sj.bjc.6606028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang XY, Wu MH, Liu F, Li Y, Li N, Li GY, Shen SR. Differential miRNA expression and their target genes between NGX6-positive and negative colon cancer cells. Mol Cell Biochem. 2010;345:283–290. doi: 10.1007/s11010-010-0582-7. [DOI] [PubMed] [Google Scholar]
- 33.Chien W, Pei L. A novel binding factor facilitates nuclear translocation and transcriptional activation function of the pituitary tumor-transforming gene product. J Biol Chem. 2000;275:19422–19427. doi: 10.1074/jbc.M910105199. [DOI] [PubMed] [Google Scholar]
- 34.Boelaert K, Tannahill LA, Bulmer JN, Kachilele S, Chan SY, Kim D, Gittoes NJ, Franklyn JA, Kilby MD, McCabe CJ. A potential role for PTTG/securin in the developing human fetal brain. FASEB J. 2003;17:1631–1639. doi: 10.1096/fj.02-0948com. [DOI] [PubMed] [Google Scholar]
- 35.Yaspo ML, Aaltonen J, Horelli-Kuitunen N, Peltonen L, Lehrach H. Cloning of a novel human putative type Ia integral membrane protein mapping to 21q22.3. Genomics. 1998;49:133–136. doi: 10.1006/geno.1998.5217. [DOI] [PubMed] [Google Scholar]
- 36.Watkins RJ, Read ML, Smith VE, Sharma N, Reynolds GM, Buckley L, Doig C, Campbell MJ, Lewy G, Eggo MC, Loubiere LS, Franklyn JA, Boelaert K, McCabe CJ. Pituitary tumor transforming gene binding factor: a new gene in breast cancer. Cancer Res. 2010;70:3739–3749. doi: 10.1158/0008-5472.CAN-09-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]


